Abstract
To completely understand the ecology of a bacterial community, we need to identify its ecologically distinct populations (ecotypes). The greatest promise for enumerating a community's constituent ecotypes is held by molecular approaches that identify bacterial ecotypes as DNA sequence clusters. These approaches succeed when ecotypes correspond with sequence clusters, but some models of bacterial speciation predict a one-to-many and others a many-to-one relationship between ecotypes and sequence clusters. A further challenge is that sequence-based phylogenies often contain a hierarchy of clusters and subclusters within clusters, and there is no widely accepted theory to guide systematists and ecologists to the size of cluster most likely to correspond to ecotypes. While present systematics attempts to use universal thresholds of sequence divergence to help demarcate species, the recently developed ‘community phylogeny’ approach assumes no universal thresholds, but demarcates ecotypes based on the analysis of a lineage's evolutionary dynamics. Theory-based approaches like this one can give a conceptual framework as well as operational criteria for hypothesizing the identity and membership of ecotypes from sequence data; ecology-based approaches can then confirm that the putative ecotypes are actually ecologically distinct. Bacterial ecotypes that are demonstrated to have a history of coexistence as ecologically distinct lineages (based on sequence analysis) and as a prognosis of future coexistence (based on ecological differences), are the fundamental units of bacterial ecology and evolution, and should be recognized by bacterial systematics.
Keywords: ecotype, community phylogeny, periodic selection, species concept, bacterial diversity
1. Introduction
How many ecologically distinct populations are in a natural bacterial community, who are they, and what differences allow them to coexist or perform different ecosystem functions? Bacterial systematics has recognized that a molecular approach is needed to address these central questions of ecology and evolution (Hutchinson 1968; Mayr 1982; Blaxter 2004). Indeed, molecular methods developed in recent decades have enabled identification of many new taxa, from divisions to species, with and without the benefit of culturing the bacteria (Murray & Stackebrandt 1995; Pace 1997). Nevertheless, I will argue that the species recognized and sought by bacterial systematics are generally too broadly defined to yield what is deeply needed by microbial ecologists and evolutionary biologists—a general, sequence-based systematics that delivers the ecologically distinct players within a community. I will describe a new approach to the systematics of bacterial species, based on the theory of bacterial evolutionary dynamics, and will argue that this approach can yield the ecologically distinct populations within a community.
2. The challenge of characterizing bacterial diversity
Characterizing a community's ecological diversity is difficult enough for zoologists and botanists, but this challenge is especially vexing for those who study bacteria. Most bacterial species are known only by the sequence of a single gene (usually 16S rRNA; Harris et al. 2004) because much less than 1% of species are cultivable (Giovannoni & Stingl 2005). Some organisms known only by a single gene represent previously unknown divisions of bacteria, and so we have no idea what their contributions to ecosystem function might be. Moreover, even when an organism falls within a characterized bacterial group, knowing only a single gene's sequence gives limited predictive value about the organism's ecosystem function and its interactions within the community. This is because bacteria can almost instantly alter their ecology, as if by deus ex machina, through acquisition of genes from distant relatives (Gogarten et al. 2002). Thus, close relatives might infect entirely different hosts (Dobrindt 2005; Ron 2006) or can have different roles in mineral cycling (Coleman et al. 2006).
One of the greatest challenges in understanding bacterial diversity is the sheer enormity of species diversity now known to exist. The pioneering work of Vigdis Torsvik and colleagues (Torsvik et al. 1998, 2002) has opened our imaginations to the possibility that bacterial species may number in the millions or even billions. Their approach, based on the rates of reannealing of the pool of DNA from the environment, estimated that just 30 g of forest soil held about 10 000 species, assuming that species were equally abundant, and several other key assumptions. Dykhuizen (1998), Gans et al. (2005) and Curtis et al. (2006) have taken into account that species are not equally abundant (and challenged other assumptions) and have found that, in all likelihood, modest environmental samples contain millions to tens of millions of species.
After we catch our breath, we are compelled to wonder how these organisms could possibly partition an ecosystem's resources so finely as to sustain such a huge number of species. Ecological theory tells us that species cannot coexist for the long term unless they use different resources, or thrive in different conditions, or respond differently to their predators and pathogens (Chase & Leibold 2003). There are certainly many resource dimensions upon which bacteria could diversify. There are probably very few carbon sources that cannot be used by at least one bacterium; for example, some bacteria can use ordinary toxic compounds and ordinary insoluble macromolecules. In addition, each of the many eukaryotic species may have its own exclusive bacterial endosymbionts and pathogens. Finally, bacteria can coexist on the basis of different unusual conditions under which they can grow, including an extreme range of salinity, temperature and radiation levels (Hodgson 1989). Finding the basis for coexistence among close relatives will be a challenge for the next generation of bacterial community ecologists and systematists.
While the Torsvik approach addresses the issue of ‘how many’, it does not lead to the identities and functions of the ecological players within a community. Sequence-based approaches are believed to hold the greatest promise of addressing these issues (Palys et al. 1997; Blaxter 2004). For example, by sorting sequences of a single gene from a sample of organisms into clusters, one can in principle demarcate organisms into ecologically distinct groups (Palys et al. 1997; Schloter et al. 2000; Stach et al. 2003; Birky et al. 2005) and then estimate the number of such groups in the community at large (Hughes et al. 2001).
Recent studies have demarcated and counted bacterial taxa in a community by binning DNA sequences for a given gene (often 16S rRNA) into sequence clusters and then attributing each sequence cluster to an operational taxonomic unit (OTU). Some workers have defined OTUs as clusters with up to 2.5% sequence divergence in 16S rRNA (Hughes et al. 2001); the rationale is that this divergence is the level of diversity found within many named species (Stackebrandt & Goebel 1994). Other workers have considered each 16S rRNA sequence type as a distinct OTU. Both approaches have allowed comparisons of different environments for their ability to sustain microbial diversity (Bohannan & Hughes 2003).
However, no sequence-based OTU proposed either by systematists or ecologists appears to correspond to the fundamental units of bacterial ecology (Cohan 2002b; Staley 2003). More generally, even the named species of bacterial systematics, which are based on molecular as well as phenotypic criteria (Vandamme et al. 1996), fail to identify the functionally distinct populations of a community, as I will next discuss.
3. The ‘species’ of bacterial systematics
Bacterial systematics began in much the same way as the systematics of animals and plants. Systematists of both microbes and macrobes began with the observation that organisms fall into clusters of very similar organisms, and they demarcated and named these clusters. These species demarcations were originally based entirely on phenotype, principally on morphology in the case of animals and plants, and on metabolism in the case of bacteria. However, the practices of microbial and macrobial systematics diverged in the 1940s and 1950s when a theory-based concept for species was brought into the systematics of animals (Mayr 1942) and plants (Stebbins 1957). First, with Mayr's biological species concept (Mayr 1942), and later with many alternative concepts of species (de Queiroz 1998), systematists of animals and plants sought to make their species more than just clusters; systematics now aimed to identify species that represented the fundamental units of ecology and evolution. Despite the plethora of modern species concepts used today, nearly all these concepts share certain quintessential attributes, which are as follows: species are cohesive (in that some force acts to constrain divergence within species; Meglitsch 1954; Templeton 1989); they are irreversibly separate (because there is no force of cohesion that constrains divergence between species; Wiley 1978); they are ecologically distinct (and thus able to coexist within a community; VanValen 1976) and they are monophyletic (i.e. each species is invented only once; Mishler & Donoghue 1982). It has long been understood that diversity within the highly sexual animal and plant species is constrained by a powerful force of cohesion, i.e. genetic exchange. We shall see that bacterial species, which recombine sexually at a low rate, within an order of magnitude of the mutation rate (Maynard Smith et al. 1993; Cohan 2002a) can also be defined so as to meet the criterion of cohesiveness, as well as the other attributes of species (Cohan 1994, 2001; Ward 1998).
Bacterial systematics has not moved towards a theory-based concept of species, but in recent decades it has developed increasingly discerning methods to distinguish species. On the phenotypic front, bacterial species can now be distinguished by the whole cell fatty acid components (Vandamme et al. 1992) and more recently by matrix-assisted laser desorption/ionization–time of flight (MALDI–TOF) mass spectrometry, which allows characterization of high-throughput analysis of low abundance molecules (Keys et al. 2004).
For the past three decades, whole-genome DNA–DNA hybridization has allowed quantification of the fraction of genome that is not shared across individual organisms (although confounded by high divergence between genes that are shared; Johnson 1973). Early on, systematists determined a criterion of DNA–DNA hybridization that frequently corresponded to the established, phenotype-based species demarcations. Annealing of 70% or less genome became a ‘gold standard’ for demarcating organisms into different species (Johnson 1973; Wayne et al. 1987).
Systematists are currently developing sequence-based approaches for demarcating species (Gevers et al. 2005). The appeal is that any uncharacterized organism, whether cultured or not, can be readily placed onto the tree of life by its 16S rRNA sequence (Harris et al. 2004); efforts are underway to make possible a universal tree based on the protein sequences (Zeigler 2003; Santos & Ochman 2004). Moreover, Stackebrandt & Goebbel (1994) have determined an empirical relationship between levels of 16S sequence divergence and annealing by DNA–DNA hybridization. Any two organisms that are at least 2.5% divergent in their 16S sequences were shown almost universally to be members of different named species, although in some cases organisms that were less divergent were shown to fall into different species.
The infusion of molecular techniques into systematics has clearly made the demarcation of species more standard across taxa, and the universality of molecular techniques has made species demarcation accessible to a greater diversity of microbiologists. However, we must note that these molecular techniques have not been designed to infuse a theory of species into systematics. We are merely finding easier ways to classify newly isolated organisms into the old phenotype-based species; we are also demarcating newly discovered species into clusters that have about the same molecular diversity as the old phenotype-based species (Cohan 2002b). In either case, we are enshrining the species of yore with new molecular data.
What is wrong in demarcating bacterial species without a theory-based concept of species? We should note that both microbial and macrobial species are in practice demarcated simply as clusters. The difference is that while microbial systematists have not embraced a theory-based concept of species, macrobial systematists have attempted to fit their cluster-based demarcations in accordance with theory. In macrobial systematics, cluster-based and theory-based approaches have generally yielded the same species demarcations. This is because, having a theory of species, practicing macrobial systematists may continually reset their vision of how large a cluster should be to fit within a species (Godreuil et al. 2005). Because mainstream bacterial systematics does not aspire to base its species on theory, there is no opportunity for recalibrating the size of a bacterial species cluster to theory. Therefore, it is not surprising to find a huge level of diversity contained within a typical named bacterial species.
Consider next the variation in genome content, DNA sequence, phenotype and ecology typically seen within the named bacterial species. Three decades of whole-genome DNA–DNA hybridization by bacterial systematists have demonstrated that members of a named species frequently share only 80–90% of their genes (Feldgarden et al. 2003), a result corroborated by physical mapping of genomes (Bergthorsson & Ochman 1998; Thompson et al. 2005), and most recently by genome sequence comparisons (Alm et al. 1999; Bansal & Meyer 2002; Joyce et al. 2002; Boucher et al. 2004; Lindsay & Holden 2004; Nelson et al. 2004). With regard to genes that are shared among the species' members, sequence divergence within a bacterial species is far greater than that within an animal or plant species. For example, at the 16S rRNA locus, sequence diversity within a recognized bacterial species is frequently at 1%, which is the level of diversity typically found between orders of mammals at the homologous nuclear gene 18S rRNA (Staley 2003).
This is consistent with a much greater time of divergence among members of a bacterial species than in the case for an animal species. However, this conclusion must take into account a much faster rate of molecular evolution in bacteria, at least when we compare a eukaryote's nuclear genes with homologues in bacteria (Ochman & Wilson 1987; Whittam 1996; Ochman et al. 1999). The proper comparison is between a eukaryote's mitochondrial genes and their homologues in bacteria (e.g. CO II), since these have been found to evolve at about the same pace (von Dohlen et al. 2006). Therefore, the relative times of divergence within the species can be inferred directly from sequence diversity of these genes. This kind of comparison has not been carefully studied, but my impression is that the time of divergence among conspecific bacteria is about five times greater than that for eukaryotic species.
Named bacterial species hold an enormous amount of phenotypic diversity. Even though bacterial species were originally demarcated as phenotypic (usually metabolic) clusters, most species are highly diverse in their metabolic capabilities (Logan & Berkeley 1984; De Clerck et al. 2004). It would seem that even by the criteria on which bacterial species were originally demarcated, bacterial systematics has been prone to lump high levels of diversity within a named species.
Most importantly, a named bacterial species is typically an assemblage of ecologically distinct populations that are able to coexist in the same region (Schloter et al. 2000; Lopez-Lopez et al. 2005; Smith et al. 2006). For example, David Gordon and colleagues have demonstrated a great deal of genetically based ecological diversity among Australian Escherichia coli, based on the soil conditions and hosts to which they are adapted (Gordon & Cowling 2003). The various sequence clusters of Listeria monocytogenes have been shown to differ in their optimal habitats in food processing and animal production (Ward et al. 2004), and sequence clusters of the pathogen Mycobacterium tuberculosis differ in their host ranges (Smith et al. 2006). In addition, various molecular techniques (including PCR- and restriction-based methods (Berthier et al. 1993; Schloter et al. 2000) as well as multilocus sequence typing (Jolley et al. 2000; Feil et al. 2004)) have subclassified species members into multiple clusters that are frequently ecologically distinct (Giovannoni & Stingl 2005). Given the high diversity within a named bacterial species in the genome content, sequences of shared genes, phenotype and ecology, bacterial species appear to be more like the genera (or even sub-families) of animals than animal species (Cohan 2002b; Staley 2003). Thus, the species demarcated in bacteria fail to meet a fundamental goal of systematics to provide a species label that gives us precise predictions about the biology of all the members of a species (Mayr 1963; Hutchinson 1968).
4. The stable ecotype model
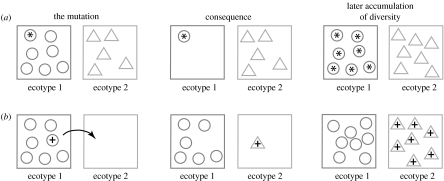
Consider next how we might infuse evolutionary models into systematic thinking. I will consider first a model based on the ‘ecotype’—a group of bacteria that are ecologically similar to one another. More specifically, member organisms of an ecotype are so similar that an adaptive mutant (or an adaptive recombinant) from one ecotype can outcompete all other individuals from the same ecotype (Cohan 1994; figure 1). Owing to the low frequency of recombination in bacteria (Maynard Smith et al. 1993; Cohan 2002a), natural selection favouring each adaptive mutant leads to purging of diversity at all loci within the ecotype, an event called ‘periodic selection’. An adaptive mutant does not outcompete to extinction members of other ecotypes owing to the ecological differences among ecotypes (Cohan 1994).
Figure 1.
The effects of adaptive mutations on diversity within and between ecotypes. (a) The effect of a periodic selection event. Here, a mutant (or recombinant) with improved ability to compete for the resources of ecotype 1, indicated by an asterisk, is able to extinguish the diversity within the same ecotype. The diversity within ecotype 2 is not affected by periodic selection occurring within ecotype 1. After the periodic selection, diversity once again accumulates within ecotype 1. (b) The effect of a niche-invasion mutation. Here, a mutant, indicated by a plus sign, obtains the ability to utilize a new set of resources and thereby founds an ecotype. Ecotype 2 begins as a clone, with no diversity, but it eventually accumulates genetic diversity by mutation and recombination (Ward & Cohan 2005). (Used with permission from the Thermal Biology Institute.)
Diversity within an ecotype is ephemeral, persisting only until the next periodic selection event, when diversity is crushed to near zero at all loci (figure 1a). What is, then, the source of permanent divergence among closely related bacteria? Divergence can become permanent when a mutation (or recombination event) places the organism into a new ecological niche and the organism thereby founds a new ecotype. Because the new ecotype is ecologically distinct from the parental ecotype, periodic selection events in the parental ecotype cannot extinguish the founding organism and its descendants (figure 1b). Thus, the new ecotype escapes the periodic selection of the parental ecotype, and the two new ecotypes are free to diverge indefinitely.
Each ecotype so defined is a cohesive group, in the sense that its diversity is capped by a force of evolution, in this case periodic selection. Different ecotypes are irreversibly separate because they are out of range of one another's periodic selection events, and recombination is too rare to prevent their adaptive divergence (Cohan 2002b; Lawrence 2002). By definition, different ecotypes are ecologically distinct, which allows them to coexist into the future. Finally, such ecotypes are monophyletic groups because they were founded by a single individual (except in the ‘recurrent niche invasion’ model described subsequently). These properties of ecotypes constitute the dynamic properties expected for species under all modern concepts of species (de Queiroz 1998; Cohan 2001).
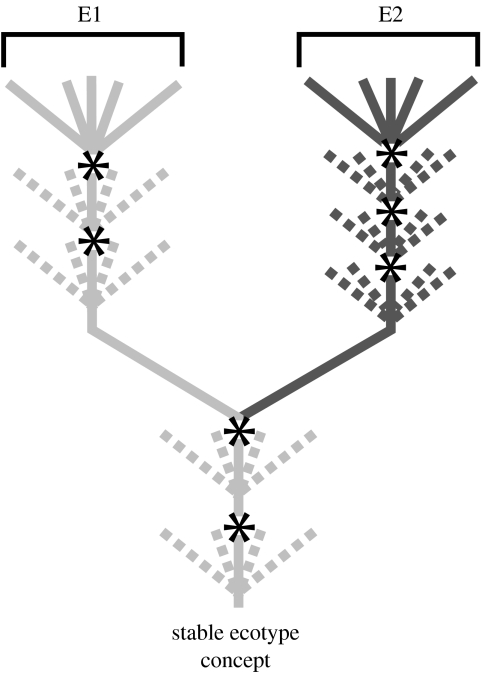
Guided by the ecotype model, we can outline a general method for using sequence-based approaches to find cohesive, irreversibly separate, ecologically distinct and monophyletic groups of bacteria. The key is that periodic selection events recurrently purge sequence diversity at all loci within an ecotype, but different ecotypes are free to accumulate sequence divergence at all loci. Thus, at least under certain circumstances I will enumerate, each ecotype can be identified in principle as one sequence cluster (figure 2; Palys et al. 1997; Cohan 2004).
Figure 2.
The phylogenetic history of two closely related ecotypes under the stable ecotype model. After each periodic selection event, indicated by an asterisk, only one variant from an ecotype survives. After periodic selection, the descendants of the surviving variant diverge (indicated by dashed lines), but with the next periodic selection event, again only one variant survives. Note that a sequence-based phylogeny of two ecotypes will indicate very limited sequence diversity within an ecotype, with much greater sequence divergence between members of different ecotypes. This model yields a one-to-one correspondence between ecotypes and sequence clusters. (Used with permission from the Thermal Biology Institute.)
A one-to-one correspondence between ecotypes and sequence clusters is most probable under the ‘stable ecotype’ model. Here, ecotypes are created and extinguished at a very low rate, and during its long lifetime, an ecotype is recurrently purged of its diversity by periodic selection events (Gevers et al. 2005; Godreuil et al. 2005; Ward & Cohan 2005). Most such ecotypes (of sufficient age) should be distinguishable from other ecotypes as separate sequence clusters under most rates of recombination encountered in bacteria (Cohan 1995; Palys et al. 1997; Lawrence 2002). In principle, then, we may identify ecotypes as DNA sequence clusters, provided the stable ecotype model applies.
5. Identifying the sequence clusters corresponding to ecotypes
Even if we assume the stable ecotype model, ecological interpretation of sequence-based phylogenies is not straightforward. Any sequence-based phylogeny is likely to contain a hierarchy of subclusters within clusters, and it is not clear which level of cluster corresponds to ecotypes (Cohan 2002b). Therefore, we have recently proposed and tested a conceptual framework, based on the evolutionary dynamics of bacterial populations, for estimating the number of ecotypes within a community and identifying them (Cohan et al. submitted).
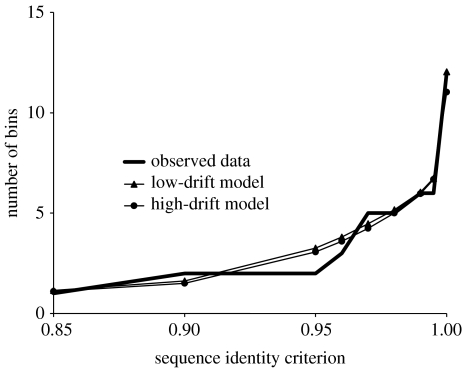
This ‘community phylogeny’ approach begins by characterizing the ‘community sequence diversity’ for a clade as the number of sequence clusters (or bins) present for different sequence-identity criteria, following Martin (2002) and Acinas et al. (2004) (figure 3). The number of sequence clusters at a particular sequence identity level represents the number of lineages at some point in the past that have survived to the present in the focus community (Martin 2002). The evolutionary history of a clade is modelled using four parameters so as to yield the community sequence diversity pattern of figure 3 with maximum likelihood (figure 4).
Figure 3.
Observed and predicted community sequence diversity pattern for gyrA sequences of the strains of B. licheniformis–B. sonorensis clade isolated from ‘Evolution Canyon’ III. Complete linkage clustering was used to bin the sequences into clusters with different levels of minimum pairwise identity ranging from 0.85 to 1.00. The model curves are based on the mean number of bins for each sequence-identity criterion, over 1000 replicate runs of the high-drift and low-drift parameter solutions (Cohan et al. submitted).
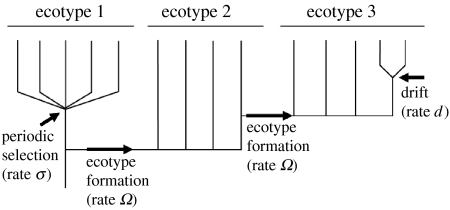
Figure 4.
The community phylogeny simulation. To begin a simulation with a given quartet of values for the net ecotype formation rate (Ω), the periodic selection rate (σ), the genetic drift rate (d) and the number of ecotypes (n), the υ contemporary organisms are distributed randomly among the n ecotypes. (In the case of this figure, υ=14 and n=3.) Working backwards from the present, the processes of ecotype formation, periodic selection and drift occur stochastically in time according to their respective rates. The backward phase of the simulation ends when only a single lineage remains; this represents the most recent common ancestor of the community. The forward simulation begins when a sequence is assigned to the most recent common ancestor. Substitutions then occur stochastically, going forward in time, between each pair of nodes in the phylogeny, according to the time between the events determining the nodes (Cohan et al. submitted).
In this model (Cohan et al. submitted), genome-wide diversity within each ecotype is purged recurrently by periodic selection at rate σ (figure 4). Diversity within ecotypes is also limited by genetic drift, occurring at a rate d. New ecotypes are formed at a net rate of Ω, taking into account the rate of extinction of ecotypes. The model also includes the number of ecotypes, n, represented in the sample of sequences. The community phylogeny analysis evaluates different quartets of parameter values for their likelihood of yielding an evolutionary history consistent with the observed community sequence diversity of figure 3 and produces a maximum likelihood estimate of the parameter values.
We have employed the community phylogeny approach to examine three clades whose ecological diversity and habitats have been intensively studied (Cohan et al. submitted), including isolates of the Bacillus subtilis–Bacillus licheniformis clade (Palmisano et al. 2001) from ‘Evolution Canyon’ III of the Negev Desert (Grishkan et al. in press), sequences from uncultured members of the Synechococcus A–A′ clade from Yellowstone hot springs (Ferris et al. 2003; Ward & Cohan 2005; Ward et al. 2006) and world-wide clinical and environmental isolates of Legionella pneumophila (Ratcliff 2006).
We note that community phylogeny analysis generally yields both a low-drift and a high-drift solution that fit the community sequence diversity pattern (figure 3; Cohan et al. submitted). In the low-drift solution, periodic selection dominates the clearing of sequence diversity within an ecotype with negligible effect of drift (d≈0); in the high-drift solution, sequence diversity within an ecotype is dominated by drift with negligible effect of periodic selection (σ≈0). The high-drift solution requires an effective population size of 108 or less, which is improbably low for the organisms analysed so far. Therefore, our analyses of Bacillus, Synechococcus and Legionella have focused on the low-drift solutions. Nevertheless, for many species with much more limited population sizes (e.g. a pathogen whose effective population size is about the same as its host), the high-drift solutions might be more appropriate.
In each system analysed, the community phylogeny estimated several ecotypes per traditional species (Ward & Cohan 2005; Cohan et al. submitted). Thus community phylogeny provides a theory-based method to detect a multiplicity of ecotypes within a named species, even before the ecological differences between ecotypes can be confirmed.
We extended the community phylogeny analysis to identify all the individual ecotypes of a clade (Cohan et al. submitted). The rationale was first to quantify the community sequence diversity within a given subclade and then to determine the number of ecotypes (n) in the four-parameter solution that yields the subclade's community sequence diversity pattern with maximum likelihood. We then demarcated putative ecotypes as the largest clades that were each consistent with a single ecotype.
Ecotype demarcations corresponded most closely to the existing species and subspecies boundaries in the case of Bacillus (figure 5); in contrast, community phylogeny estimated 14 ecotypes within L. pneumophila and eight ecotypes within the Synechococcus A–A′ clade, which contains less 16S sequence diversity than a typical named species (Cohan et al. submitted). These results corroborate previous evidence that the demarcations of bacterial systematics frequently lump many ecologically distinct populations into a single species (Schloter et al. 2000; Gordon & Cowling 2003; Sikorski & Nevo 2005; Ward & Cohan 2005; Smith et al. 2006).
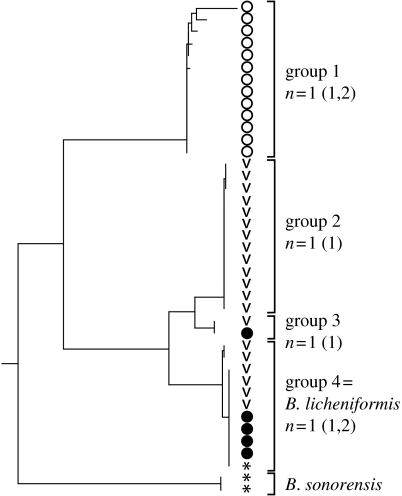
Figure 5.
Phylogeny of the Bacillus licheniformis subclade of the B. subtilis–B. licheniformis clade based on the neighbour-joining analysis of sequence diversity at rpoB. The putative ecotypes demarcated by community phylogeny are listed as the ‘Groups’ in the figure. The ecotypes were demarcated as the largest clades that were each consistent with being a single ecotype (i.e. such that 95% CI included n=1 ecotype). The phylogeny is rooted by B. halodurans. Microhabitat sources were the south-facing slope (open circles), the north-facing slope (filled circles) and the canyon bottom (V-shaped) within ‘Evolution Canyon’; asterisks indicate reference strains isolated outside of ‘Evolution Canyon’. The phylogeny of the B. subtilis subclade, which was included in the community phylogeny analysis, is not shown here.
In each clade we have analysed, community phylogeny has demarcated some putative ecotypes that appear ecologically distinct in nature (Cohan et al. submitted). Most of these ecological distinctions were inferred from differences in the microhabitats where the organisms (or sequences) were most frequently isolated. In the Synechococcus A′ clade, microhabitat distribution suggests that one putative ecotype is specialized to the upper photic zone at 68°C, while another group is specialized to the lower photic zone at this temperature (Ward et al. 2006; Cohan et al. submitted). In Bacillus, one putative ecotype (group 1) appears to be specialized to the high solar insolation of the south-facing slope of the canyon (and/or the other organisms adapted to this microhabitat), and a closely related group appears specialized to the canyon bottom (figure 5). In the case of Legionella, putative ecotypes differ in the species of amoebae they can infect (Berk et al. 1998; Cohan et al. submitted). In addition, the two ecotypes differ in the time-courses of gene expression during infection of one amoeba species (Brüggemann et al. 2006; Cohan et al. 2006). Community phylogeny appears to be an efficient approach for discovering the ecologically distinct populations within a community.
6. Systematics and the diversity of models of bacterial evolution
We next consider alternatives to the stable ecotype model, where the correspondence between ecotypes and sequence clusters is not expected to be one-to-one. In some alternative models of bacterial evolution, a single ecotype may correspond to multiple sequence clusters. Consider first the ‘geotype’ model (Papke et al. 2003) in which ecotypes are long-lived as in the stable ecotype model but there is only rare migration among the geographical regions of the ecotype. Thus, ecologically identical populations in different regions can diverge into different sequence clusters called ‘geotypes’.
Indeed, migration is rare enough to foster geotypes, at least in certain circumstances. Some extremophiles appear to have shown only rare dispersal across uninhabitable, mesic habitats to other favourable extreme locales. Indeed, marine psychrophiles appear to have formed distinct geotypes associated with each pole (Staley 2003). Likewise, hot spring thermophiles appear to have produced distinct geotypes in similar hot springs on different continents (Papke et al. 2003; Whitaker et al. 2003). Low migration is not expected to be limited to extremophiles. For example, any fastidious pathogen lacking a significant environmental phase, such as the syphilis-causing Treponema pallidum (Knell 2004), can be only as mobile as its hosts. In addition, obligate endosymbionts, such as the Buchnera mutualists of aphids, have cospeciated with their hosts and so do not ‘travel’ from one host species to another (Funk et al. 2000). Therefore, the Buchnera populations associated with different hosts can diverge into different clusters, like geotypes, even if they are not ecologically distinct.
Some environments are expected to be exceptionally prohibitive of dispersal. The highly viscous substrate of deep-rock bacteria is likely to constrain long-distance migration (van Waasbergen et al. 2000). Likewise, the perennially frozen lakes of Antarctica are not likely to be conducive to migration (Vincent et al. 2000; Tindall 2004).
There are, of course, celebrated examples of very high dispersal, supporting the classic view promoted by Baas-Becking that ‘everything is everywhere and the environment selects’ (Beijerinck 1913; Baas-Becking 1934). Bacteria with spores resistant to environmental stress have been found to migrate readily across continents (Roberts & Cohan 1995). In addition, human and agricultural pathogens are found to migrate at very high rates (Selander & Musser 1990). It remains to be seen which paradigm for dispersal is the more typical of bacteria, i.e. jet-set human pathogens or the deeply entombed bacteria of rocks.
A history of geographical separation among ecologically interchangeable populations can lead to difficulties for sequence-based taxonomy. Consider in particular the implications of a rapid increase in the rate of migration of human pathogens following the advent of jet planes (or perhaps transoceanic shipping). In the geotype-plus-Boeing model (Gevers et al. 2005; Godreuil et al. 2005; Ward & Cohan 2005), geographically isolated populations of the same ecotype were formerly able to diverge into separate sequence clusters during the time before rapid human transport; in the recent decades, jet planes have been able to carry all the endemic clusters of a single ecotype into each region of the world. In this transitional era when air travel (and even transoceanic sea travel) is still relatively new, we may see multiple sequence clusters (the pre-Boeing geotypes) within one ecotype at one place. Therefore, we cannot conclude from sequence clustering alone that two sympatric clusters are separate ecotypes.
Consider next the effect of genetic drift on the correspondence between ecotypes and sequence clusters. Genetic drift can be a significant force in bacterial populations of modest size, and genetic drift will tend to yield subclusters of closely related organisms of the same ecotype. This subclustering is most probable for pathogens and endosymbionts, especially when the number of bacteria transmitted from host to host approaches one. Thus, under either the geotype-plus-Boeing or the drift models, the relationship between ecotypes and sequence clusters is expected to be one-to-many.
Consider next models yielding a many-to-one correspondence between ecotypes and sequence clusters. In the species-less model (Lawrence 2002; Cohan 2004; Gevers et al. 2005; Godreuil et al. 2005; Ward & Cohan 2005), there is frequent invention and extinction of ecologically distinct populations. Frequent invention of new niches is especially plausible in the context of frequent horizontal genetic transfer, which can donate entire working biochemical pathways to a recipient (Gogarten et al. 2002). In this model, the diversity within an ecotype need not be constrained into the indefinite future by any force of cohesion (such as periodic selection); instead, diversity may be constrained principally by the small amount of time from the ecotype's founding from a single mutant (or recombinant) to the time the ecotype goes extinct. Here, a single sequence cluster might contain multiple very young ecotypes.
Another possibility is that recurrent recombination prevents separate ecotypes from diverging into distinct sequence clusters for most genes (cohesive recombination model). Recombination may be frequent enough to retard sequence divergence at loci that are not responsible for adaptive divergence, while recombination is not sufficient to hinder adaptive divergence (Cohan 1994, 1996). If this model is correct, multiple ecotypes would be contained indefinitely within a single sequence cluster for all genes not involved in the adaptive divergence between ecotypes. However, recombination between ecotypes is expected to decrease over time, owing to a positive feedback between sexual isolation and sequence divergence among ecotypes (Cohan 1995; Lawrence 2002).
While ecotypes are expected eventually to diverge into distinct sequence clusters (Cohan 1995; Lawrence 2002), we should note that recombination can retard this process. Indeed, the sequence of any given gene is an unreliable indicator of an individual's ecotype in frequently recombining taxa such as Neisseria meningitidis (Feil et al. 2001) and Thermotoga (Nesbo et al. 2006). Nevertheless, Hanage et al. (2006) have shown that a concatenation of a modest number of genes can reliably distinguish into separate sequence clusters frequently recombining Neisseria taxa, which is known to be ecologically distinct. We may conclude that frequent recombination is unlikely to prevent newly divergent ecotypes from appearing as separate sequence clusters, provided that enough genes are analysed.
A many-to-one correspondence between ecologically distinct populations and sequence clusters is also possible under the ‘recurrent niche invasion’ model (Godreuil et al. 2005). In this model, members of each ecotype frequently lose the adaptations of their present ecotype and acquire the adaptations of another. This is most probable when the populations owe their ecological distinctness entirely to the facile gain or loss of a plasmid. In an ‘ecological conversion’ process, members of a plasmid-free population (e.g. a plasmid-free Rhizobium population adapted to soil) can acquire a plasmid and thereby become converted to another population (e.g. a plasmid-bearing Rhizobium population adapted to mutualism with its legume host), and the reverse conversion can occur with the loss of the plasmid. If these reciprocal ecological conversions recur, then the populations are not irreversibly separate lineages, and they may never appear as separate sequence clusters.
A many-to-one correspondence between ecologically distinct populations is also possible under the ‘nano-niche model’. Here, we postulate a great diversity of types of ephemeral habitats, e.g. small particles in the marine water column (M. Polz 2005, personal communication). In this model, each of the subgroups within one ecotype become adapted in nuanced ways to the subtleties of their own habitats; they may even have their own separate periodic selection events. Nevertheless, it may be possible for one especially competitive adaptive mutant to outcompete to extinction all the ecological diversity among the ecotype's subgroups (previously called a ‘speciation-quashing’ event; Cohan 2005). In this case, the various ecologically distinct subgroups within an ecotype are not irreversibly separate and do not have a chance to diverge into separate sequence clusters.
Clearly, any model that attempts to identify ecotypes from sequence clusters will have to take into account the diversity of models yielding a many-to-one or a one-to-many correspondence between ecotypes and sequence clusters.
7. A paradigm for incorporating evolution and ecology into a systematics of ecotypes
I suggest an approach for demarcation of bacterial ecotypes that takes into account the great diversity of models of bacterial evolution. To this end, I suggest that ecotypes should be demarcated as the smallest groups that (i) show a history of coexistence as separate, ecologically distinct lineages, as inferred from community phylogeny (or an equivalent sequence-based approach) and (ii) show a prognosis for future coexistence, as inferred from the ecological distinctness of the groups in nature.
Consider why ecological distinctness alone is not sufficient to demarcate ecotypes. First, given the potential for horizontal genetic transfer, any two closely related isolates or populations are likely to differ somewhat in their ecology (Joyce et al. 2002; Lawrence & Hendrickson 2003). Clearly, what we want to know goes beyond the assessment of physiological differences that have no bearing on ecological niche in nature. Rather, we need to ascertain that populations are ecologically distinct in a way that allows them to partition resources in nature and thereby coexist into the future. Sequence data provide a means for inferring that ecological differences observed in the laboratory are important in nature. When two ecologically distinct populations fall into distinct sequence clusters, we may infer that the populations are longstanding in their coexistence, possibly owing to their ecological differences (alternatively to previous geographical separation; Godreuil et al. 2005).
There is a second problem in identifying populations as different ecotypes when they are not yet separate sequence clusters. The populations might not be irreversibly separate, a case most probable when populations owe their ecological distinctness entirely to the gain or loss of a plasmid (the recurrent niche invasion model), or when ecological differences between populations are not sufficient to indefinitely evade one another's periodic selection events (nano-niche model). However, if we demarcate ecotypes only when ecologically distinct groups form separate sequence clusters, we can be assured that the populations have had a history of divergence as separate lineages.
Why should we not demarcate ecotypes solely by sequence clustering? To the extent that the stable ecotype model is correct, different sequence clusters are indeed likely to represent different ecotypes. However, to the extent that the geotype-plus-Boeing model applies, different clusters could represent formerly isolated populations of the same ecotype that have recently been flown or shipped to the same locations. Alternatively, in cases where drift is likely to be an important force, an ecotype could contain multiple sequence clusters caused by genetic drift. Therefore, sequence clusters must be verified to be ecologically distinct before they can be declared ecotypes.
Finally, we must take into account models like the species-less model, in which ecologically distinct populations are frequently too new to be distinguishable as sequence clusters. I believe that we will not normally want to grant ecotype status to a new population that has not yet demonstrated its ability to coexist with others (by forming a separate sequence cluster), but there are clearly some cases where we would want to waive the sequence cluster requirement. Some newly arisen pathogens, for example, are difficult to distinguish from closely related populations by sequences of protein-coding genes (e.g. Bacillus anthracis versus Bacillus cereus; Keim & Smith 2002), but the ecological distinctness we observe (regarding virulence) is clearly relevant to the ways that the bacteria make a living in nature, and the ecological distinctness of these groups is not readily reversible (e.g. with the gain or loss of a plasmid; Welkos et al. 1993). Therefore, it is reasonable to give a prognosis for the continued coexistence of such populations as separate lineages.
In summary, to accommodate models yielding a one-to-many correspondence between ecotypes and sequence clusters, systematists would need to confirm that putative ecotypes identified as sequence clusters are ecologically distinct from one another. To accommodate models with a many-to-one correspondence, we will need to confirm that each putative ecotype is ecologically homogeneous within itself.
Community phylogeny promises to be an effective way to begin the process of identifying the putative ecotypes within a community. In all the systems we have analysed, community phylogeny has succeeded in identifying ecotypes with both a history of coexistence (indicated by sequence clustering) and a prognosis for future coexistence (indicated by ecological distinctness in nature; Cohan et al. submitted). By identifying these fundamental units, community phylogeny should allow ecologists to focus on groups most likely to differ in adaptations of physiology, genome content and gene expression, an important step towards identifying the myriad ecological interactions within a community.
Some important pragmatic issues remain. First, how do we determine that putative ecotypes identified by community phylogeny (or a similar sequence-based approach) are indeed ecologically distinct? Some critical evidence will come from the microhabitats from which the organisms are isolated, especially if ecologists and systematists are dedicated to providing highly detailed information about the habitat. Other critical evidence will surely come from post-genomic approaches, such as comparing genome content or genome-wide gene expression, either at the transcription level or the proteosystematic level (Godreuil et al. 2005). Much of the evidence would draw on the existing skills of polyphasic taxonomists who are trained in testing the capabilities of the growth of organisms with different resources and under different physical conditions. Therefore, I would recommend a new charge for polyphasic taxonomy to move from finding diagnostic phenotypic characters (Vandamme et al. 1996) to using the broadest diversity of techniques to assess ecological differences.
Giving taxonomic status to ecotypes would encourage the routine practice of identifying pathogens to go a step further than the named species. For example, identifying an unknown pathogen to ecotype may yield more specific information about the strain's properties of carriage, transmitting disease and treatment. Granting taxonomic status to ecotypes is most straightforward in cases when a newly described ecotype falls outside any named species. For example, we have used community phylogeny to determine that some sequence clusters within the genus Legionella are not in the same ecotype as the most closely related named species (Cohan et al. 2006). In these cases, the ecotypes can simply be labelled as new species.
Consider next the case when a named species is found to contain multiple ecotypes, each with a history of coexistence and a prognosis for further coexistence. While it is acknowledged that there are probably many such named species, there is understandably little enthusiasm to rename the existing species (Gevers et al. 2005). A reasonable solution is to keep the demarcations of legacy named species as is, but to give a trinomial name with an ecovar label to infraspecific groups confirmed to have a history of past coexistence and a prognosis for future coexistence (e.g. perhaps L. pneumophila ecovar pneumophila; Gevers et al. 2005).
A more precise taxonomy, providing names to all the long-coexisting and ecologically distinct groups within a community, will allow systematics to uphold its obligation to ecologists and epidemiologists to allow accurate predictions of the properties of newly isolated organisms (Mayr 1963; Hutchinson 1968).
Acknowledgments
This work was supported by grants from the National Science Foundation (EF-0328698) and the National Aeronautics and Space Administration (NAG5-8824).
Footnotes
One contribution of 15 to a Discussion Meeting Issue ‘Species and speciation in micro-organisms’.
References
- Acinas S.G, Klepac-Ceraj V, Hunt D.E, Pharino C, Ceraj I, Distel D.L, Polz M.F. Fine-scale phylogenetic architecture of a complex bacterial community. Nature. 2004;430:551–554. doi: 10.1038/nature02649. doi:10.1038/nature02649 [DOI] [PubMed] [Google Scholar]
- Alm R.A, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. doi:10.1038/16495 [DOI] [PubMed] [Google Scholar]
- Baas-Becking L.G.M. The Hague. W.P. Van Stockum & Zoon; The Netherlands: 1934. Geobiologie of Inleiding tot de Milieukunde. [Google Scholar]
- Bansal A.K, Meyer T.E. Evolutionary analysis by whole-genome comparisons. J. Bacteriol. 2002;184:2260–2272. doi: 10.1128/JB.184.8.2260-2272.2002. doi:10.1128/JB.184.8.2260-2272.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beijerinck M.W. Müller; Amsterdam, The Netherlands: 1913. Jaarboek van de Koninklijke Akademie v. Wetenschappen. [Google Scholar]
- Bergthorsson U, Ochman H. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol. Biol. Evol. 1998;15:6–16. doi: 10.1093/oxfordjournals.molbev.a025847. [DOI] [PubMed] [Google Scholar]
- Berk S.G, Ting R.S, Turner G.W, Ashburn R.J. Production of respirable vesicles containing live Legionella pneumophila cells by two Acanthamoeba spp. Appl. Environ. Microbiol. 1998;64:279–286. doi: 10.1128/aem.64.1.279-286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthier Y, Verdier V, Guesdon J.L, Chevrier D, Denis J.B, Decoux G, Lemattre M. Characterization of Xanthomonas campestris pathovars by rRNA gene restriction patterns. Appl. Environ. Microbiol. 1993;59:851–859. doi: 10.1128/aem.59.3.851-859.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky C.W, Wolf C, Maughan H, Herbertson L, Henry E. Speciation and selection without sex. Hydrobiologia. 2005;546:29–45. doi:10.1007/s10750-005-4097-2 [Google Scholar]
- Blaxter M.L. The promise of a DNA taxonomy. Phil. Trans. R. Soc. B. 2004;359:669–679. doi: 10.1098/rstb.2003.1447. doi:10.1098/rstb.2003.1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohannan B.J, Hughes J. New approaches to analyzing microbial biodiversity data. Curr. Opin. Microbiol. 2003;6:282–287. doi: 10.1016/s1369-5274(03)00055-9. doi:10.1016/S1369-5274(03)00055-9 [DOI] [PubMed] [Google Scholar]
- Boucher Y, Douady C.J, Sharma A.K, Kamekura M, Doolittle W.F. Intragenomic heterogeneity and intergenomic recombination among haloarchaeal rRNA genes. J. Bacteriol. 2004;186:3980–3990. doi: 10.1128/JB.186.12.3980-3990.2004. doi:10.1128/JB.186.12.3980-3990.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüggemann H, Hagman A, Coppée J, Dillies M, Heuner K, Steinert M, Gouyette C, Buchrieser C. Bringing the genome of L. pneumophila to life: gene expression patterns of the replicative and transmissive phase in vitro. In: Cianciotto N.P, et al., editors. Legionella. ASM Press; Washington, DC: 2006. [Google Scholar]
- Chase J.M, Leibold M.A. University of Chicago; Chicago, IL: 2003. Ecological niches: linking classical and contemporary approaches. [Google Scholar]
- Cohan F.M. The effects of rare but promiscuous genetic exchange on evolutionary divergence in prokaryotes. Am. Nat. 1994;143:965–986. doi: 10.1016/0169-5347(94)90081-7. doi:10.1086/285644 [DOI] [PubMed] [Google Scholar]
- Cohan F.M. Does recombination constrain neutral divergence among bacterial taxa? Evolution. 1995;49:164–175. doi: 10.1111/j.1558-5646.1995.tb05968.x. doi:10.2307/2410302 [DOI] [PubMed] [Google Scholar]
- Cohan F.M. The role of genetic exchange in bacterial evolution. ASM News. 1996;62:631–636. [Google Scholar]
- Cohan F.M. Bacterial species and speciation. Syst. Biol. 2001;50:513–524. doi: 10.1080/10635150118398. [DOI] [PubMed] [Google Scholar]
- Cohan F.M. Population structure and clonality of bacteria. In: Pagel M, editor. Encyclopedia of evolution. vol. 1. Oxford University Press; New York, NY: 2002a. pp. 161–163. [Google Scholar]
- Cohan F.M. What are bacterial species? Annu. Rev. Microbiol. 2002b;56:457–487. doi: 10.1146/annurev.micro.56.012302.160634. doi:10.1146/annurev.micro.56.012302.160634 [DOI] [PubMed] [Google Scholar]
- Cohan F.M. Concepts of bacterial biodiversity for the age of genomics. In: Fraser C.M, Read T.D, Nelson K.E, editors. Microbial genomes. Humana; New Jersey, NJ: 2004. pp. 175–194. [Google Scholar]
- Cohan F.M. Periodic selection and ecological diversity in bacteria. In: Nurminsky D, editor. Selective sweep. Landes Bioscience; Georgetown, TX: 2005. [Google Scholar]
- Cohan F.M, Koeppel A, Krizanc D. Sequence-based discovery of ecological diversity within Legionella. In: Cianciotto N.P, et al., editors. Legionella. ASM Press; Washington, DC: 2006. [Google Scholar]
- Cohan, F. M. et al Submitted. Identifying the fundamental units of bacterial diversity.
- Coleman M.L, Sullivan M.B, Martiny A.C, Steglich C, Barry K, Delong E.F, Chisholm S.W. Genomic islands and the ecology and evolution of Prochlorococcus. Science. 2006;311:1768–1770. doi: 10.1126/science.1122050. doi:10.1126/science.1122050 [DOI] [PubMed] [Google Scholar]
- Curtis T.P, Head I.M, Lunn M, Woodcock S, Schloss P.D, Sloan W.T. What is the extent of prokaryotic diversity? Phil. Trans. R. Soc. 2006;361:2023–2038. doi: 10.1098/rstb.2006.1921. doi:10.1098/rstb.2006.1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clerck E, Rodriguez-Diaz M, Forsyth G, Lebbe L, Logan N.A, DeVos P. Polyphasic characterization of Bacillus coagulans strains, illustrating heterogeneity within this species, and amended description of the species. Syst. Appl. Microbiol. 2004;27:50–60. doi: 10.1078/0723-2020-00250. doi:10.1078/0723-2020-00250 [DOI] [PubMed] [Google Scholar]
- de Queiroz K. The general lineage concept of species, species criteria, and the process of speciation. In: Howard D.J, Berlocher S.H, editors. Endless forms: species and speciation. Oxford University Press; Oxford, UK: 1998. [Google Scholar]
- Dobrindt U. (Patho-)Genomics of Escherichia coli. Int. J. Med. Microbiol. 2005;295:357–371. doi: 10.1016/j.ijmm.2005.07.009. doi:10.1016/j.ijmm.2005.07.009 [DOI] [PubMed] [Google Scholar]
- Dykhuizen D.E. Santa Rosalia revisited: why are there so many species of bacteria? Antonie Van Leeuwenhoek. 1998;73:25–33. doi: 10.1023/a:1000665216662. doi:10.1023/A:1000665216662 [DOI] [PubMed] [Google Scholar]
- Feil E.J, et al. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl Acad. Sci. USA. 2001;98:182–187. doi: 10.1073/pnas.98.1.182. doi:10.1073/pnas.98.1.182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil E.J, Li B.C, Aanensen D.M, Hanage W.P, Spratt B.G. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. doi:10.1128/JB.186.5.1518-1530.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldgarden M, Byrd N, Cohan F.M. Gradual evolution in bacteria: evidence from Bacillus systematics. Microbiology. 2003;149:3565–3573. doi: 10.1099/mic.0.26457-0. doi:10.1099/mic.0.26457-0 [DOI] [PubMed] [Google Scholar]
- Ferris M.J, Kuhl M, Wieland A, Ward D.M. Cyanobacterial ecotypes in different optical microenvironments of a 68 degrees C hot spring mat community revealed by 16S–23S rRNA internal transcribed spacer region variation. Appl. Environ. Microbiol. 2003;69:2893–2898. doi: 10.1128/AEM.69.5.2893-2898.2003. doi:10.1128/AEM.69.5.2893-2898.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk D.J, Helbling L, Wernegreen J.J, Moran N.A. Intraspecific phylogenetic congruence among multiple symbiont genomes. Proc. Biol. Sci. 2000;267:2517–2521. doi: 10.1098/rspb.2000.1314. doi:10.1098/rspb.2000.1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gans J, Wolinsky M, Dunbar J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science. 2005;309:1387–1390. doi: 10.1126/science.1112665. doi:10.1126/science.1112665 [DOI] [PubMed] [Google Scholar]
- Gevers D, et al. Opinion: re-evaluating prokaryotic species. Nat. Rev. Microbiol. 2005;3:733–739. doi: 10.1038/nrmicro1236. doi:10.1038/nrmicro1236 [DOI] [PubMed] [Google Scholar]
- Giovannoni S.J, Stingl U. Molecular diversity and ecology of microbial plankton. Nature. 2005;437:343–348. doi: 10.1038/nature04158. doi:10.1038/nature04158 [DOI] [PubMed] [Google Scholar]
- Godreuil S, Cohan F, Shah H, Tibayrenc M. Which species concept for pathogenic bacteria? An e-debate. Infect. Genet. Evol. 2005;5:375–387. doi: 10.1016/j.meegid.2004.03.004. doi:10.1016/j.meegid.2004.03.004 [DOI] [PubMed] [Google Scholar]
- Gogarten J.P, Doolittle W.F, Lawrence J.G. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- Gordon D.M, Cowling A. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology. 2003;149:3575–3586. doi: 10.1099/mic.0.26486-0. doi:10.1099/mic.0.26486-0 [DOI] [PubMed] [Google Scholar]
- Grishkan, I., Beharav, A., Kirzhner, V. & Nevo, E. In press Adaptive spatiotemporal distribution of soil microfungi in “Evolution Canyon” III, Nahal Shaharut, extreme southern Negev Desert, Israel. Biol. J. Linn. Soc
- Hanage W.P, Fraser C, Spratt B.G. Sequences, sequence clusters and bacterial species. Phil. Trans. R. Soc. B. 2006;361:1917–1927. doi: 10.1098/rstb.2006.1917. doi:10.1098/rstb.2006.1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J.K, Kelley S.T, Pace N.R. New perspective on uncultured bacterial phylogenetic division OP11. Appl. Environ. Microbiol. 2004;70:845–849. doi: 10.1128/AEM.70.2.845-849.2004. doi:10.1128/AEM.70.2.845-849.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson D.A. Bacterial diversity: the range of interesting things that bacteria do. In: Hopwood D.A, Chater K.F, editors. Genetics of bacterial diversity. Academic Press; London, UK: 1989. pp. 4–22. [Google Scholar]
- Hughes J.B, Hellmann J.J, Ricketts T.H, Bohannan B.J. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 2001;67:4399–4406. doi: 10.1128/AEM.67.10.4399-4406.2001. doi:10.1128/AEM.67.10.4399-4406.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson G.E. When are species necessary? In: Lewontin R.C, editor. Population biology and evolution. Syracuse University Press; Syracuse, NY: 1968. pp. 177–186. [Google Scholar]
- Johnson J. Use of nucleic-acid homologies in the taxonomy of anaerobic bacteria. Int. J. Syst. Bacteriol. 1973;23:308–315. [Google Scholar]
- Jolley K.A, Kalmusova J, Feil E.J, Gupta S, Musilek M, Kriz P, Maiden M.C. Carried meningococci in the Czech Republic: a diverse recombining population. J. Clin. Microbiol. 2000;38:4492–4498. doi: 10.1128/jcm.38.12.4492-4498.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce E.A, Chan K, Salama N.R, Falkow S. Redefining bacterial populations: a post-genomic reformation. Nat. Rev. Genet. 2002;3:462–473. doi: 10.1038/nrg820. [DOI] [PubMed] [Google Scholar]
- Keim P, Smith K.L. Bacillus anthracis evolution and epidemiology. Curr. Top. Microbiol. Immunol. 2002;271:21–32. doi: 10.1007/978-3-662-05767-4_2. [DOI] [PubMed] [Google Scholar]
- Keys C.J, Dare D.J, Sutton H, Wells G, Lunt M, McKenna T, McDowall M, Shah H.N. Compilation of a MALDI-TOF mass spectral database for the rapid screening and characterisation of bacteria implicated in human infectious diseases. Infect. Genet. Evol. 2004;4:221–242. doi: 10.1016/j.meegid.2004.02.004. doi:10.1016/j.meegid.2004.02.004 [DOI] [PubMed] [Google Scholar]
- Knell R.J. Syphilis in renaissance Europe: rapid evolution of an introduced sexually transmitted disease? Proc. R. Soc. B. 2004;271(Suppl. 4):S174–S176. doi: 10.1098/rsbl.2003.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J.G. Gene transfer in bacteria: speciation without species? Theor. Popul. Biol. 2002;61:449–460. doi: 10.1006/tpbi.2002.1587. doi:10.1006/tpbi.2002.1587 [DOI] [PubMed] [Google Scholar]
- Lawrence J.G, Hendrickson H. Lateral gene transfer: when will adolescence end? Mol. Microbiol. 2003;50:739–749. doi: 10.1046/j.1365-2958.2003.03778.x. doi:10.1046/j.1365-2958.2003.03778.x [DOI] [PubMed] [Google Scholar]
- Lindsay J.A, Holden M.T. Staphylococcus aureus: superbug, super genome? Trends Microbiol. 2004;12:378–385. doi: 10.1016/j.tim.2004.06.004. doi:10.1016/j.tim.2004.06.004 [DOI] [PubMed] [Google Scholar]
- Logan N.A, Berkeley R.C. Identification of Bacillus strains using the API system. J. Gen. Microbiol. 1984;130:1871–1882. doi: 10.1099/00221287-130-7-1871. [DOI] [PubMed] [Google Scholar]
- Lopez-Lopez A, Bartual S.G, Stal L, Onyshchenko O, Rodriguez-Valera F. Genetic analysis of housekeeping genes reveals a deep-sea ecotype of Alteromonas macleodii in the Mediterranean Sea. Environ. Microbiol. 2005;7:649–659. doi: 10.1111/j.1462-2920.2005.00733.x. doi:10.1111/j.1462-2920.2005.00733.x [DOI] [PubMed] [Google Scholar]
- Martin A.P. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 2002;68:3673–3682. doi: 10.1128/AEM.68.8.3673-3682.2002. doi:10.1128/AEM.68.8.3673-3682.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J, Smith N, O'Rourke M, Spratt B.G. How clonal are bacteria? Proc. Natl Acad. Sci. USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. doi:10.1073/pnas.90.10.4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. Columbia University; New York, NY: 1942. Systematics and the origin of species from the viewpoint of a zoologist. [Google Scholar]
- Mayr E. Harvard University Press; Cambridge, MA: 1963. Animal species and evolution. [Google Scholar]
- Mayr E. Harvard University Press; Cambridge, MA: 1982. The growth of biological thought: diversity, evolution, and inheritance. [Google Scholar]
- Meglitsch P. On the nature of species. Syst. Zool. 1954;3:491–503. doi:10.2307/2411836 [Google Scholar]
- Mishler B.D, Donoghue M.J. Species concepts: a case for pluralism. Syst. Zool. 1982;31:491–503. doi:10.2307/2413371 [Google Scholar]
- Murray R.G, Stackebrandt E. Taxonomic note: implementation of the provisional status Candidatus for incompletely described procaryotes. Int. J. Syst. Bacteriol. 1995;45:186–187. doi: 10.1099/00207713-45-1-186. [DOI] [PubMed] [Google Scholar]
- Nelson K.E, et al. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 2004;32:2386–2395. doi: 10.1093/nar/gkh562. doi:10.1093/nar/gkh562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbo C.L, Dlutek M, Doolittle W.F. Recombination in Thermotoga: implications for species concepts and biogeography. Genetics. 2006;172:759–769. doi: 10.1534/genetics.105.049312. doi:10.1534/genetics.105.049312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Wilson A.C. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J. Mol. Evol. 1987;26:74–86. doi: 10.1007/BF02111283. doi:10.1007/BF02111283 [DOI] [PubMed] [Google Scholar]
- Ochman H, Elwyn S, Moran N.A. Calibrating bacterial evolution. Proc. Natl Acad. Sci. USA. 1999;96:12 638–12 643. doi: 10.1073/pnas.96.22.12638. doi:10.1073/pnas.96.22.12638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N.R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. doi:10.1126/science.276.5313.734 [DOI] [PubMed] [Google Scholar]
- Palmisano M.M, Nakamura L.K, Duncan K.E, Istock C.A, Cohan F.M. Bacillus sonorensis sp. nov. a close relative of Bacillus licheniformis, isolated from soil in the Sonoran Desert, Arizona. Int. J. Syst. Evol. Microbiol. 2001;51:1671–1679. doi: 10.1099/00207713-51-5-1671. [DOI] [PubMed] [Google Scholar]
- Palys T, Nakamura L.K, Cohan F.M. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Bacteriol. 1997;47:1145–1156. doi: 10.1099/00207713-47-4-1145. [DOI] [PubMed] [Google Scholar]
- Papke R.T, Ramsing N.B, Bateson M.M, Ward D.M. Geographical isolation in hot spring cyanobacteria. Environ. Microbiol. 2003;5:650–659. doi: 10.1046/j.1462-2920.2003.00460.x. doi:10.1046/j.1462-2920.2003.00460.x [DOI] [PubMed] [Google Scholar]
- Ratcliff R.M. The problem of complexity. In: Cianciotto N.P, et al., editors. Legionella. ASM Press; Washington, DC: 2006. [Google Scholar]
- Roberts M.S, Cohan F.M. Recombination and migration rates in natural populations of Bacillus subtilis and Bacillus mojavensis. Evolution. 1995;49:1081–1094. doi: 10.1111/j.1558-5646.1995.tb04435.x. doi:10.2307/2410433 [DOI] [PubMed] [Google Scholar]
- Ron E.Z. Host specificity of septicemic Escherichia coli: human and avian pathogens. Curr. Opin. Microbiol. 2006;9:28–32. doi: 10.1016/j.mib.2005.12.001. doi:10.1016/j.mib.2005.12.001 [DOI] [PubMed] [Google Scholar]
- Santos S.R, Ochman H. Identification and phylogenetic sorting of bacterial lineages with universally conserved genes and proteins. Environ. Microbiol. 2004;6:754–759. doi: 10.1111/j.1462-2920.2004.00617.x. doi:10.1111/j.1462-2920.2004.00617.x [DOI] [PubMed] [Google Scholar]
- Schloter M, Lebuhn M, Heulin T, Hartmann A. Ecology and evolution of bacterial microdiversity. FEMS Microbiol. Rev. 2000;24:647–660. doi: 10.1111/j.1574-6976.2000.tb00564.x. doi:10.1111/j.1574-6976.2000.tb00564.x [DOI] [PubMed] [Google Scholar]
- Selander R, Musser J. Population genetics of bacterial pathogenesis. In: Iglewski B, Iglewsk B, Clark V, editors. Molecular basis of bacterial pathogenesis. Academic Press; San Diego, CA: 1990. pp. 11–36. [Google Scholar]
- Sikorski J, Nevo E. Adaptive and incipient sympatric speciation of Bacilllus simplex under microclimatic contrast at “Evolution Canyons” I and II, Israel. Proc. Natl Acad. Sci. 2005;102:15 924–15 929. doi: 10.1073/pnas.0507944102. doi:10.1073/pnas.0507944102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N.H, Kremer K, Inwald J, Dale J, Driscoll J.R, Gordon S.V, van Soolingen D, Hewinson R.G, Smith J.M. Ecotypes of the Mycobacterium tuberculosis complex. J. Theor. Biol. 2006;239:220–225. doi: 10.1016/j.jtbi.2005.08.036. doi:10.1016/j.jtbi.2005.08.036 [DOI] [PubMed] [Google Scholar]
- Stach J.E, Maldonado L.A, Masson D.G, Ward A.C, Goodfellow M, Bull A.T. Statistical approaches for estimating actinobacterial diversity in marine sediments. Appl. Environ. Microbiol. 2003;69:6189–6200. doi: 10.1128/AEM.69.10.6189-6200.2003. doi:10.1128/AEM.69.10.6189-6200.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E, Goebel B.M. Taxonomic note: a place for DNA : DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 1994;44:846–849. [Google Scholar]
- Staley J.T. Speciation and bacterial phylospecies. In: Bull A.T, editor. Microbial diversity and bioprospecting. American Society for Microbiology Press; Washington, DC: 2003. pp. 40–48. [Google Scholar]
- Stebbins G. Columbia University Press; Irvington, NY: 1957. Variation and evolution in plants. [Google Scholar]
- Templeton A. The meaning of species and speciation: a genetic perspective. In: Otte D, Endler J, editors. Speciation and its consequences. Sinauer Associates; Sunderland, MA: 1989. [Google Scholar]
- Thompson J.R, Pacocha S, Pharino C, Klepac-Ceraj V, Hunt D.E, Benoit J, Sarma-Rupavtarm R, Distel D.L, Polz M.F. Genotypic diversity within a natural coastal bacterioplankton population. Science. 2005;307:1311–1313. doi: 10.1126/science.1106028. doi:10.1126/science.1106028 [DOI] [PubMed] [Google Scholar]
- Tindall B.J. Prokaryotic diversity in the Antarctic: the tip of the iceberg. Microb. Ecol. 2004;47:271–283. doi: 10.1007/s00248-003-1050-7. doi:10.1007/s00248-003-1050-7 [DOI] [PubMed] [Google Scholar]
- Torsvik V, Daae F.L, Sandaa R.A, Ovreas L. Novel techniques for analysing microbial diversity in natural and perturbed environments. J. Biotechnol. 1998;64:53–62. doi: 10.1016/s0168-1656(98)00103-5. doi:10.1016/S0168-1656(98)00103-5 [DOI] [PubMed] [Google Scholar]
- Torsvik V, Ovreås L, Thingstad T.F. Prokaryotic diversity–magnitude, dynamics, and controlling factors. Science. 2002;296:1064–1066. doi: 10.1126/science.1071698. doi:10.1126/science.1071698 [DOI] [PubMed] [Google Scholar]
- van Waasbergen L.G, Balkwill D.L, Crocker F.H, Bjornstad B.N, Miller R.V. Genetic diversity among Arthrobacter species collected across a heterogeneous series of terrestrial deep-subsurface sediments as determined on the basis of 16S rRNA and recA gene sequences. Appl. Environ. Microbiol. 2000;66:3454–3463. doi: 10.1128/aem.66.8.3454-3463.2000. doi:10.1128/AEM.66.8.3454-3463.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme P, Pot B, Gillis M, de Vos P, Kersters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme P, et al. Polyphasic taxonomic study of the emended genus Arcobacter with Arcobacter butzleri comb. nov. and Arcobacter skirrowii sp. nov. an aerotolerant bacterium isolated from veterinary specimens. Int. J. Syst. Bacteriol. 1992;42:344–356. doi: 10.1099/00207713-42-3-344. [DOI] [PubMed] [Google Scholar]
- VanValen L. Ecological species, multispecies, and oaks. Taxon. 1976;25:233–239. doi:10.2307/1219444 [Google Scholar]
- Vincent W.F, Bowman J.P, Rankin L.M, McMeekin T.A. Phylogenetic diversity of picocyanobacteria in Arctic and Antarctic ecosystems. In: Bell C.R, Brylinsky M, Johnson-Green P, editors. Microbial systems: new frontiers. Atlantic Canada Society for Microbial Ecology; Halifax, Nova Scotia: 2000. [Google Scholar]
- von Dohlen C.D, Rowe C.A, Heie O.E. A test of morphological hypotheses for tribal and subtribal relationships of Aphidinae (Insecta: Hemiptera: Aphididae) using DNA sequences. Mol. Phylogenet. Evol. 2006;38:316–329. doi: 10.1016/j.ympev.2005.04.035. doi:10.1016/j.ympev.2005.04.035 [DOI] [PubMed] [Google Scholar]
- Ward D.M. A natural species concept for prokaryotes. Curr. Opin. Microbiol. 1998;1:271–277. doi: 10.1016/s1369-5274(98)80029-5. doi:10.1016/S1369-5274(98)80029-5 [DOI] [PubMed] [Google Scholar]
- Ward D.M, Cohan F.M. Microbial diversity in hot spring cyanobacterial mats: pattern and prediction. In: Inskeep W.P, McDermott T, editors. Geothermal biology and geochemistry in Yellowstone National Park. Thermal Biology Institute; Bozeman, MT: 2005. pp. 185–202. [Google Scholar]
- Ward T.J, Gorski L, Borucki M.K, Mandrell R.E, Hutchins J, Pupedis K. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J. Bacteriol. 2004;186:4994–5002. doi: 10.1128/JB.186.15.4994-5002.2004. doi:10.1128/JB.186.15.4994-5002.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D.M, Bateson M.M, Ferris M.J, Kühl M, Wieland A, Koeppel A, Cohan F.M. Cyanobacterial ecotypes in the microbial mat community of Mushroom Spring (Yellowstone National Park, Wyoming) as species-like units linking microbial community composition, structure and function. Phil. Trans. R. Soc. B. 2006;361:1997–2008. doi: 10.1098/rstb.2006.1919. doi:10.1098/rstb.2006.1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne L, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 1987;37:463–464. [Google Scholar]
- Welkos S.L, Vietri N.J, Gibbs P.H. Non-toxigenic derivatives of the Ames strain of Bacillus anthracis are fully virulent for mice: role of plasmid pX02 and chromosome in strain-dependent virulence. Microb. Pathog. 1993;14:381–388. doi: 10.1006/mpat.1993.1037. doi:10.1006/mpat.1993.1037 [DOI] [PubMed] [Google Scholar]
- Whitaker R.J, Grogan D.W, Taylor J.W. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science. 2003;301:976–978. doi: 10.1126/science.1086909. doi:10.1126/science.1086909 [DOI] [PubMed] [Google Scholar]
- Whittam T.S. Genetic variation and evolutionary processes in natural populations of E. coli. In: Curtiss R, Ingraham J.L, Lin E.C.C, Low K.B, Magasanik B, Reznikoff W.S, Riley M, Schaechter M, Umbarger H.E, editors. E. coli and Salmonella. ASM Press; Washington, DC: 1996. pp. 2708–2720. [Google Scholar]
- Wiley E. The evolutionary species concept reconsidered. Syst. Zool. 1978;27:17–26. doi:10.2307/2412809 [Google Scholar]
- Zeigler D.R. Gene sequences useful for predicting relatedness of whole genomes in bacteria. Int. J. Syst. Evol. Microbiol. 2003;53:1893–1900. doi: 10.1099/ijs.0.02713-0. doi:10.1099/ijs.0.02713-0 [DOI] [PubMed] [Google Scholar]