Abstract
Peptidoglycans from bacterial cell walls trigger immune responses in insects and mammals. A peptidoglycan recognition protein, PGRP, has been cloned from moths as well as vertebrates and has been shown to participate in peptidoglycan-mediated activation of prophenoloxidase in the silk moth. Here we report that Drosophila expresses 12 PGRP genes, distributed in 8 chromosomal loci on the 3 major chromosomes. By analyzing cDNA clones and genomic databases, we grouped them into two classes: PGRP-SA, SB1, SB2, SC1A, SC1B, SC2, and SD, with short transcripts and short 5′-untranslated regions; and PGRP-LA, LB, LC, LD, and LE, with long transcripts and long 5′-untranslated regions. The predicted structures indicate that the first group encodes extracellular proteins and the second group, intracellular and membrane-spanning proteins. Most PGRP genes are expressed in all postembryonic stages. Peptidoglycan injections strongly induce five of the genes. Transcripts from the different PGRP genes were found in immune competent organs such as fat body, gut, and hemocytes. We demonstrate that at least PGRP-SA and SC1B can bind peptidoglycan, and a function in immunity is likely for this family.
In recent years, our knowledge of innate immunity in mammals and insects has increased dramatically. Insects and mammals have developed similar mechanisms and molecular pathways to recognize and eliminate invaders (1–4). Insects are therefore good models for innate immunity in vertebrates. In Drosophila, the activation of the immune response is mediated by membrane receptors of the Toll family and by transcription factors related to NF-κB in mammals (5). They signal the presence of invading microorganisms and induce the production of antimicrobial peptides and proteins, more than 500 of which have been described from different animal species.
Although we have a relatively good understanding of the transcriptional activation of the immune genes in insects, this cannot be said of the initial recognition events. As in mammals, lipopolysaccharide, peptidoglycan, and β-1,3-glucans are good elicitors of the innate immune system and are therefore likely targets for recognition molecules in insects (6–9). Toll and 18-wheeler are membrane receptors that can mediate immune responses in Drosophila (10–12). Toll-like receptors have also been found in mammals, where they mediate reactions to lipopolysaccharide and to Gram-positive bacteria (13–17). However, it is uncertain whether the Toll-like receptors bind directly to the microbial substances, at least in Drosophila, where the signal is mediated to Toll via an endogenous ligand Spätzle (11). At least two classes of proteins bind directly to Gram-negative bacteria in insects and may serve as recognition molecules: the Gram-negative-binding protein GNBP and a C-type lectin (18, 19). It was recently shown that GNBP actually mediates an immune response to lipopolysaccharide in Drosophila (20). However, other recognition molecules must be used to recognize Gram-positive bacteria, because they lack lipopolysaccharides. One interesting candidate is the peptidoglycan recognition protein, PGRP (21, 22).
Peptidoglycans are found in the cell walls of almost all bacteria. In many insects, they induce a strong antibacterial response, and they also trigger the activation of phenoloxidase, leading to the formation of melanin in infected wounds. Humoral PGRPs with a high affinity for peptidoglycan (23) were recently found in the moths Trichoplusia ni (21) and Bombyx mori (22, 24). They are produced in the fat body and in hemocytes (21, 22). The B. mori PGRP is required for the activation of the phenoloxidase cascade (24). A PGRP was also isolated from the cuticle of another lepidopteran, Calpodes ethlius (25), and a homologous expressed sequence tag (EST) found in Anopheles (26). Interestingly, homologs were also found in mouse and humans, where they are strongly expressed in hematopoietic tissue (21).
Recent data from the Drosophila genome project have identified several PGRP homologs. Here we show that they constitute a highly diversified family with at least 12 members. We have characterized these genes in more detail.
Methods
General molecular techniques follow Sambrook et al. (27).
Molecular Cloning of PGRP-SA.
A PGRP-SA-specific primer (Dm1: 5′-AGATCTGCTGGCCTGCGGAGTG) and a vector primer (M13–20) were used to amplify a PGRP-SA cDNA fragment, with a λ DNA library (28) as a template. The PCR product was cloned and sequenced. The insert was used as a probe to screen 5 × 105 plaque-forming units of the cDNA library to obtain a full-length cDNA clone according to standard methods. Three identical clones were found, except for length variations. The longest one was named DmVc16.1.9.1.
Sequencing of Plasmid DNA.
cDNA clones corresponding to PGRP-LA (GH04960, GH05009, GH10945, GH18280, SD09180), LB (GH11251, GH21008), LC (GH11582, SD04722, LP06704), LD (GH14535, GH13671), LE (GH01554), and SC1B (GH07464) were obtained from the Berkeley Drosophila Genome Project/Howard Hughes Medical Institute EST Project (29). We used the Thermo Sequenase II dye terminator cycle sequencing premix kit from Amersham Pharmacia Biotech and 17- to 18-bases-long primers from Interactiva (Ulm, Germany).
PCR of PGRP Domains.
Templates for Northern probes were made with the PCR Supermix kit (GIBCO), 17- to 18-bases-long primers, and genomic fly DNA of the wild-type Canton S strain. Primers were designed to amplify the PGRP domains.
Flies, Cells, and Bacterial Strains.
Canton S flies were used for most experiments and kept on a standard mashed-potato diet. Homozygous l(3)mbn larvae (30) with blood-cell tumors were used to collect hemocytes. The mbn-2 cell line (31) was grown in Schneider's medium (44) with 10% FCS. The bacteria Micrococcus luteus Ml 11, Bacillus megaterium Bm11, and Enterobacter cloacae β12 were originally obtained from Hans G. Boman (Karolinska Institute, Stockholm) (see ref. 32 and refs. therein). The Bacillus subtilis wild-type strain was obtained from Sven Bergström (Department of Microbiology, Umeå University).
Collection of Different Developmental Stages; Preparation of Fat Body, Gut, and Hemocytes, and Collection of mbn-2 Cells.
All samples were frozen on dry ice and kept at −80°C until further use. To obtain synchronized developmental stages, newly hatched Canton S flies were kept at 25°C and fed with additional yeast for 6 days before they were allowed to lay eggs on apple juice plates with yeast. Embryos were collected at 5-h intervals. Larvae were kept on fly food. Eggs and larvae were washed in Drosophila Ringer's solution (182 mM KCl/46 mM NaCl/3 mM CaCl2/10 mM Tris⋅HCl) and frozen. Prepupae, pupae, and 1-day-old flies were frozen without prior washing.
Fat bodies and intestinal tracts (mid- and hindgut) were dissected from third instar Canton S larvae in ice-chilled Drosophila Ringer's solution. The separated tissues and remaining carcasses were frozen. For collection of hemocytes, batches of 100 l(3)mbn larvae were washed in Schneider's medium and rapidly dried on tissue paper. The larvae were opened in 200 μl Schneider's medium containing a small amount of phenylthiocarbamide on ice. After removal of the dead larvae, the hemocyte suspension was frozen. The mbn-2 cells were centrifuged (500 × g) for 10 min before freezing.
Injection of Bacteria and Peptidoglycan.
Newly hatched flies were kept for 6 days at 25°C without additional yeast, washed in 70% ethanol, dried, and injected with approximately 0.1 μl of suspension of M. luteus, B. subtilis, B. megaterium, E. cloacae, or insoluble peptidoglycan isolated from the same bacteria except E. cloacae. Overnight cultures in LB were diluted 1:10 in sterile Ringer's solution for injection. Insoluble peptidoglycan was prepared according to Morishima (33). A 10-μg/μl stock suspension of peptidoglycan was diluted 1:10 in sterile Ringer's solution, and approximately 100 ng per fly was injected. Controls were: (i) no injection, (ii) sterile Ringer's solution, and (iii) sterile Ringer's solution containing 10% LB.
RNA Preparation, Northern Blot, and Hybridization.
Total RNA was prepared with Trizol (GIBCO/BRL) according to the manufacturer. For Northern blots, 15 μg total RNA per lane was run on a 1% agarose gel containing formaldehyde (27). Hybridization was performed under high stringency conditions (50% formamide, 42°C). EST plasmid DNA, cDNA, or PCR fragments were used as templates, and the probes were labeled with the Rediprime II kit from Amersham Pharmacia, according to the manufacturer. The RNA marker was from Promega (0.28–6.25 kb). Radioactivity was monitored by PhosphorImager (Storm 860, Molecular Dynamics).
Baculovirus Expression, Protein Purification, and Antibody Production.
Constructs with six histidine residues at the C terminus of PGRP-SA and SC1B were amplified by PCR with PGRP-SA clone DmVc16.1.9.1 by using primers 5′- CCGCTCGAGTTAATGATGATGATGATGATGGGGATTTGAGAGCCAGTGC- 3′ and 5′-GCGGATAACAATTTCACACAGGAA-3′, and SC1B clone GH07464 by using primers 5′- CCGCTCGAGTTAATGATGATGATGATGATGACCGCCAGACCAGTGGGACCAGCC-3′ and 5′-TAATACGACTCACTATAGGG-3′. The PCR products were sequenced, cleaved with EcoRI and XhoI, and inserted into the baculovirus transfer vector pBackPAK9 (CLONTECH). Amplification of the purified plaques yielded recombinant viruses vPGRP-SA-his and vPGRP-SC1B-his encoding his-tagged PGRP-SA and SC1B, respectively. Proteins were purified by using FPLC Ni-affinity chromatography (Novagen). The purified PGRP-SA-his protein was resolved on a 15% SDS/PAGE gel. After staining with Coomassie brilliant blue (Sigma), a 20-kDa band, corresponding to PGRP-SA-his, was excised from the gel and homogenized in PBS. Rabbit immunization was performed with three injections, each containing 50 μg of protein homogenate.
Amino Acid Sequence Analysis.
After resolution on SDS/PAGE, blotting to Immobilon P membranes (Millipore), and visualization by Coomassie brilliant blue staining, the 20-kDa PGRP-SA band was excised and sequenced by using an Applied Biosystems model 437A Protein Sequencer.
Peptidoglycan-Binding Assay.
Insoluble peptidoglycan was prepared from M. luteus as described (34). The peptidoglycan-binding assay was essentially according to Yoshida et al. (24), incubating peptidoglycan with PGRP in 10 mM maleate buffer, pH 6.5/0.15 M NaCl for 30 min at 4°C. After centrifugation at 13,000 × g for 10 min, the supernatant and the pellet were analyzed on a 15% SDS/PAGE gel. The binding of PGRP-SA was assayed by using Coomassie brilliant blue staining of the SDS/PAGE gel. SC1B was detected by Western blot analysis. Proteins were transferred onto a Hybond-C pure membrane (Amersham). The antibody was a rabbit anti-PGRP-SA, and the second antibody and the detection were carried out as before (21).
Results
PGRP Genes Are Widespread Over the Genome.
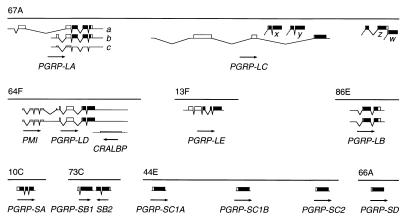
Using tblastn search (35), we routinely checked databases for homologs to the T. ni PGRP. The sequences found were used to search for additional members of the PGRP gene family. We identified 12 homologs in the Drosophila genome, distributed over eight chromosomal loci on the three major chromosomes (Fig. 1). The PGRP-SA gene was first identified in RpII215 flanking sequence (36), the PGRP-SC1B, LA, LB, and LC genes in EST sequences (29), the PGRP-SB1, SB2, SC1A, and SC2 genes in genomic sequence from the Berkeley Drosophila Genome Project, and the PGRP-SD, LD, and LE genes from Celera Genomics (Rockville, MD) (37). We sequenced all relevant cDNA clones from the EST project (29), and for PGRP-SA, we isolated a cDNA clone. By comparison to the genomic sequences, we determined the splicing patterns for all genes except PGRP-SB, SC1A, SC2, and SD. For the PGRP-SB genes, we predicted a splicing pattern, based on conserved ORFs and splice signals. Finally, we assume that PGRP-SC1A, SC2, and SD lack introns, by analogy to PGRP-SC1B. The cDNA sequences identify multiple alternative splicing patterns for the PGRP-LA, LB, and LD genes (Fig. 1).
Figure 1.
The Drosophila PGRP genes. Maps of PGRP transcripts found at eight chromosomal positions. ORFs are shown as boxes, the PGRP domain in black. Lower boxes are ORFs without PGRP domain. The 5′ region of the PGRP-LD transcript has an ORF related to the human PMI gene, and the antisense strand of the same gene has an ORF similar to cellular retinaldehyde-binding protein (CRALBP). The direction of transcription is indicated by arrows.
Near the PGRP-LC gene are four additional blocks of PGRP sequence homology (x, y, z, and w in Fig. 1), which are not represented among the cDNA clones. They could be exons of alternative splice forms of PGRP-LC, independent genes, or pseudogenes.
At Least Two Classes of PGRP Genes.
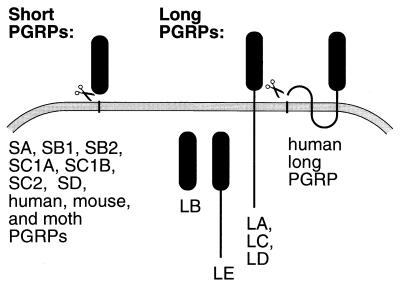
All PGRP genes share a PGRP domain of approximately 160 amino acids, conserved from insects to humans (Fig. 2). Outside the PGRP domain, there is little sequence similarity. Based on their general structure, the PGRP genes can be separated into two classes: the short and the long PGRPs.
Figure 2.
Alignment of the PGRP proteins. Amino acids are color coded by chemical properties. Predicted (38) or experimentally determined signal peptides and transmembrane regions (39) are underlined. Intron positions are indicated by arrowheads. Sequences from T. ni, B. mori, C. ethlius, and human are from refs. 21, 22, and 25. The human long PGRP is from chromosome 19 clone CTB-187L3 (AC011492) and the splicing pattern deduced by comparison to the cDNA of the mouse homolog Tagl-α (AF149837).
The short PGRPs are very similar to the genes previously found in moths and in mammals. The PGRP domain is immediately preceded by a typical signal peptide (processing confirmed for PGRP-SA), and the mature products are predicted to be exported proteins that essentially consist of a single PGRP domain. This class includes PGRP-SA, SB1, SB2, SC1A, SC1B, SC2, and SD. The PGRP-SA gene has two introns in the PGRP domain, and the B. mori and vertebrate PGRP genes also have introns in the corresponding positions (Fig. 2). One of these introns is retained in the PGRP-SB2 gene, whereas all other short genes appear to have lost these introns.
The long PGRP genes, PGRP-LA to LE, are more complex. They all give rise to longer transcripts, in many cases with several splice forms. None of them encodes a typical signal peptide, but PGRP-LA, LC, and LD have a predicted transmembrane region, indicating that they are likely to encode membrane proteins. PGRP-LE has a highly charged N-terminal domain connected to the PGRP domain but no obvious export signal or transmembrane region. Finally, PGRP-LB encodes only the PGRP domain, attached to a relatively long 5′-untranslated region. The splicing pattern in the long genes differs completely from that seen in the short genes. The PGRP domains of the long genes are interrupted by introns at various positions, all different from those seen in the short genes. The exception is PGRP-LB, which shares one intron position with the short genes.
For PGRP-LA, the alternative splice forms encode different proteins. PGRP-LAa has a long N-terminal region with weak sequence similarity to the corresponding region in PGRP-LC. This region is absent in PGRP-LAb. Finally, PGRP-LAc has an alternative splice acceptor site for the second intron, which introduces a stop codon upstream of the PGRP domain. The predicted product would be a short transmembrane protein, similar to PGRP-LAb but without a PGRP domain.
The two PGRP-LD cDNAs have an additional ORF in the 5′-flanking sequence (Fig. 1). The encoded protein is homologous to a human gene of unknown function, PMI (40), and to related ESTs from other species (not shown). Thus, the 3.0- to 3.6-kb cDNA inserts we sequenced appear to correspond to dicistronic transcripts. The situation is further complicated by the fact that the long 3′-untranslated region of PGRP-LD overlaps with a third long ORF on the opposite strand (Fig. 1), homologous to cellular retinaldehyde-binding protein and to α-tocopherol transfer protein.
The previously described human and mouse homologs are typical short PGRPs. However, at least one long gene can also be found in the databases for these species. The human long (L) PGRP gene has one intron position in common with the short genes. The protein is predicted to have two transmembrane regions (Fig. 2) as well as an N-terminal signal peptide (not shown).
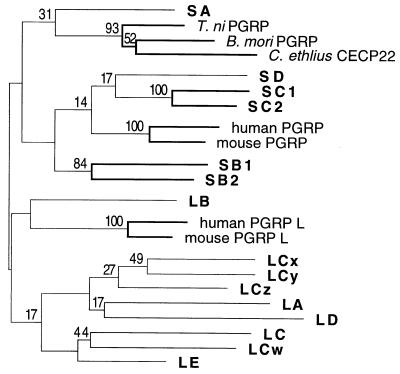
We attempted to deduce the phylogenetic relationship between the different PGRP domains, by using maximum parsimony analysis (41), but most of the relationships found are uncertain and are not supported by bootstrap analysis (Fig. 3), except that the genes within the PGRP-SB and SC clusters are closely related. However, there is a tendency for the long and short genes to form separate monophyletic groups. Again, PGRP-LB is an exception and, depending on which sequences are included in the comparison, it may cluster with the short or with the long genes. Similarly, the position of the vertebrate long PGRP genes is uncertain.
Figure 3.
Phylogenetic tree of the PGRP genes. A maximum parsimony tree was constructed with the amino acid sequences, by using the paup program (41). For branches supported by bootstrap analysis, the percentage of 1,000 replications that support the branch is indicated. Branches supported by at least 50% of the replications are drawn with thick lines.
Developmental Expression and Response to Infection of the PGRP Genes.
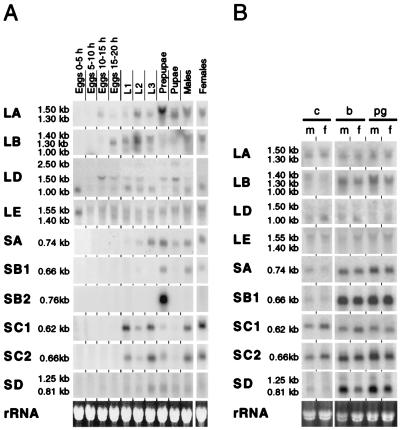
We followed the expression of the PGRP genes during development by using Northern blot analysis (Fig. 4A). PGRP-SC1A and B crosshybridize, because they differ in two bases only, and therefore the panel marked PGRP-SC1 shows the sum of the two transcripts.
Figure 4.
Expression of the PGRP genes. Northern blot analysis by using 15 μg of total RNA per lane. Identical samples were loaded on several gels and probed for the different genes. Equal loading was checked by ethidium bromide staining; one example is shown (Bottom). Because of crosshybridization between the PGRP-SC1A and SC1B genes, the PGRP-SC1B probe used will detect both transcripts. (A) Developmental expression. L1, L2, L3: first, second, and third instar larvae. (B) Bacterial induction of the PGRP genes. Untreated control flies (c) are compared with flies induced by injection of bacteria (b) of purified peptidoglycan (pg). m, males; f, females.
All short genes gave rise to short transcripts of 0.6–0.8 kb. In addition, we could detect a longer transcript of 1.25 kb from the PGRP-SD gene. The long genes gave rise to several transcripts of varying length. Most of the PGRP genes could be detected in all postembryonic stages. A striking exception is the PGRP-SB2 gene, which is exclusively expressed in prepupae. PGRP-LD and LE transcripts are highly expressed in 0- to 5-h-old embryos and in adult females, as they are probably maternally derived. PGRP-LA, LB, SD, and the longer transcripts of PGRP-LD become active later in the embryonic development. In the early larval stages, additional PGRP genes become constitutively expressed and remain active at variable levels until the adult stage. PGRP-SA, SB1, and SD transcripts were found only at very low levels in uninduced animals, and we were not able to detect PGRP-LC transcripts at all under these conditions (see below).
Because the PGRP genes are thought to play an important role in the innate immune response, we wanted to investigate their inducibility after immune stimulation. We injected 6-day-old flies with different Gram-positive and Gram-negative bacteria and peptidoglycan preparations. Five genes were strongly induced: PGRP-LB, SA, SB1, SC2, and SD. In Fig. 4B, we show the results with whole cells or purified peptidoglycan from B. subtilis (B. megaterium was used for PGRP-LD), but similar effects were seen with all bacteria and peptidoglycans tested. Even sterile Ringer's solution induced these genes to a variable extent (not shown), and for this reason it is difficult to draw firm conclusions about the specificity of different bacteria or peptidoglycans. Most of the small genes are strongly inducible. PGRP-SC1 and SC2 also show a high constitutive expression. PGRP-SB2 is not expressed at all in adults. Of the long genes, only PGRP-LB is inducible.
PGRP Transcripts Are Expressed in Tissues Known to Play a Role in the Immune Response.
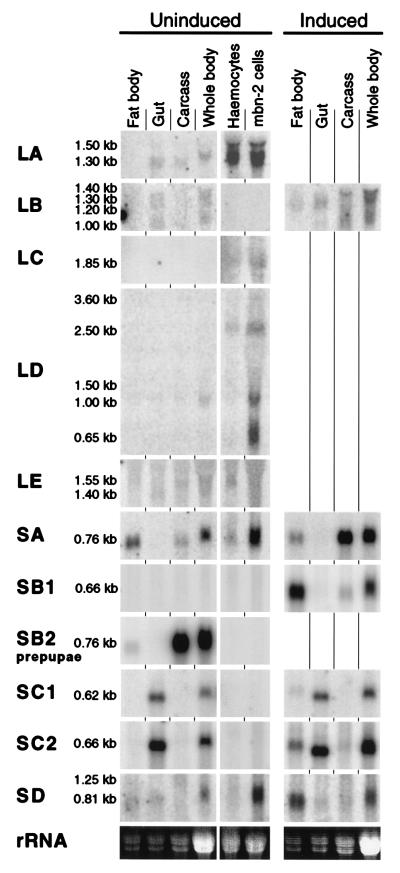
We compared the tissue distribution of the different PGRP gene transcripts by using Northern blots of RNA from dissected third instar larvae (Fig. 5). We also included RNA from hemocytes, isolated from the hemocyte-overproducing mutant l(3)mbn, and from the hemocyte-like cell line mbn-2. For PGRP-SB2, we dissected the corresponding tissues from early prepupae.
Figure 5.
Tissue distribution of the PGRP gene transcripts. Transcripts are analyzed by Northern blot as in Fig. 4. We also included RNA from hemocytes, isolated from the hemocyte-overproducing mutant l(3)mbn and from the hemocyte-like cell line mbn-2. In each lane, 15 μg of total RNA was loaded. For the samples from fat body, gut, and remaining carcass, we loaded a smaller amount of RNA, corresponding to the same number of larval equivalents as in the whole-body preparation. For PGRP-SB2, we used tissues from prepupae.
A few distinct patterns of gene expression can be observed (Fig. 5). First, inducible genes such as PGRP-SB1 and SD are mainly expressed in the fat body. PGRP-SA is also expressed in the fat body, although strong induction is seen only in the remaining carcass, including all epidermal layers. In addition, we detected PGRP-SA and SD in uninduced hemocytes and much more strongly in mbn-2 cells. The high level seen in the mbn-2 cells may be because the cultures of this cell line are often more or less induced. The inducible PGRP-LB gene differs from this pattern. It appears to be ubiquitously expressed with several different transcripts, but we cannot detect it in hemocytes or mbn-2 cells.
Unlike the other short PGRP genes, the PGRP-SC genes are constitutively expressed at high levels in the gut, in addition to their induced expression in the fat body. Yet another pattern is seen for some of the long PGRP genes, including LA, LC, and LD, which appear to be enriched in hemocytes and in mbn-2 cells. Transcripts from the latter genes can also be detected in other tissues, possibly because of associated hemocytes. Particularly PGRP-LD shows a complex pattern with several transcripts of different length. The major 0.65- to 2.5-kb transcripts are much shorter than the 3.0- to 3.6-kb dicistronic PGRP-LD cDNAs we have sequenced, and they are probably monocistronic. The fact that as many as three independent gene products may come from the PGRP-LD complex makes the interpretation of the Northern pattern difficult for this gene. Finally, PGRP-LE is very weakly expressed with a 1.4-kb transcript in the gut and a 1.55-kb transcript in hemocytes and in the larval carcass.
Peptidoglycan Affinity of the PGRP Proteins.
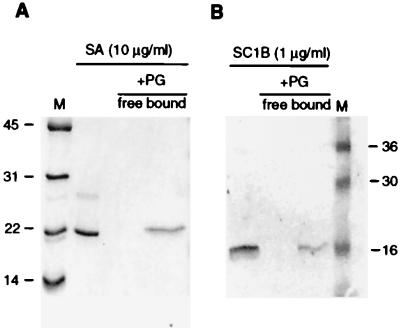
On the basis of previous results with the moth homologs, we assumed that the PGRP domain is a peptidoglycan-binding motif. We tested this directly for two of the Drosophila gene products expressed in the baculovirus system, one of which is expressed in the fat body, PGRP-SA, and one gut specific, PGRP-SC1B (Fig. 6). After incubation with insoluble peptidoglycan, both proteins are quantitatively removed from the free solution. They are retained in the peptidoglycan fraction even after extensive washing, indicating that both PGRP proteins bind peptidoglycan with high affinity. We were not able to isolate any of the long PGRPs in sufficient quantity to test peptidoglycan binding. At least PGRP-LC and PGRP-LA remain insoluble after expression in the baculovirus system, consistent with the proposal that they are membrane proteins.
Figure 6.
Binding of recombinant PGRP-SA and PGRP-SC1B to peptidoglycan from M. luteus. Purified PGRP was incubated with peptidoglycan (PG) at 0.5 mg/ml, and free protein was isolated from bound protein and analyzed on a 15% SDS/PAGE (see Methods). Molecular weights in kilodaltons of the marker proteins (M) are indicated. (A) Binding of recombinant PGRP-SA to peptidoglycan shown with Coomassie brilliant blue staining. (B) Binding of recombinant PGRP-SC1B to peptidoglycan shown by Western blot.
Discussion
The data presented here show that the PGRP family is highly diversified. Previously described PGRPs from insects and vertebrates are small exported proteins, and seven PGRPs in Drosophila belong to this class. Other PGRP genes encode longer proteins, and at least three of them, PGRP-LA, LC, and LD, are likely to be membrane proteins (Fig. 7). A localization in the membrane is likely also for the human long PGRP, although two long internal hydrophobic regions and a likely signal peptide indicate a different membrane topology (Fig. 7). The predicted PGRP-LB and LE have neither obvious signal peptides nor hydrophobic transmembrane regions, and it is possible that they remain in the cytoplasm. However, these genes give rise to several different transcripts, not all of which correspond to sequenced cDNAs. It can therefore not be excluded that signal peptides or transmembrane regions are present in some of these forms.
Figure 7.
Predicted cellular distribution of the PGRP proteins. All short PGRPs are predicted to be exported after removal of the N-terminal signal peptide. Some of the long PGRPs are likely to be membrane proteins: PGRP-LA, LC, LD, and the human L protein. Others may be retained in the cytoplasm: PGRP-LB and LE. Membrane topology is according to Sipos and von Heijne (39) and signal peptidase cleavage sites according to von Heijne (38).
Several lines of evidence indicate that the divergence of the PGRP family is of ancient origin. First, the Drosophila PGRPs are not obviously more closely related to each other than they are to the vertebrate PGRPs. Only genes within a cluster, such as the PGRP-SC genes, are similar in sequence and may derive from more recent gene duplications (Fig. 3). Second, the PGRP genes are widely dispersed over the Drosophila genome. The generation of a gene family by duplication typically results in a cluster of closely linked genes, which can remain linked over considerable time before they are broken up by chromosomal rearrangements. The highly dispersed localization is therefore an indication that this gene family is old. Third, the intron positions differ widely between the PGRP genes. All short PGRP genes could in principle be derived from an ancestral pattern with two introns, as in PGRP-SA, assuming that one or both of them may be lost in evolution. PGRP-LB and the human long PGRP also fit this pattern. However, introns are found in five different positions in the other long PGRP genes, none of which corresponds to the introns of the short genes. These facts point to long separate evolutionary histories.
The short PGRPs are a structurally homogenous group and are likely to be exported proteins, secreted in different parts of the body. Three of them, PGRP-SB1, SC2, and SD, are strongly induced in the fat body after infection, which is typical for inducible hemolymph proteins such as the cecropins and many other antibacterial peptides (42). PGRP-SA is also inducible, but not in the fat body or gut. Interestingly, this gene is most closely related to the lepidopteran PGRPs (Fig. 3), at least two of which are known to be expressed in the epidermis (22, 25). The PGRP from C. ethlius was even originally isolated as a cuticular protein (25). It is possible that these genes, including Drosophila PGRP-SA, play a specific role in the epidermis. The epidermis is a barrier to infections and has its own inducible antibacterial defense (43). Similarly, the PGRP-SC genes are specifically expressed in the gut, another possible route of infection.
The short PGRPs in moths and vertebrates have affinity for bacterial peptidoglycans, and they have been suggested to act as pattern recognition molecules, initiating the innate immune response (21, 22). Direct evidence for such a role was shown for B. mori PGRP, which is required for the activation of the phenoloxidase cascade by peptidoglycan (24). Alternatively, short PGRPs may function as opsonins or as effector molecules in the antibacterial defense. The PGRP domain has sequence similarity to phage lysozymes, and although no enzymatic activity was found in PGRPs (21, 22), they can inhibit bacterial growth (23). Some of the PGRP genes could even be specialized for different functions. At least two of the Drosophila PGRPs bind peptidoglycans. Furthermore, several of the short PGRPs are induced after bacterial infection, and they are all secreted in places such as hemolymph, gut, and epidermis, where they could easily interact with invading bacteria.
We know less about the long PGRPs, but their structures and patterns of expression give hints about possible functions. Their well conserved PGRP domain is likely to bind peptidoglycan. PGRP-LA, LC, and probably also LD are likely to be hemocyte transmembrane proteins. This is consistent with a role as pattern recognition molecules, signaling the presence of bacteria, or as receptors that mediate the uptake of bacteria by these phagocytically active cells.
Finally, PGRP-LB and LE show a more generalized tissue distribution, and they are likely to be retained in the cytoplasm. A role in immunity is indicated, at least for PGRP-LB, which is induced after infection. These proteins could interact with bacteria that invade the cytoplasmic compartment, or they are exported by alternative routes.
The long PGRPs have complex transcription patterns with several alternative transcripts. For PGRP-LC, there is an interesting possibility that the PGRP homology domains denoted x, y, z, and w could be spliced to the intracellular and transmembrane parts of PGRP-LC. They all have potential acceptor splice sites in the correct position for an in-frame fusion to PGRP-LC. In this way, the peptidoglycan recognition domain cassette could be exchanged on the PGRP-LC membrane anchor. However, we have so far not been able to obtain experimental evidence for this hypothesis.
In conclusion, we have shown that the Drosophila PGRPs constitute a highly diversified gene family, and that much of this diversity probably preceded the protostome–deuterostome split over half a billion years ago. Both short and long PGRPs are present in the human genome, and additional PGRP genes can now be found in the accumulating databases of the human genome project (unpublished observation). It is likely that many, if not all, members of this family play an important role in the interaction with invading bacteria, both in insects and in humans.
Acknowledgments
We thank Michael Williams, Svenja Stöven, Calle Zettervall, Marika Hedengren, and Peter Mellroth for technical advice and helpful discussions, Svenja Stöven for mbn-2 cells, and Anna-Karin Kronhamn for help with the flies. Our initial PGRP-SA genomic sequence was based on clones isolated by Ulrika Troedsson. This research was supported by grants from the Göran Gustafsson Foundation for Scientific Research, the Swedish Natural Science Research Council, and the Swedish Medical Research Council.
Abbreviations
- PGRP
peptidoglycan recognition protein
- EST
expressed sequence tag
Note Added in Proof.
The sequences reported in this article have been deposited in GenBank under accession nos. AF207535–AF207542 and AF313389–AF313393. The sequences of PGRP-SB1 (CG9681), PGRP-SB2 (CG9697), PGRP-SC1A (CG14746), PGRP-SC2 (CG14745), and PGRP-SD (CG7496) are predicted by the Berkeley Drosophila Genome Project. The a transcript of PGRP-LA was inferred from partial sequences of cDNA clones GH18280 and GH10945. Full sequencing of GH18280 later showed that the ORF is interrupted in this clone, as in the c transcript.
Footnotes
References
- 1.Hultmark D. Nature (London) 1994;367:116–117. doi: 10.1038/367116a0. [DOI] [PubMed] [Google Scholar]
- 2.Brey P T, Hultmark D. Molecular Mechanisms of Immune Responses in Insects. London: Chapman & Hall; 1998. [Google Scholar]
- 3.Engström Y. Dev Comp Immunol. 1999;23:345–358. doi: 10.1016/s0145-305x(99)00016-6. [DOI] [PubMed] [Google Scholar]
- 4.Hoffmann J A, Kafatos F C, Janeway C A, Ezekowitz R A B. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh S, May M J, Kopp E B. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 6.Samakovlis C, Åsling B, Boman H G, Gateff E, Hultmark D. Biochem Biophys Res Commun. 1992;188:1169–1175. doi: 10.1016/0006-291x(92)91354-s. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida H, Ochiai M, Ashida M. Biochem Biophys Res Commun. 1986;141:1177–1184. doi: 10.1016/s0006-291x(86)80168-1. [DOI] [PubMed] [Google Scholar]
- 8.Taniai K, Furukawa S, Shono T, Yamakawa M. Biochem Biophys Res Commun. 1996;226:783–790. doi: 10.1006/bbrc.1996.1429. [DOI] [PubMed] [Google Scholar]
- 9.Iketani M, Nishimura H, Akayama K, Yamano Y, Morishima I. Insect Biochem Mol Biol. 1999;29:19–24. doi: 10.1016/s0965-1748(98)00099-x. [DOI] [PubMed] [Google Scholar]
- 10.Rosetto M, Engström Y, Baldari C T, Telford J L, Hultmark D. Biochem Biophys Res Commun. 1995;209:111–116. doi: 10.1006/bbrc.1995.1477. [DOI] [PubMed] [Google Scholar]
- 11.Lemaitre B, Nicolas E, Michaut L, Reichhart J-M, Hoffmann J A. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 12.Williams M J, Rodriguez A, Kimbrell D A, Eldon E D. EMBO J. 1997;16:6120–6130. doi: 10.1093/emboj/16.20.6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medzhitov R, Preston-Hurlburt P, Janeway C A., Jr Nature (London) 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 14.Rock F L, Hardiman G, Timans J C, Kastelein R A, Bazan J F. Proc Natl Acad Sci USA. 1998;95:588–593. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang R-B, Mark M R, Gray A, A, H, Xie M H, Zhang M, Goddard A, Wood W I, Gurney A L, Godowski P J. Nature (London) 1998;395:284–288. doi: 10.1038/26239. [DOI] [PubMed] [Google Scholar]
- 16.Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning C J. J Biol Chem. 1999;274:17406–17409. doi: 10.1074/jbc.274.25.17406. [DOI] [PubMed] [Google Scholar]
- 17.Yoshimura A, Lien E, Ingalls R R, Tuomanen E, Dziarski R, Golenbock D. J Immunol. 1999;163:1–5. [PubMed] [Google Scholar]
- 18.Lee W J, Lee J D, Kravchenko V V, Ulevitch R J, Brey P T. Proc Natl Acad Sci USA. 1996;93:7888–7893. doi: 10.1073/pnas.93.15.7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jomori T, Natori S. J Biol Chem. 1991;266:13318–13323. [PubMed] [Google Scholar]
- 20.Kim Y-s, Ryu J-h, Han S-j, Choi K-H, Nam K, Jang I, Lemaitre B, Brey P T, Lee W-J. J Biol Chem. 2000;275:32721–32727. doi: 10.1074/jbc.M003934200. [DOI] [PubMed] [Google Scholar]
- 21.Kang D, Liu G, Lundström A, Gelius E, Steiner H. Proc Natl Acad Sci USA. 1998;95:10078–10082. doi: 10.1073/pnas.95.17.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ochiai M, Ashida M. J Biol Chem. 1999;274:11854–11858. doi: 10.1074/jbc.274.17.11854. [DOI] [PubMed] [Google Scholar]
- 23.Liu C, Gelius E, Liu G, Steiner H, Dziarski R. J Biol Chem. 2000;275:24490–24499. doi: 10.1074/jbc.M001239200. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida H, Kinoshida K, Ashida M. J Biol Chem. 1996;271:13854–13860. doi: 10.1074/jbc.271.23.13854. [DOI] [PubMed] [Google Scholar]
- 25.Marcu O, Locke M. Insect Biochem Mol Biol. 1998;28:659–669. doi: 10.1016/s0965-1748(98)00048-4. [DOI] [PubMed] [Google Scholar]
- 26.Dimopoulos G, Casavant T L, Chang S, Scheetz T, Roberts C, Donohue M, Schultz J, Benes V, Bork P, Ansorge W, et al. Proc Natl Acad Sci USA. 2000;97:6619–6624. doi: 10.1073/pnas.97.12.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 28.Dushay M S, Åsling B, Hultmark D. Proc Natl Acad Sci USA. 1996;93:10343–10347. doi: 10.1073/pnas.93.19.10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rubin G M, Hong L, Brokstein P, Evans-Holm M, Frise E, Stapleton M, Harvey D A. Science. 2000;287:2222–2224. doi: 10.1126/science.287.5461.2222. [DOI] [PubMed] [Google Scholar]
- 30.Gateff E. Science. 1978;200:1448–1459. doi: 10.1126/science.96525. [DOI] [PubMed] [Google Scholar]
- 31.Gateff E, Gissmann L, Shrestha R, Plus N, Pfister H, Schröder J, zur Hausen H. In: Invertebrate Systems in Vitro. Kurstak E, Maramorosch K, Dübendorfer A, editors. Amsterdam: Elsevier; 1980. pp. 517–533. [Google Scholar]
- 32.Hultmark D, Engström Å, Bennich H, Kapur R, Boman H G. Eur J Biochem. 1982;127:207–217. doi: 10.1111/j.1432-1033.1982.tb06857.x. [DOI] [PubMed] [Google Scholar]
- 33.Morishima I. In: Techniques in Insect Immunology. Wiesner A, Dunphy G B, Marmaras V J, Morishima I, Sugumaran M, Yamakawa M, editors. Fair Haven, NJ: SOS; 1998. pp. 59–64. [Google Scholar]
- 34.Araki Y, Nakatani T, Nakayama K, Ito E. J Biol Chem. 1972;247:6312–6322. [PubMed] [Google Scholar]
- 35.Gish W, States D J. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 36.Jokerst R S, Weeks J R, Zehring W A, Greenleaf A L. Mol Gen Genet. 1989;215:266–275. doi: 10.1007/BF00339727. [DOI] [PubMed] [Google Scholar]
- 37.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2182–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 38.von Heijne G. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sipos L, von Heijne G. Eur J Biochem. 1993;213:1333–1340. doi: 10.1111/j.1432-1033.1993.tb17885.x. [DOI] [PubMed] [Google Scholar]
- 40.Murphy P M, Malech H L. Nucleic Acids Res. 1990;18:1896. doi: 10.1093/nar/18.7.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swofford D L. paup: Phylogenetic Analysis Using Parsimony. Champaign, IL: Illinois National History Survey; 1991. [Google Scholar]
- 42.Hetru C, Hoffmann D, Bulet P. In: Molecular Mechanisms of Immune Responses in Insects. Brey P T, Hultmark D, editors. London: Chapman & Hall; 1998. pp. 40–66. [Google Scholar]
- 43.Brey P T, Lee W-J, Yamakawa M, Koizumi Y, Perrot S, François M, Ashida M. Proc Natl Acad Sci USA. 1993;90:6275–6279. doi: 10.1073/pnas.90.13.6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider I, Blumenthal A B. In: The Genetics and Biology of Drosophila. Ashburner M, Wright T R F, editors. 2a. London: Academic; 1978. pp. 265–315. [Google Scholar]