Abstract
The evolution of tryptophan-to-heme (W/heme) distance distributions extracted from analysis of fluorescence energy transfer kinetics during the refolding of Rhodopseudomonas palustris cytochrome c′ reveals dramatic differences between two variants [W32 (Q1A/F32W/W72F) and W72 (Q1A)]. Both W32/heme and W72/heme distance distributions measured at the earliest time point attainable with a continuous-flow mixer (150 μs) confirm that the polypeptide ensemble is not uniformly collapsed and that native structure is not formed. Time-resolved fluorescence spectra indicate that W32 is sequestered from the aqueous solution during the first 700 μs of folding, whereas W72 remains exposed to solvent. The first moment of the W32/heme distance distribution evolves to its native value faster than that of W72, suggesting that the approach of W32 to the heme precedes that of W72.
Keywords: fluorescence energy transfer, protein folding, ultrafast mixer
Understanding exactly how an unfolded polypeptide forms a discrete native structure is a central goal in biophysics. Characterization of the conformational heterogeneity of the protein ensemble is particularly important in detecting the formation of intermediates and clarifying the roles of diverse folding pathways (1–11). Energy landscape theory (3, 5, 6, 12–17) suggests that multiple parallel folding routes can lead to native structures, yet most experimental studies are interpreted in terms of chemical kinetics models involving relatively few discrete states (1, 18–22). In an effort to provide deeper mechanistic insights and make stronger connections to theoretical folding models, we have developed methods to measure fluorescence energy transfer (FET) kinetics between a fluorescent donor (D) and an energy acceptor (A) during folding reactions. These measurements provide nanosecond snapshots of D–A distances distributions [P(r)] as a polypeptide evolves from denatured to native states. Our initial studies of yeast cytochrome (cyt) c folding triggered with a stopped-flow mixer revealed the presence of both extended and collapsed conformations in the intermediates formed during the instrument dead time (burst phase). These data show clearly that cyt c does not fold by a two-state mechanism (23–25).
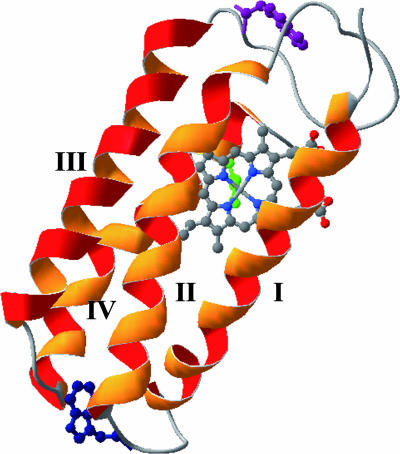
Empirical correlations (26–29) and theoretical (5, 6, 16) analyses point to a strong link between native-state topology and folding kinetics. Variations in folding kinetics among structurally homologous proteins, then, are attributable to the fine details of protein amino acid sequences. Kinetics studies of structurally homologous proteins have revealed situations in which folding pathways and transition-state structures are conserved, and others in which the folding bottlenecks differ substantially (30–42). In our studies of heme proteins, we have found that despite nearly identical four-helix bundle motifs, the refolding rates of cyt c-b562, c556, and c′ differ by many orders of magnitude (43–46). We report here results from FET experiments that shed light on the folding mechanism of one of these heme-containing proteins. The four-helix bundle of Rhodopseudomonas palustris cyt c′, which folds much more slowly than c-b562 and c556, is comprised of 125 aa with a heme prosthetic group bound to the polypeptide through thioether linkages to Cys-113 and Cys-116, and an axial imidazole ligand from His-117 (the four helices are constructed from the following residues: I, 4–26; II, 34–54; III, 75–95; and IV, 99–120) (44, 47). In our prior study of cyt c′, we examined tryptophan fluorescence from the native W72 residue in a pseudo-WT protein (Q1A) and W32 fluorescence in a less stable mutant (Q1A/F32W/W72F) (44). Measurements of tryptophan-to-heme (W/heme) FET kinetics from the two sites [Fig. 1: W72 (native) in the large loop between helix II and III in Q1A, and W32 in the small loop between helix I and II in Q1A/F32W/W72F] revealed underlying structural heterogeneity in chemically denatured cyt c′. Substantial populations of compact conformations in the denatured protein, and extensive tryptophan fluorescence quenching within the dead time of the stopped-flow mixer (≈5 ms) in refolding experiments, suggested that nonnative compact states may be responsible for the slow refolding kinetics (43, 44). We have extended these investigations by using a continuous-flow (CTF) fast mixer (τmixing ≤ 100 μs) (48, 49) to probe early events in cyt c′ folding. Using measurements of W/heme (W72 and W32) FET kinetics in cyt c′ variants, we have obtained the time dependence of P(r) for two distinct sites on the four-helix bundle.
Fig. 1.
Structure of R. palustris cyt c′ (Q1A/F32W/W72F) (Protein Data Bank ID code 1MQV) (44). The side chains of W72 (purple) and W32 (blue) are highlighted [the W72 orientation was modeled on the basis of the WT structure (47)]. The heme iron is axially ligated to H117 shown in green. Helices I, II, III, and IV are constructed from the residues of 4–26, 34–54, 75–95, and 99–120, respectively.
Results
Burst-Phase Ensemble.
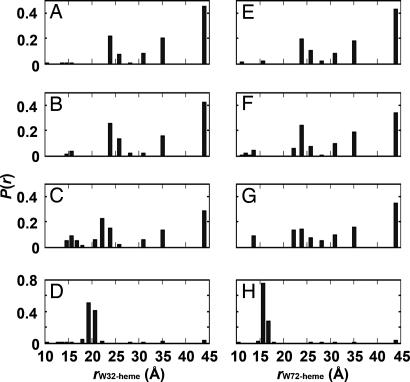
Analysis of the W/heme FET kinetics from two distinct sites on the polypeptide chain reveals structural heterogeneity during cyt c′ folding. In the unfolded states of W32 and W72 [2.0 and 3.5 M guanidine hydrochloride (GuHCl), respectively], distributions of both proteins feature extended (E; r > 30 Å, ≈70%) and relatively compact (M; 20 < r < 30 Å, ≈30%) conformations, as reported (44) (Fig. 2A and E). This type of heterogeneity is expected for an unstructured heteropolymer; fits of the W72 and W32 FET kinetics to a freely jointed chain model (50) suggest similar W/heme distance distributions, but different persistence lengths for the two sites [supporting information (SI) Table 1]. Immediately after the dead time of our CTF mixer (≈150 μs), we observed small decreases in W32 and W72 fluorescence intensity, consistent with compaction of the protein ensemble. Distance distributions extracted from W32/heme and W72/heme FET kinetics indicate that the populations of extended polypeptides decrease, largely in favor of M (Figs. 2 and 3). Interestingly, we observed a small component (<5%) in the tryptophan-decay kinetics that suggests the presence of compact structures (C; r ≤ 15 Å) with D–A distances shorter those found in the native conformation (W32, 18–21 Å; W72, 15–17 Å). This fast early collapse is consistent with a response of the polypeptide to the change in solvent conditions upon denaturant dilution, but not extensive population of specific compact intermediates. It is clear that 150 μs after dilution of denaturant, the cyt c′ polypeptide ensemble is highly heterogeneous, with a considerable range of W/heme distances.
Fig. 2.
Probability distance distributions [P(r)] extracted from maximum entropy analyses of the FET kinetics data. Folding of cyt c′ was triggered by dilution of GuHCl in an ultrafast mixer (pH 7.0; 18°C; W32 [GuHCl] = 2.0–0.33 M; W72 [GuHCl] = 3.5–0.58 M). Only 4 of the 31 (W32, Left) or 52 (W72, Right) transient P(r) distributions are shown for clarity, corresponding to the unfolded state (A and E), folding times of 150 μs (B and F) and 2.5 ms (C and G), and the native state (D and H).
Fig. 3.
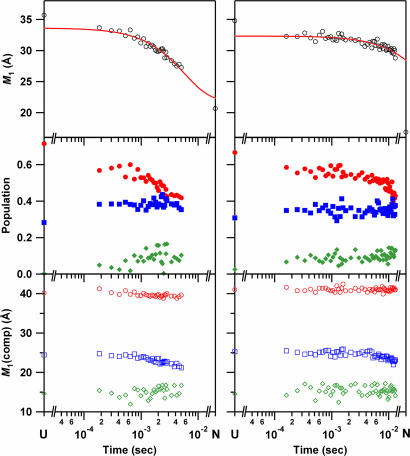
Folding kinetics of cyt c′ probed by W32 (Left) and W72 (Right) FET kinetics. (Top) Mean distance between tryptophan and heme, M1, as a function of refolding time. The solid line is a fit a single-exponential function: the rate constant for W32 is 1.7 × 102 s−1; that for W72 is 15 s−1 [determined by fixing infinite-time value to match that of the native state (16.9 Å)]. (Middle) Time course of the population of each component [●, extended conformers (>30 Å, E); ■, conformations with intermediate D–A distances (20 < r < 30 Å, M); and ◆, nonnative collapsed conformers (< 18 and 15 Å for W32 and W72, respectively, C)]. (Bottom) Time course of the average distance of each component (○, E; □, M; and ◇, C).
The large burst-phase populations of structures with long W32/heme and W72/heme distances argue against the presence of native-like structures. Of the four helices in cyt c′, helix III (residues 75–95) has the greatest propensity to form an unsupported α helix, and the modest reduction in average W72/heme distances found in the burst-phase ensemble could arise from partial formation of helix III (51). Overall, the long W32/heme and W72/heme distances extracted from the 150-μs FET kinetics data clearly preclude formation of the helix III/turn/helix IV and helix II/turn/helix III/turn/helix IV motifs of cyt c′.
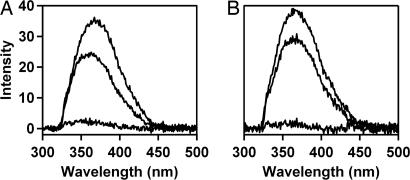
To characterize further the burst-phase ensemble, we measured the W32 and W72 fluorescence spectra averaged over the 200- to 700-μs time period. The mutant W32 side chain has greater solvent-accessible surface area (46.4 Å2) than W72 (11.72 Å2) (52), yet both proteins exhibit tryptophan fluorescence maxima at ≈325 nm, consistent with hydrophobic environments for the indole groups. Relative to the spectrum of the unfolded protein, W32 fluorescence (200–700 μs) exhibits a modest blue shift and a 30% decrease in integrated intensity (Fig. 4A), suggesting that the W32 indole side chain has moved into a more hydrophobic environment. The reduced fluorescence intensity can be attributed to the modest burst-phase collapse indicated by FET kinetics measurements. The data suggest that, upon denaturant dilution, W32 becomes incorporated in a hydrophobic cluster that produces a blue shift in W32 fluorescence. The x-ray crystal structure of folded (Q1A/F32W/W72F) cyt c′ (Protein Data Bank ID code 1MQV) (44) reveals that the W32 residue forms short hydrophobic contacts (<5 Å) with nearby residues in helices I and II (M25 and V37) and with residues near the origin of helix IV (I96 and L102) (47). The early appearance of a hydrophobic environment around W32, then, may indicate partial formation of the helix I–helix II turn through development of native contacts with nearby nonpolar residues. Burst-phase formation of tertiary contacts with helix IV appears less likely, because the population of shorter W32/heme distances does not increase substantially in the first 700 μs after denaturant dilution.
Fig. 4.
Fluorescence spectra during cyt c′ folding. Conditions were the same as in Fig. 3. (A) W32. (B) W72. Upper to lower curves: unfolded, average over the 200- to 700-μs folding interval, and native states.
The behavior of W32 fluorescence contrasts with that of W72. The fluorescence maximum of W72 shifts very little during the 200- to 700-μs time interval, although the intensity decreases by ≈25% (Fig. 4B), the latter effect likely caused by contraction of the polypeptide. Apparently W72 remains exposed to the aqueous solvent after GuHCl dilution. The hydrophobic interactions with W72 in the native protein structure (47) primarily involve contacts (<5 Å) with residues in the large loop region between helices II and III (F55, P56, A57, A67, L68, P69, and I71). The absence of a burst-phase blue shift of the W72 fluorescence suggests that the loop structure between helices II and III does not develop during the earliest stages of cyt c′ folding.
Time Evolution of W32/Heme and W72/Heme Distance Distributions.
FET kinetics measurements for the two tryptophan residues exhibit distinct behavior during the first 5 ms of folding. This difference is apparent in the time evolution of the first moments (M1) of the W/heme distance distributions (Fig. 3). The finding that M1(W32) decreases from 33 to 27 Å in the 0.15- to 5-ms time window contrasts with that of M1(W72), which remains nearly constant at 32 Å throughout this time interval. The time dependence of the change in M1(W32) is reasonably described by an exponential function with a 6-ms time constant. We only begin to observe the first stages of W72/heme approach in the 5- to 7.5-ms range; we estimate that M1(W72) changes with a time constant of 70 ms. This result is particularly unexpected given that under our refolding conditions the folding driving force (−ΔG = 23 kJ·mol−1) is twice that for the W32 protein. The W32 and W72 FET kinetics obtained with the CTF mixer are consistent with results from our earlier stopped-flow study of cyt c′ in which tryptophan fluorescence intensity measurements revealed a larger burst-phase amplitude and folding rate constant for W32 than W72 at similar folding driving forces (≈11 kJ·mol−1) (44). It is noteworthy that the final development of the native heme environment, as probed by absorption spectroscopy under our experimental conditions, is faster in W72 than in W32.
It is informative to consider the redistributions of D–A distances that give rise to the observed first-moment changes (Fig. 3). For W32, the population of extended (E) conformations decreases in favor of compact (C) conformations: E decreases from 60% to 45% of the total population, and C increases from 0% to 15%. Although the relative population of intermediate conformations (M) does not change in this time interval, the first moment for that subpopulation [M1(M)] undergoes a modest shift from 25 to 23 Å. The first moments for the other two subpopulations (E and C) do not change substantially during the first 5 ms of folding. For W72, the FET kinetics during the first 5 ms of cyt c′ folding change very little, remaining similar to those of the unfolded protein. On the other hand, in this time period the average W32/heme distance decreases to ≈77% of its unfolded value, largely because of the population of conformations in which the W32/heme distance is shorter than in the folded protein.
Discussion
Our analysis of W/heme energy transfer kinetics provides several insights into the folding mechanism of R. palustris cyt c′. The conformational properties of the denatured protein are expected to have an impact on the refolding kinetics, particularly in the early loop formation steps (53–56). The W32/heme and W72/heme distributions in unfolded cyt c′ are remarkably similar, which is surprising because W72 is separated from the heme (Cys-113) by many fewer residues (41 residues) than W32 (81 residues). In denatured cyt c, for example, mean D–A distances generally increase with the number of residues separating the dye and the heme (57). Apparently, the denatured protein does not adopt a random coil structure and some interactions in the unfolded state keep W32 closer to the heme than expected. These interactions might strengthen after dilution of denaturant, leading to the early development of a hydrophobic environment around W32 and its increased proximity to the heme. Notably, the faster-folding cyts c-b562 (45, 46) and c556 (58) both contain greater proportions of polar amino acids so that early hydrophobic clustering in these proteins may not be as prevalent.
The different time-resolved fluorescence spectra and FET kinetics for W32 and W72 may be a consequence of a sequential folding mechanism in which different regions of the protein develop native structures at different rates (59). Behavior of this type also is consistent with low-barrier energy-landscape models (5, 6, 12), although it is typically associated with ultrafast folding proteins (60–63). In our earlier study of stopped-flow triggered refolding of cyt c′, we found large burst-phase reductions in W32 and W72 fluorescence (44). In addition, residual W32 fluorescence decayed faster (30 s−1, −ΔG = 11 kJ·mol−1) than that of W72 (1.4 s−1, −ΔG = 12 kJ·mol−1), which exhibited a time constant similar to that found when probing folding kinetics with heme absorption spectroscopy (1 s−1, −ΔG = 11 kJ·mol−1). Using an ultrafast mixer with measurements of time-resolved tryptophan fluorescence spectra and decay kinetics, we have found a similar pattern with important additional details. The dynamics of the large reductions in W32 and W72 fluorescence occurring during the stopped-flow dead time have been resolved. W32 becomes incorporated into a hydrophobic environment and approaches the heme on a substantially shorter time scale than W72. The absence of any change in the W72 fluorescence spectrum in the first 700 μs of folding and the constancy of M1(W72) between 150 μs and 5 ms show clearly that neither the helix II–III loop nor the helix III–IV coiled-coil develops during this time period. The data are consistent with early formation of the helix I–II bend, but relatively late formation of the helix II–III–IV bundle. The late formation of native structure around W72 may reflect difficulty in assembly of the large loop between helices II and III.
We cannot rule out the possibility that the relative folding times in the W32 and W72 proteins arise from variations in folding pathway that result from the differences in primary sequence in the two mutant proteins. The kinetics measured by heme absorption spectroscopy are consistent with the finding that the W32 mutant is substantially less stable than W72; development of native environment around the porphyrin is slower in the W32 mutant (under our solvent conditions). The unusually short W32/heme distances in the denatured protein may be caused by the presence of the F32W and W72F mutations, and this denatured state structural preference may lead to the development of short W/heme distances on a faster time scale. If the W72 protein does not have a similar denatured-state conformation, then its folding pathway may differ as well. Hence, it is possible that the faster M1(W32) kinetics in the Q1A/F32W/W72F mutant arise from an altered folding landscape rather than a folding sequence in which W32 collapse precedes that of W72.
It is interesting to compare our observations on cyt c′ folding to results from studies of cyt b562. In the early stage of apo-cyt b562 folding, partial native-like secondary structure and backbone topology are thought to develop in helices II, III, and the N-terminal part of helix IV, whereas helix I and the C terminus of helix IV remain relatively unstructured (64–67). Moreover, the results obtained from electron transfer triggered folding of Fe(II)–cyt c–b562 suggest that the heme is sequestered from the solvent on very short time scales (150 μs) (45). This finding contrasts with the modest collapse observed for Fe(III)–cyt c′. It appears that these structural homologs may have substantially different folding bottlenecks.
Our analysis of cyt c′ FET kinetics sheds light on the burst-phase collapse of these four-helix bundles. Theoretical investigations of lattice model proteins have identified two different behaviors: fast-track folding in which the native state forms without population of collapsed intermediates, and slow-track folding where collapsed intermediates form before the native state (2, 68–71). The W32/heme and W72/heme distance distributions at 150 μs after the initiation of the folding reaction are highly heterogeneous; they reveal that the cyt c′ folding ensemble is composed of at least three subpopulations whose W/heme distances are distinct from those of the native state. It is clear from our data that there are no fast-track cyt c′ folders. Rather, the transient distributions reveal rapid formation of collapsed intermediates (C components) whose W/heme distances are shorter than that of the native conformation. Nonnative compact structures also have been detected 1 ms after the initiation of yeast cyt c folding, but FET kinetics did not provide evidence for structures more compact than in the folded protein (24). The fact that W32/heme distances in the compact conformers of cyt c′ are shorter than that of the folded protein is consistent with a globular collapse of the polypeptide upon denaturant dilution. In the folded protein, W32 and the heme are at opposite ends of the four-helix bundle, so shorter distances would be expected if denaturant dilution initially forms a collapsed globular structure. The time courses of M1(W32, C) and its population show a gradual shift of the W/heme distances to values consistent with the native state. These results suggest a mechanism in which polypeptides evolve to the native structure within a compact nonnative globule. Hindered diffusion, owing to topological and energetic frustration in the collapsed polypeptide ensemble (12, 70, 71), may then be responsible for the sluggish and more complex folding dynamics observed in cyt c′ (43, 44).
Materials and Methods
Materials.
N-acetyl-tryptophanamide, N-bromosuccinimide, and GuHCl Ultra grade were used as received from Sigma (St. Louis, MO). A pseudo-WT variant of R. palustris cyt c′ (Q1A) served as a base mutant with a tryptophan residue at position 72 (W72). A second variant (W32) contained three point mutations (Q1A/F32W/W72F). Both proteins were expressed and purified as described (43, 44).
Experimental Conditions.
The folding reactions were initiated by mixing a solution of GuHCl-denatured cyt c′ (100 mM sodium phosphate, pH 7.0) with phosphate buffer (100 mM, pH 7.0) at a volume ratio of 1:5. The initial GuHCl concentrations for W72 and W32 were 3.5 and 2.0 M, respectively. Protein concentrations were confirmed by absorption spectroscopy (ε398 = 8.5 × 104 M−1·cm−1) (72), and the final concentrations of the cyt c′ variants were 12–16 μM. All measurements were conducted at ambient temperature (≈18°C). GuHCl concentrations were determined by refractive-index measurements (73).
Experimental Configuration for FET Kinetics.
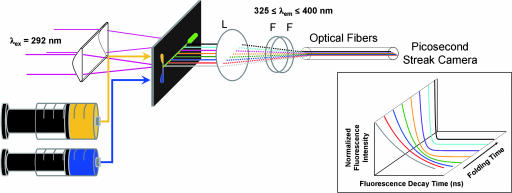
A schematic diagram of the apparatus used for FET kinetics measurements is illustrated in Fig. 5. The mixing cell was based on a previously published design (48). We used a 200-μm-thick stainless steel mixing plate (Toray Precision Inc., Shiga, Japan) etched with two flow channels (width = 35 μm; depth = 100 μm) and an observation channel (200 × 200 μm). The mixing plate was sandwiched between synthetic fused silica windows in a stainless-steel holder. The dead time of the mixer and folding reaction times were determined by using the reaction of N-acteyl-tryptophanamide with N-bromosuccinimide for calibration (SI Fig. 6).
Fig. 5.
Schematic of the system used for FET kinetics measurements during protein folding. A 200-μm-thick stainless mixing plate is placed between two 3-mm-thick quartz windows (which are compressed by a stainless holder, not shown). Fluorescence from the flow channel is focused onto a bundle of optical fibers (≈0.1-mm diameter) and directed to a picosecond streak camera. L, lens; F, dielectric filter. (Inset) Image of the experimental data detected by a picosecond streak camera.
Tryptophan fluorescence-decay kinetics measurements were carried out as described (44, 74). The third harmonic (292 nm) of a regeneratively amplified femtosecond Ti:sapphire laser (Spectra-Physics, Mountain View, CA) was used as an excitation source. A combination of spherical and cylindrical lenses was used to expand and then focus the excitation beam into a narrow strip (200 μm × 1.5 cm) on the center of the observation channel. Fluorescence was collected from the back of the flow cell and focused onto a linear array of 25 optical fibers (≈100-μm diameter). Excitation and extraneous wavelengths of light were removed with a combination of dielectric and color filters (325 ≤ λobsd ≤ 400 nm) positioned after the collection lens. The output from the optical fiber array was directed onto the entrance slit of a picosecond streak camera (C5680; Hamamatsu Photonics, Hamamatsu, Japan) operated in the analog-integration mode. Tryptophan fluorescence decay kinetics were measured on both short (1 ns) and long (20 ns) time scales; weak fluorescence from the buffer solution was subtracted before data analysis. The resulting short and long time-scale data were spliced together, and the combined traces were compressed logarithmically before fitting (100 points per decade). We confirmed that the compression did not alter the interpretation of data.
Fluorescence Spectra.
The excitation configuration for measurements of tryptophan fluorescence spectra during folding was the same as that used for FET kinetics. For collection, however, fluorescence from the CTF mixer was focused onto a single optical fiber (1 mm diameter), and the emission spectrum (λ ≥ 325 nm) was detected with a CCD-coupled fluorimeter (USB2000; Ocean Optics, Dunedin, FL).
Data Fitting and Analysis.
Analysis of FET kinetics involves the numerical inversion of a Laplace transform [I(t) = Σk P(k) exp(−kt)] (75, 76). We have used two algorithms to invert our kinetics data with regularization methods that impose additional constraints on the properties of P(k). The simplest constraint that applies to the FET kinetics data are that P(k) ≥ 0 (∀ k). We have fit the kinetics data by using a MATLAB algorithm (LSQNONNEG) (Mathworks, Natick, MA) that minimizes the sum of the squared deviations (χ2) between observed and calculated values of I(t), subject to a nonnegativity constraint. It is our experience that LSQNONNEG produces the narrowest P(k) distributions and smallest values of χ2 with relatively few nonzero components. Information theory suggests that the least-biased solution to this inversion problem minimizes χ2 and maximizes the breadth of P(k) (77). This regularization condition can be met by maximizing the Shannon–Jaynes entropy of the rate-constant distribution {S = −Σk P(k)ln[P(k)]}, implicitly requiring that P(k) ≥ 0 (∀ k) (78). Maximum-entropy (ME) fitting produces stable and reproducible numerical inversions of the kinetics data. The balance between χ2 minimization and entropy maximization is determined by graphical L-curve analysis (79). This approach yields upper limits for the widths of P(k) consistent with our experimental data. The P(k) distributions from ME fitting are broader than those obtained with LSQNNONEG fitting, but exhibit maxima in similar locations. A simple coordinate transformation using the Förster relation (Eq. 1)
recasts the probability distribution of the decay rates, P(k), obtained by LSQNONNEG or ME fitting as probability distributions over r (74, 80) (SI Fig. 7). The Förster critical length, R0, for the W/heme pair in cyt c′ is 34 Å under both native and unfolded conditions (44). The value of k0 (3.2 × 108 s−1) was obtained from luminescence-decay measurements with N-acetyl-tryptophanamide (20 μM) in the CTF mixer under various solvent conditions [100 mM sodium phosphate with and without GuHCl (0, 0.58, and 3.5 M) at pH 7.0]. At distances > 1.5 R0, energy transfer quenching of D is not competitive with excited-state decay, and, at distances < ≈10 Å, the Förster model does not reliably describe FET kinetics (81, 82); accordingly, our cyt c′ FET measurements can provide information about D–A distances only in the range 10 ≤ r ≤ 44 Å.
FET kinetics for the unfolded proteins have been fit to a freely jointed chain distribution of D–A distances [PFJC(r); Eq. 2] with a single adjustable parameter, the mean-squared end-to-end distance, 〈r2〉 (50):
The moments (Mn) and variance (V) of the P(r) distributions (SI Fig. 8) were calculated according to Eqs. 3 and 4:
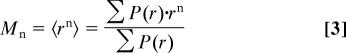 |
Supplementary Material
Acknowledgments
We thank Dr. Linda Thöny-Meyer (Eidgenössiche Technische Hochschule, Zürich, Switzerland) for the ccm plasmid pEC86 and Ekaterina V. Pletneva for numerous discussions. This work was supported by National Institutes of Health Grant GM068461 (to J.R.W.), the Arnold and Mabel Beckman Foundation (Beckman Senior Research Fellowship to J.C.L.), and the Ellison Medical Foundation (Senior Scholar Award in Aging to H.B.G.). T.K. was supported by a Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists.
Abbreviations
- W/heme
tryptophan-to-heme
- FET
fluorescence energy transfer
- GuHCl
guanidine hydrochloride
- cyt
cytochrome
- CTF
continuous flow.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609413103/DC1.
References
- 1.Fersht AR, Itzhaki LS, ElMasry NF, Matthews JM, Otzen DE. Proc Natl Acad Sci USA. 1994;91:10426–10429. doi: 10.1073/pnas.91.22.10426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo Z, Thirumalai D. Biopolymers. 1995;36:83–102. [Google Scholar]
- 3.Dill KA, Chan HS. Nat Struct Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 4.Bilsel O, Matthews CR. Curr Opin Struct Biol. 2006;16:86–93. doi: 10.1016/j.sbi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Plotkin SS, Onuchic JN. Q Rev Biophys. 2002;35:111–167. doi: 10.1017/s0033583502003761. [DOI] [PubMed] [Google Scholar]
- 6.Plotkin SS, Onuchic JN. Q Rev Biophys. 2002;35:205–286. doi: 10.1017/s0033583502003785. [DOI] [PubMed] [Google Scholar]
- 7.Salvatella X, Dobson CM, Fersht AR, Vendruscolo M. Proc Natl Acad Sci USA. 2005;102:12389–12394. doi: 10.1073/pnas.0408226102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellison PA, Cavagnero S. Protein Sci. 2006;15:564–582. doi: 10.1110/ps.051758206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang WY, Gruebele M. J Am Chem Soc. 2004;126:7758–7759. doi: 10.1021/ja0493751. [DOI] [PubMed] [Google Scholar]
- 10.Bren KL, Kellogg JA, Kaur R, Wen X. Inorg Chem. 2004;43:7934–7944. doi: 10.1021/ic048925t. [DOI] [PubMed] [Google Scholar]
- 11.Travaglini-Allocatelli C, Gianni S, Dubey VK, Borgia A, Di Matteo A, Bonivento D, Cutruzzola F, Bren KL, Brunori M. J Biol Chem. 2005;280:25729–25734. doi: 10.1074/jbc.M502628200. [DOI] [PubMed] [Google Scholar]
- 12.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 13.Onuchic JN, Luthey-Schulten Z, Wolynes PG. Annu Rev Phys Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 14.Portman JJ, Takada S, Wolynes PG. J Chem Phys. 2001;114:5069–5081. [Google Scholar]
- 15.Portman JJ, Takada S, Wolynes PG. J Chem Phys. 2001;114:5082–5096. [Google Scholar]
- 16.Onuchic JN, Wolynes PG. Curr Opin Struct Biol. 2004;14:70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 17.Naganathan AN, Doshi U, Fung A, Sadqi M, Munoz V. Biochemistry. 2006;45:8466–8475. doi: 10.1021/bi060643c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matouschek A, Kellis JT, Serrano L, Fersht AR. Nature. 1989;340:122–126. doi: 10.1038/340122a0. [DOI] [PubMed] [Google Scholar]
- 19.Fersht AR, Matouschek A, Serrano L. J Mol Biol. 1992;224:771–782. doi: 10.1016/0022-2836(92)90561-w. [DOI] [PubMed] [Google Scholar]
- 20.Matouschek A, Fersht AR. Proc Natl Acad Sci USA. 1993;90:7814–7818. doi: 10.1073/pnas.90.16.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matouschek A, Otzen DE, Itzhaki LS, Jackson SE, Fersht AR. Biochemistry. 1995;34:13656–13662. doi: 10.1021/bi00041a047. [DOI] [PubMed] [Google Scholar]
- 22.Ferguson N, Sharpe TD, Schartau PJ, Sato S, Allen MD, Johnson CM, Rutherford TJ, Fersht AR. J Mol Biol. 2005;353:427–446. doi: 10.1016/j.jmb.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 23.Lyubovitsky JG, Gray HB, Winkler JR. J Am Chem Soc. 2002;124:5481–5485. doi: 10.1021/ja017399r. [DOI] [PubMed] [Google Scholar]
- 24.Pletneva EV, Gray HB, Winkler JR. Proc Natl Acad Sci USA. 2005;102:18397–18402. doi: 10.1073/pnas.0509076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pletneva EV, Gray HB, Winkler JR. J Am Chem Soc. 2005;127:15370–15371. doi: 10.1021/ja0555318. [DOI] [PubMed] [Google Scholar]
- 26.Plaxco KW, Simons KT, Baker D. J Mol Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 27.Baker D. Nature. 2000;405:39–42. doi: 10.1038/35011000. [DOI] [PubMed] [Google Scholar]
- 28.Plaxco KW, Simons KT, Ruczinski I, Baker D. Biochemistry. 2000;39:11177–11183. doi: 10.1021/bi000200n. [DOI] [PubMed] [Google Scholar]
- 29.Ivankov DN, Garbuzynskiy SO, Alm E, Plaxco KW, Baker D, Finkelstein AV. Protein Sci. 2003;12:2057–2062. doi: 10.1110/ps.0302503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Viguera AR, Serrano L, Wilmanns M. Nat Struct Biol. 1996;3:874–880. doi: 10.1038/nsb1096-874. [DOI] [PubMed] [Google Scholar]
- 31.Friel CT, Capaldi AP, Radford SE. J Mol Biol. 2003;326:293–305. doi: 10.1016/s0022-2836(02)01249-4. [DOI] [PubMed] [Google Scholar]
- 32.Paci E, Friel CT, Lindorff-Larsen K, Radford SE, Karplus M, Vendruscolo M. Proteins Struct Funct Genet. 2004;54:513–525. doi: 10.1002/prot.10595. [DOI] [PubMed] [Google Scholar]
- 33.Chiti F, Taddei N, White PM, Bucciantini M, Magherini F, Stefani M, Dobson CM. Nat Struct Biol. 1999;6:1005–1009. doi: 10.1038/14890. [DOI] [PubMed] [Google Scholar]
- 34.Villegas V, Martinez JC, Aviles FX, Serrano L. J Mol Biol. 1998;283:1027–1036. doi: 10.1006/jmbi.1998.2158. [DOI] [PubMed] [Google Scholar]
- 35.Martinez JC, Serrano L. Nat Struct Biol. 1999;6:1010–1016. doi: 10.1038/14896. [DOI] [PubMed] [Google Scholar]
- 36.Northey JGB, Di Nardo AA, Davidson AR. Nat Struct Biol. 2002;9:126–130. doi: 10.1038/nsb748. [DOI] [PubMed] [Google Scholar]
- 37.Grantcharova VP, Riddle DS, Santiago JV, Baker D. Nat Struct Biol. 1998;5:714–720. doi: 10.1038/1412. [DOI] [PubMed] [Google Scholar]
- 38.Riddle DS, Grantcharova VP, Santiago JV, Alm E, Ruczinski I, Baker D. Nat Struct Biol. 1999;6:1016–1024. doi: 10.1038/14901. [DOI] [PubMed] [Google Scholar]
- 39.Guerois R, Serrano L. J Mol Biol. 2000;304:967–982. doi: 10.1006/jmbi.2000.4234. [DOI] [PubMed] [Google Scholar]
- 40.McCallister EL, Alm E, Baker D. Nat Struct Biol. 2000;7:669–673. doi: 10.1038/77971. [DOI] [PubMed] [Google Scholar]
- 41.Nishimura C, Prytulla S, Dyson HJ, Wright PE. Nat Struct Biol. 2000;7:679–686. doi: 10.1038/77985. [DOI] [PubMed] [Google Scholar]
- 42.Teilum K, Thormann T, Caterer NR, Poulsen HI, Jensen PH, Knudsen J, Kragelund BB, Poulsen FM. Proteins Struct Funct Bioinf. 2005;59:80–90. doi: 10.1002/prot.20340. [DOI] [PubMed] [Google Scholar]
- 43.Lee JC, Gray HB, Winkler JR. Proc Natl Acad Sci USA. 2001;98:7760–7764. doi: 10.1073/pnas.141235198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee JC, Engman KC, Tezcan FA, Gray HB, Winkler JR. Proc Natl Acad Sci USA. 2002;99:14778–14782. doi: 10.1073/pnas.192574099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faraone-Mennella J, Gray HB, Winkler JR. Proc Natl Acad Sci USA. 2005;102:6315–6319. doi: 10.1073/pnas.0502301102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Faraone-Mennella J, Tezcan FA, Gray HB, Winkler JR. Biochemistry. 2006;45:10504–10511. doi: 10.1021/bi060242x. [DOI] [PubMed] [Google Scholar]
- 47.Shibata N, Iba S, Misaki S, Meyer TE, Bartsch RG, Cusanovich MA, Morimoto Y, Higuchi Y, Yasuoka N. J Mol Biol. 1998;284:751–760. doi: 10.1006/jmbi.1998.2190. [DOI] [PubMed] [Google Scholar]
- 48.Akiyama S, Takahashi S, Kimura T, Ishimori K, Morishima I, Nishikawa Y, Fujisawa T. Proc Natl Acad Sci USA. 2002;99:1329–1334. doi: 10.1073/pnas.012458999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kimura T, Uzawa T, Ishimori K, Morishima I, Takahashi S, Konno T, Akiyama S, Fujisawa T. Proc Natl Acad Sci USA. 2005;102:2748–2753. doi: 10.1073/pnas.0407982102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flory PJ. Statistical Mechanics of Chain Molecules. New York: Interscience; 1969. [Google Scholar]
- 51.Chakrabartty A, Kortemme T, Baldwin RL. Protein Sci. 1994;3:843–852. doi: 10.1002/pro.5560030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fraczkiewicz R, Braun W. J Comput Chem. 1998;19:319–333. [Google Scholar]
- 53.Kubelka J, Hofrichter J, Eaton WA. Curr Opin Struct Biol. 2004;14:76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 54.Godbole S, Hammack B, Bowler BE. J Mol Biol. 2000;296:217–228. doi: 10.1006/jmbi.1999.3454. [DOI] [PubMed] [Google Scholar]
- 55.Kurchan E, Roder H, Bowler BE. J Mol Biol. 2005;353:730–743. doi: 10.1016/j.jmb.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 56.Wandschneider E, Bowler BE. J Mol Biol. 2004;339:185–197. doi: 10.1016/S0022-2836(04)00331-6. [DOI] [PubMed] [Google Scholar]
- 57.Pletneva EV, Gray HB, Winkler JR. J Mol Biol. 2005;345:855–867. doi: 10.1016/j.jmb.2004.10.085. [DOI] [PubMed] [Google Scholar]
- 58.McGuirl MA, Lee JC, Lyubovitsky JG, Thanyakoop C, Richards JH, Gray HB, Winkler JR. Biochim Biophys Acta. 2003;1619:23–28. doi: 10.1016/s0304-4165(02)00437-3. [DOI] [PubMed] [Google Scholar]
- 59.Weinkam P, Zong CH, Wolynes PG. Proc Natl Acad Sci USA. 2005;102:12401–12406. doi: 10.1073/pnas.0505274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma HR, Gruebele M. J Comp Chem. 2006;27:125–134. doi: 10.1002/jcc.20311. [DOI] [PubMed] [Google Scholar]
- 61.Larios E, Li JS, Schulten K, Kihara H, Gruebele M. J Mol Biol. 2004;340:115–125. doi: 10.1016/j.jmb.2004.04.048. [DOI] [PubMed] [Google Scholar]
- 62.Ma HR, Gruebele M. Proc Natl Acad Sci USA. 2005;102:2283–2287. doi: 10.1073/pnas.0409270102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garcia-Mira MM, Sadqi M, Fischer N, Sanchez-Ruiz JM, Munoz V. Science. 2002;298:2191–2195. doi: 10.1126/science.1077809. [DOI] [PubMed] [Google Scholar]
- 64.Chu R, Pei W, Takei J, Bai Y. Biochemistry. 2002;41:7998–8003. doi: 10.1021/bi025872n. [DOI] [PubMed] [Google Scholar]
- 65.Feng H, Takei J, Lipsitz R, Tjandra N, Bai Y. Biochemistry. 2003;42:12461–12465. doi: 10.1021/bi035561s. [DOI] [PubMed] [Google Scholar]
- 66.Feng H, Zhou Z, Bai Y. Proc Natl Acad Sci USA. 2005;102:5026–5031. doi: 10.1073/pnas.0501372102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcia P, Bruix M, Rico M, Ciofi-Baffoni S, Banci L, Ramachandra Shastry MC, Roder H, de Lumley Woodyear T, Johnson CM, et al. J Mol Biol. 2005;346:331–344. doi: 10.1016/j.jmb.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 68.Gutin AM, Abkevich VI, Shakhnovich EI. Biochemistry. 1995;34:3066–3076. doi: 10.1021/bi00009a038. [DOI] [PubMed] [Google Scholar]
- 69.Chahine J, Nymeyer H, Leite VB, Socci ND, Onuchic JN. Phys Rev Lett. 2002;88:168101. doi: 10.1103/PhysRevLett.88.168101. [DOI] [PubMed] [Google Scholar]
- 70.Hardin C, Luthey-Schulten Z, Wolynes PG. Proteins Struct Funct Genet. 1999;34:281–294. [PubMed] [Google Scholar]
- 71.Zhou Y, Karplus M. Nature. 1999;401:400–403. doi: 10.1038/43937. [DOI] [PubMed] [Google Scholar]
- 72.Bartsch RG. Cytochromes. New York: Plenum; 1978. [Google Scholar]
- 73.Nozaki Y. Methods Enzymol. 1972;26:43–50. doi: 10.1016/s0076-6879(72)26005-0. [DOI] [PubMed] [Google Scholar]
- 74.Wu P, Brand L. Anal Biochem. 1994;218:1–13. doi: 10.1006/abio.1994.1134. [DOI] [PubMed] [Google Scholar]
- 75.Beecham JM, Haas E. Biophys J. 1989;55:1225–1236. doi: 10.1016/S0006-3495(89)82918-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beals JM, Haas E, Krausz S, Scheraga HA. Biochemistry. 1991;30:7680–7692. doi: 10.1021/bi00245a004. [DOI] [PubMed] [Google Scholar]
- 77.Istratov AA, Vyvenko OF. Rev Sci Instrum. 1999;70:1233–1257. [Google Scholar]
- 78.Livesey AK, Delaye M, Licinio P, Brochon JC. Faraday Discuss Chem Soc. 1987:247–258. [Google Scholar]
- 79.Lawson CL, Hanson RJ. Solving Least Squares Problems. Englewood Cliffs, NJ: Prentice–Hall; 1974. [Google Scholar]
- 80.Förster T. Ann Phys (Leipzig) 1948;2:55–75. [Google Scholar]
- 81.Jordanides XJ, Scholes GD, Shapley WA, Reimers JR, Fleming GR. J Phys Chem B. 2004;108:1753–1765. [Google Scholar]
- 82.Scholes GD, Jordanides XJ, Fleming GR. J Phys Chem B. 2001;105:1640–1651. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.