Abstract
Technological developments provide new insights into prokaryotic evolution and diversity and provoke a continuous need to update taxonomy and revise classification schemes. Our present species concept and definition are being challenged by the growing amount of whole genomic information, which should allow improvements in the natural species definition. The continuous quest for an objective and stable method for sorting strains into coherent homogeneous groups is inherent to prokaryotic systematics and nomenclature. Morphological, biochemical, physiological, phenotypic and chemotaxonomic criteria have been complemented by molecular data and pragmatic, purpose built, species definitions are being replaced by more natural ones based on evolutionary insights. It is imperative to give due consideration to both fundamental and applied aspects of future species concepts and definitions. The present paper discusses the present practice in prokaryotic taxonomy of how this system developed and how it may evolve in the future.
Keywords: prokaryotes, taxonomy, landscaping prokaryotic systems, species definition
1. Introduction
Taxonomy is an essential discipline in biology, as it provides a reference system for all biological knowledge. For prokaryotes, it essentially comprises: (i) classification, i.e. the organization of large numbers of individual strains into an orderly framework based upon the similarities of their biochemical, physiological, genetic and morphological characteristics, (ii) creation of a satisfactory phylogenetic and evolutionary framework, (iii) nomenclature, i.e. the labelling of individual groups in the framework with a binomial name according to strict rules and (iv) identification, i.e. determination of discriminating properties for rapid recognition of new isolates. Ever since the pioneering days of microbiology, scientists have been on a quest for a satisfactory classification system for prokaryotes. The system sought after is based on objective methods that allow discrimination of species on the basis of natural relationships and to reconstruct a hierarchical evolutionary scheme.
Even to date, there is no official classification system for prokaryotes that fulfils these requirements and is followed by all microbiologists. This has led to the remark of Brenner et al. (2001) that the closest to an official classification is the one that is widely accepted by the community. The main reason for this lack is that any effort to produce a robust species definition is hindered by the lack of a solid theoretical basis explaining the effect of biological processes on cohesion within and divergence between prokaryotic species (Cohan 2002, 2006). The Linnean binomial nomenclature followed in prokaryotic classification suggests analogy between prokaryotic and eukaryotic species which are completely different biological systems. Moreover, it is being questioned whether a hierarchical evolutionary scheme can even be constructed, as this ignores genomic variability as a consequence of horizontal gene transfer (Arber 2000). Classification schemes based on only vertical inheritance can have little claim of being natural. Therefore, it has been suggested that the phylogeny of microbial species might be better described as a network rather than by the use of hierarchical trees (Kunin et al. 2005).
2. Historical foundations
Throughout the history of prokaryotic systematics, a number of technological developments were introduced to address the theoretical void and develop a more natural species definition. Old prokaryotic classification schemes relied heavily upon morphological criteria at first and physiological characteristics thereafter. Subsequent and contemporary schemes have introduced evolutionary information extracted from DNA, RNA and protein sequences by using methods that measure genetic relatedness, including DNA–DNA hybridization (DDH), DNA–rRNA hybridization, rRNA oligo-cataloguing, rRNA gene sequencing and protein sequencing (Brenner et al. 2001). Today, DDH and sequencing are the corner-stones of prokaryotic taxonomy (Stackebrandt et al. 2002). This is based on the recommendations of ad hoc committees of experts (Wayne et al. 1987; Stackebrandt et al. 2002). In 1987, it was agreed that the complete DNA sequence should be the reference standard to determine phylogeny and that phylogeny should determine taxonomy. The committee also concluded that the best applicable procedure which approximated this agreement was genomic DDH. In 2002, the committee acknowledged the value of this method and recommended that it should remain the standard for species delineation owing to the lack of a better alternative; but at the same time, investigators were encouraged to develop new methods and demonstrate their congruence with DDH. It should be emphasized that DDH was recommended as a reference standard and not a ‘gold standard’. It has its weaknesses (Vandamme et al. 1996), but it cannot be replaced until another approach has been evaluated as equivalent or superior.
Major technological shifts in the period between these two recommendation reports have been the accuracy and speed in analysing phenotypic properties and the adaptation of molecular techniques to prokaryotic systematics. One of these molecular techniques, i.e. comparative sequence analysis of 16S rRNA genes, allows determination of the phylogenetic position of novel isolates and the establishment of a more objective classification of prokaryotes (Rossello-Mora & Amann 2001). The advent of rapid and cost-effective DNA sequence analysis and the accumulation of rRNA sequence data into a comprehensive reference framework (GenBank–EMBL–DDBJ) limit the need for cumbersome physico-chemical measurements of genomic similarity. Strains showing less than 97% 16S rRNA sequence similarity to all known taxa are considered to belong to a new species, as there are hardly any examples in which strains with this extent of divergence in 16S rRNA sequence are defined as one species (Rossello-Mora & Amann 2001). Nowadays, in line with the recommendations of the last ad hoc committee (Stackebrandt et al. 2002), species descriptions routinely include a 16S rRNA gene sequence, assuring an up-to-date reference framework. It is not wise to base the prokaryotic species delineation on 16S rRNA gene sequence comparisons alone (Stackebrandt & Goebel 1994). Indeed, its lack of resolving power at the species level often hinders the recognition of groups of strains that are otherwise genetically well separated from their phylogenetic neighbours (Fox et al. 1992).
3. Present taxonomical practice
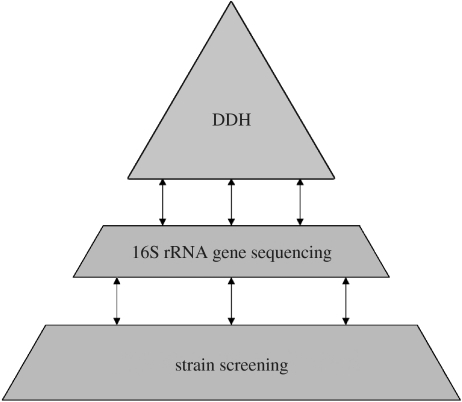
A pragmatic polyphasic approach aims to attain a consensus classification by integrating different kinds of data and information into a classification of minimal contradictions (Vandamme et al. 1996). This approach includes phenotypic data (e.g. biochemical tests), chemotaxonomic data (e.g. fatty acid composition), genotypic data (e.g. DNA fingerprints) and phylogenetic information (e.g. rRNA gene sequences). As depicted in figure 1, the typical characterization of a collection of isolates can be subdivided into three major levels, with each higher level representing a gain in information and a reduction in the number of strains to be included in further analyses. It starts with a screening using different techniques that allows the more closely related isolates to be clustered and to be distinguished from less closely related isolates. On the basis of this clustering, a set of representatives of the different clusters can be selected, and 16S rRNA gene sequencing can be carried out on these representatives to determine the phylogenetic position of each cluster. Based on the results of this analysis, organisms are selected for DDH experiments if required and species are defined using the 70% DDH cut-off criterion (Wayne et al. 1987). The advantage of a polyphasic approach is twofold: (i) it allows a more meaningful definition of the borders demarcated by DDH (especially in the twilight zone between 60 and 80% DDH), and (ii) if calibrated against DDH using well-characterized reference strains, then the use of DDH and rRNA sequencing can be limited when studying a large dataset.
Figure 1.
The present polyphasic practice consists of screening the isolates using different techniques to cluster-relate isolates. On the basis of the consensus clustering, a set of representative strains is subjected to 16S rRNA gene sequencing to determine the phylogenetic position. The second level thus represents a gain in information for a reduced set of strains. If necessary, DNA–DNA hybridization (DDH) is performed on a selection of strains.
For describing new species, one needs to show phenetic and genomic coherence among the members of the new species and to find a diagnostic phenotype that discriminates a given species from its closest relatives. According to the recommendations, a group of strains that is solely separated on the basis of genotypic data (genospecies) should not be named if it cannot be differentiated from other genospecies by a phenotypic (including chemotaxonomic) property (Wayne et al. 1987). The requirement for discriminatory phenotypes underlines the importance of phenotypic and chemotaxonomic analyses in our present descriptive taxonomic practice. Unfortunately, it is the most tedious and time-consuming task in the classification of micro-organisms. It necessitates bringing a set of representative members of a given species into pure culture and studying them independently. Complete genome sequences may help us to understand the link between genotype and phenotype, thereby making phenotypic testing obsolete.
As W. Heisenberg stated, ‘what we observe is not nature itself, but nature exposed to our methods of questioning’. This indicates why, along the pathway of searching for methods for a natural definition of organismal space, we have obtained new insights that influence the prokaryotic species definition. The latter has been improved, towards an operational and universally applicable system, but remains to some extent subjective, artificial and pragmatic, and therefore subject to recurrent controversies (Rossello-Mora & Amann 2001). It is subjective, because the objective data are interpreted by the subjective opinion of a taxonomist. It is artificial, because many named species delineated on the basis of the present taxonomic system are not real entities, but useful groupings that are not necessarily meaningful from an evolutionary standpoint (Lan & Reeves 2001). It is, and should be, pragmatic, as the ultimate function of the definition is to serve as a tool for the identification of individual isolates and to allow communication, which is essential when dealing with bacteria that have a vital role in clinical settings, food processing, industry, agriculture, bioremediation and public health. It should be kept in mind that most biologists today are the end-users of the classification systems and nomenclature produced by a small group of taxonomists. The taxonomic reality is well phrased by Staley and Krieg (1984): ‘a classification that is of little use to microbiologists, no matter how fine a scheme or who devised it will soon be ignored or significantly modified’.
4. Reflections
The present practice does raise some topics for reflection.
Although more advanced molecular methods are used, the cut-offs used for demarcating species have been calibrated to yield the species groupings previously determined by phenotypic clustering (Cohan 2002). The 3% cut-off for 16S rRNA divergence was calibrated to yield the species previously determined by DDH, and the 70% cut-off for DDH was calibrated on the basis of the phenotypic clusters previously recognized as separate species, mostly in the Enterobacteriaceae. Essentially, each new molecular technique is calibrated to yield the clusters previously determined by phenotypic criteria owing to the lack of a theory-based species concept. Microbiologists need to adopt a more natural view of the organisms they study and cope with the fact that a grounding in principles of ecology and evolution is presently lacking (Ward 1998). Moreover, the large uncultured majority (Rappe & Giovannoni 2003) pose an enormous challenge to any approach to describe and catalogue prokaryotic diversity, driving the future in the direction of a sequence-based taxonomy. Moving in the direction of more comprehensive genomic comparisons, multilocus sequence analysis (MLSA) was proposed as a next logical development (Stackebrandt et al. 2002; Gevers et al. 2005). Large-scale MLSA studies of different genera could provide a framework for in-depth study of the biological, ecological and genetic differentiation between clusters (Gevers et al. 2005). Such studies could provide the criteria to determine how we should define species and subdivisions within species. But the requirement that new species are described without sequencing their complete genome (or at least a draft version thereof) seems not at all that far away any more. The technology to do so is likely to become sufficiently widespread and accessible within a short time-frame. How this will influence our vision on the prokaryotic species is something we should start considering today (Konstantinidis & Tiedje 2005; Konstantinidis et al. 2006).
A second reflection is based on the main recommendation of the 1987 ad hoc committee, which states that the complete genome should be used as the reference standard for taxonomy (Wayne et al. 1987). During the last decade, it became apparent that the evolutionary history of the prokaryotic genome does not simply reflect the history of the organism (nucleotide changes or mutational history), but that different genes can indicate a different phylogenetic history (Coenye et al. 2005). Other processes such as gene loss, gene duplication, horizontal gene transfer, homologous recombination and chromosomal rearrangements shape the prokaryotic genome and are far more widespread than previously thought. Why rampant gene transfer has not eradicated ancient groups, and why we can obtain meaningful phylogenies, is an ongoing debate that is rooted in the prokaryotic paradox: higher-ordered taxonomy in prokaryotes (i.e. above species level) is supported both by rRNA and whole genome-based phylogenies; yet, DNA is transferred between and among distantly related taxa, resulting in individual gene trees that do not always match the rRNA tree. Therefore, the question of whether a hierarchical evolution can be reconstructed is still open (Creevey et al. 2004; Susko et al. 2006). If we want to understand what a prokaryotic species represents, and thereby improve our methods for demarcating species, we need to focus on the prokaryotic paradox and the fundamental questions: how do species emerge, become distinct and remain distinct?. But to escape this circular reasoning, which presupposes that we know what a prokaryotic species is, we should rephrase this as: how did the different prokaryotic systems emerge, separate and come into being as distinct entities? Genomic data will guide us to find the answers.
5. Towards landscaping of the prokaryotic systems
There is a growing need to reinvent prokaryotic taxonomy as a twenty-first century information science (Godfray 2002). The polyphasic approach has proven its value in prokaryotic taxonomy, but obviously it is not satisfactory to many end-users. At the present rate and with present methods, taxonomists cannot cope with the huge microbial diversity that remains to be revealed. First, it will take too much additional time and effort until most diversity will be described, named and arranged in a satisfactory classification scheme. Second, a tedious manual approach of the integration of dozens of information sources has restricted the number of strains per taxon included in most taxonomic studies to numbers not representative of the diversity within the taxon. This makes the definition of the taxon boundaries straightforward at first, but often ineffective when extending the use of the boundary to larger datasets. Third, a great deal of the decision-making for deriving a consensus view of the data is left to the microbiologist's personal interpretation. This turns the validation of the existing species definition against new empirical information into a slow and long process. Finally, many of the end-users are inclined to circumvent this time-consuming descriptive taxonomy and prefer a single-step phylogenetic taxonomy. An increasingly common practice is the unofficial allocation of isolates to new species solely on the basis of 16S rRNA gene sequencing, with many examples in recent community genomics studies (Allen & Banfield 2005). The latter method is rapid, less laborious and portable. Portability is particularly useful because it has allowed the implementation of a centralized database and online placement of new isolates in a universal context, even when the isolates cannot be cultured. As a consequence, some species are described poorly, with no attempt to relate the new taxon to existing species and classification systems through an in-depth description, not at all according to the guidelines.
Addressing the above taxonomic impediments will revolutionize the field into a global information system, i.e. an integrated comprehensive information gateway. Such a system stipulates that the data are captured by standardized methods in a reproducible and fast way yielding digitized and portable data of high resolution. The resulting increase in information will enable an enlargement of our understanding of natural phenomena and our capacities to conceptualize them. A sequence-based method is the most promising future for microbial systematics and taxonomy, fulfilling the above requirements. The method that can replace the need for combining a set of screening methods and copes with the vagaries of the single-gene approach of 16S rRNA gene sequencing is the MLSA approach (Gevers et al. 2005); it is discussed in more detail elsewhere in this issue (Hanage et al. 2006). This indeed allows objective clustering of strains within a genus at the inter- and intraspecific level, which previously required a combination of DNA fingerprinting, chemotaxonomic and other methods. But going from objective clusters towards the delineation of biologically meaningful taxonomical groups requires more than just the sequences provided by MLSA, or even whole genome sequences. Efforts should also be made to collect phenotypic, biological and ecological data for the isolates and capture the vast amount of experimental data that are generated in a structured and uniform way.
The added value of such a global information system relies on: (i) the extent to which all data are then integrated, and (ii) the potential of the system to adapt in a flexible way to the advent of new incoming data. The intelligent application of well-founded data mining techniques on this information system will allow the recognition of recurrent patterns in the process of diversification, as a means to discover objective taxonomic consensus models in a more dynamic and a more automated manner. The different or even contradictory viewpoints for the prokaryotic species concepts will inevitably give rise to diverse models and thus a parallel taxonomy. At least, the old-fashioned style of repetitive species descriptions is expected to become obsolete, and can then basically be taken over by the global information system and large-scale computational modelling of prokaryotic systems. The latter is referred to as ‘landscaping of the prokaryotic systems’ based on genomic and all other experimental data (Dawyndt 2004). As a result, the taxonomist can focus on resolving the more interesting evolutionary questions and other fundamental biological issues. The future taxonomic needs can be best illustrated by an example from the field of environmental microbiology. The biggest challenge in metagenomics from a taxonomic point of view is how to extract comprehensive taxonomic data from the samples of complex communities, and perform qualitative and quantitative comparative analyses of the populations between samples from different locations, under different conditions, etc., and all this within a relative short time-frame. This requires taxonomy to become less complex, but more automated.
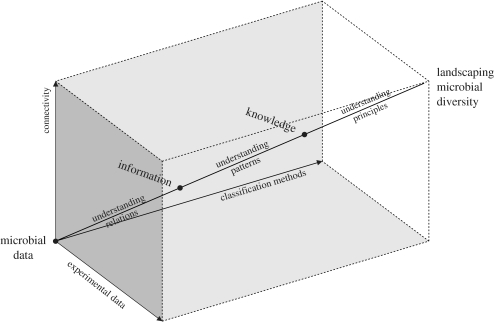
Presently, extensive taxonomic information on the prokaryotes is already available at the click of the mouse (Oren & Stackebrandt 2002), including the Approved Lists of Bacterial Names (www.bacterio.cict.fr), rRNA gene, protein and genome sequences (GenBank–EMBL–DDBJ), microbial reference material in microbial resource centres (www.wfcc.info) and scientific literature (www.ncbi.nlm.nih.gov/entrez/). But large quantities of information on micro-organisms are disseminated over diverse sources, hardly integrated and sensitive to modifications over time. The integrated strain database is a recent example of a successful integration of information on micro-organisms (Dawyndt et al. 2005). It is a central information gateway that offers systematic cross-referencing between resources that supply basic strain information and other public data sources, such as sequence databases and scientific literature databases that provide additional features of the strains. The integration and synchronization of data coming from heterogeneous and autonomous information providers are difficult to achieve and involve many aspects that need to be tackled. This will demand parallel developments of increasingly sophisticated computational, mathematical and statistical routines for data analysis, modelling and integration (Emmott et al. 2006). The future of prokaryotic taxonomy is thus situated on the crossroads between microbiology (including population genetics and microbial ecology), mathematics and computer science (figure 2). Microbiologists should constantly search for additional empirical classifiers for micro-organisms, which are more reproducible (particularly at the inter-laboratory level), have a higher resolution and meet practical requirements, such as speed, automation and cost of execution. The scope of the studies should be larger and include biological, ecological and genetic differentiation between the strains. Computer scientists can contribute by constructing easily accessible data repositories that capture the constant flow of information generated during microbiological practice, and establish the necessary cross-reference links that should enable comprehensive processing of the related pieces of the taxonomic puzzle. Research in mathematics has produced a sheer endless assortment of algorithms for finding and representing groups in datasets and should continue to design objective classification methods for grouping data in an unsupervised way. The present progress in taxonomic modelling of prokaryotic diversity has often restrictively exploited only one or two of these disciplines at the same time. However, within the envisaged microbial global information system, these disciplines should be inextricably tied together.
Figure 2.
Landscaping of the prokaryotic systems. Going from microbial data towards landscaping prokaryotic systems requires improvements in three directions: (i) microbiologists should accumulate more useful data, (ii) mathematicians should develop objective classification methods and (iii) computer scientists should contribute by constructing information gateways of integrated microbial data.
To conclude, the taxonomic future is well phrased by Godfray (2002): ‘Unless taxonomy is unitary, web-based and able to accommodate these radical new ways of doing biology, I fear it will be sidelined.’
Acknowledgments
D.G. and co-authors are indebted to the Fund for Scientific Research—Flanders (Belgium) for a position as postdoctoral fellow and research funding, respectively.
Footnotes
One contribution of 15 to a Discussion Meeting Issue ‘Species and speciation in micro-organisms’.
References
- Allen E.E, Banfield J.F. Community genomics in microbial ecology and evolution. Nat. Rev. Microbiol. 2005;3:489–498. doi: 10.1038/nrmicro1157. doi:10.1038/nrmicro1157 [DOI] [PubMed] [Google Scholar]
- Arber W. Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol. Rev. 2000;24:1–7. doi: 10.1111/j.1574-6976.2000.tb00529.x. doi:10.1111/j.1574-6976.2000.tb00529.x [DOI] [PubMed] [Google Scholar]
- Brenner D.J, Staley J.T, Krieg N.R. Classification of procaryotic organisms and the concept of bacterial speciation. In: Boone D.R, Castenholz R.W, Garrity G.M, editors. Bergey's manual of systematic bacteriology. 2nd edn. vol. 1. Springer; New York, NY: 2001. pp. 27–31. [Google Scholar]
- Coenye T, Gevers D, Van de Peer Y, Vandamme P, Swings J. Towards a prokaryotic genomic taxonomy. FEMS Microbiol. Rev. 2005;29:147–167. doi: 10.1016/j.femsre.2004.11.004. doi:10.1016/j.femsre.2004.11.004 [DOI] [PubMed] [Google Scholar]
- Cohan F.M. What are bacterial species? Annu. Rev. Microbiol. 2002;56:457–487. doi: 10.1146/annurev.micro.56.012302.160634. doi:10.1146/annurev.micro.56.012302.160634 [DOI] [PubMed] [Google Scholar]
- Cohan F.M. Toward a conceptual and operational union of bacterial systematics, ecology, and evolution. Phil. Trans. R. Soc. B. 2006;361:1985–1996. doi: 10.1098/rstb.2006.1918. doi:10.1098/rstb.2006.1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creevey C.J, Fitzpatrick D.A, Philip G.K, Kinsella R.J, O'Connell M.J, Pentony M.M, Travers S.A, Wilkinson M, McInerney J.O. Does a tree-like phylogeny only exist at the tips in the prokaryotes? Proc. Biol. Sci. 2004;271:2551–2558. doi: 10.1098/rspb.2004.2864. doi:10.1098/rspb.2004.2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawyndt P. 2004 Knowledge accumulation of microbial data aiming at a dynamic taxonomic framework. Ph.D. dissertation, Ghent University, Belgium. http://hdl.handle.net/1854/2820
- Dawyndt P, Vancanneyt M, De Meyer H, Swings J. Knowledge accumulation and resolution of data inconsistencies during the integration of microbial information sources. IEEE Trans. Knowledge Data Eng. 2005;17:1111–1126. doi:10.1109/TKDE.2005.131 [Google Scholar]
- Emmott, S. et al 2006 The 2020 Science Report. http://research.microsoft.com/towards2020science/
- Fox G.E, Wisotzkey J.D, Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- Gevers D, et al. Opinion: re-evaluating prokaryotic species. Nat. Rev. Microbiol. 2005;3:733–739. doi: 10.1038/nrmicro1236. doi:10.1038/nrmicro1236 [DOI] [PubMed] [Google Scholar]
- Godfray H.C. Challenges for taxonomy. Nature. 2002;417:17–19. doi: 10.1038/417017a. doi:10.1038/417017a [DOI] [PubMed] [Google Scholar]
- Hanage W.P, Fraser C, Spratt B.G. Sequences, sequence clusters and bacterial species. Phil. Trans. R. Soc. B. 2006;361:1917–1927. doi: 10.1098/rstb.2006.1917. doi:10.1098/rstb.2006.1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis K.T, Tiedje J.M. Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 2005;187:6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. doi:10.1128/JB.187.18.6258-6264.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis K.T, Ramette A, Tiedje J.M. The bacterial species definition in the genomic era. Phil. Trans. R. Soc. B. 2006;361:1929–1940. doi: 10.1098/rstb.2006.1920. doi:10.1098/rstb.2006.1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Goldovsky L, Darzentas N, Ouzounis C.A. The net of life: reconstructing the microbial phylogenetic network. Genome Res. 2005;15:954–959. doi: 10.1101/gr.3666505. doi:10.1101/gr.3666505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan R, Reeves P.R. When does a clone deserve a name? A perspective on bacterial species based on population genetics. Trends Microbiol. 2001;9:419–424. doi: 10.1016/s0966-842x(01)02133-3. doi:10.1016/S0966-842X(01)02133-3 [DOI] [PubMed] [Google Scholar]
- Oren A, Stackebrandt E. Prokaryote taxonomy online: challenges ahead. Nature. 2002;419:15. doi: 10.1038/419015c. doi:10.1038/419015c [DOI] [PubMed] [Google Scholar]
- Rappe M.S, Giovannoni S.J. The uncultured microbial majority. Annu. Rev. Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. doi:10.1146/annurev.micro.57.030502.090759 [DOI] [PubMed] [Google Scholar]
- Rossello-Mora R, Amann R. The species concept for prokaryotes. FEMS Microbiol. Rev. 2001;25:39–67. doi: 10.1111/j.1574-6976.2001.tb00571.x. doi:10.1111/j.1574-6976.2001.tb00571.x [DOI] [PubMed] [Google Scholar]
- Stackebrandt E, Goebel B.M. Taxonomic note: a place for DNA : DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 1994;44:846–849. [Google Scholar]
- Stackebrandt E, et al. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 2002;52:1045–1047. doi: 10.1099/00207713-52-3-1043. [DOI] [PubMed] [Google Scholar]
- Staley J.T, Krieg N.R. Classification of prokaryotic organisms: an overview. In: Krieg N.R, Holt J.G, editors. Bergey's manual of systematic bacteriology. vol. 1. Williams & Wilkins; Baltimore, MD: 1984. pp. 1–4. [Google Scholar]
- Susko E, Leigh J, Doolittle W.F, Bapteste E. Visualizing and assessing phylogenetic congruence of core gene sets: a case study of the {gamma}-Proteobacteria. Mol. Biol. Evol. 2006;23:1019–1030. doi: 10.1093/molbev/msj113. doi:10.1093/molbev/msj113 [DOI] [PubMed] [Google Scholar]
- Vandamme P, Pot B, Gillis M, de Vos P, Kersters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D.M. A natural species concept for prokaryotes. Curr. Opin. Microbiol. 1998;1:271–277. doi: 10.1016/s1369-5274(98)80029-5. doi:10.1016/S1369-5274(98)80029-5 [DOI] [PubMed] [Google Scholar]
- Wayne L.G, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 1987;37:463–464. [Google Scholar]