Abstract
Zinc-finger antiviral protein (ZAP) is a host antiviral factor that specifically inhibits the replication of Moloney murine leukemia virus (MLV) and Sindbis virus (SIN) by preventing accumulation of the viral mRNA in the cytoplasm. In previous studies, we demonstrated that ZAP directly binds to its specific target mRNAs. In this article, we provide evidence indicating that ZAP recruits the RNA processing exosome to degrade the target RNA. ZAP comigrated with the exosome in sucrose or glycerol velocity gradient centrifugation. Immunoprecipitation of ZAP coprecipitated the exosome components. In vitro pull-down assays indicated that ZAP directly interacted with the exosome component hRrp46p and that the binding region of ZAP was mapped to amino acids 224–254. Depletion of the exosome component hRrp41p or hRrp46p with small interfering RNA significantly reduced ZAP's destabilizing activity. These findings suggest that ZAP is a trans-acting factor that modulates mRNA stability.
Keywords: RNA degradation
The degradation of mRNA is an important control point in the regulation of gene expression (1–6). The general mRNA decay is initiated by removal of the poly(A) tail. The body of the RNA is degraded either from the 5′ end by exonuclease Xrn1 after decapping or from the 3′ end by an exoribonucleases complex named the exosome (7).
RNA decay mechanisms are diverse in the extent of decay and decay characteristics. This diversity presumably arises from the diverse RNA-binding factors and the diverse RNA–protein and protein–protein interactions in the decay machinery (8–11). Many cis-acting elements and trans-acting factors have been shown to engage in mRNA turnover regulation (11–20). In mammalian cells, the most common cis element is the AU-rich element (ARE) (21–24), which has been found in the 3′ UTR of a wide variety of short-lived mRNAs, such as those of growth factors, cytokines, and protooncogenes (25–27). Various ARE binding proteins (AUBPs) have been shown to modulate the stability of ARE-containing RNAs. Some AUBPs such as HuR (13, 28) and NF90 (29) stabilize the RNA, and other AUBPs such as tristetraprolin (TTP) (12, 30, 31), KRSP (32), and AUF1 (33, 34) destabilize the RNA. It has been reported that the destabilizing AUBPs recruit the exosome to degrade the ARE-containing RNAs (35).
The exosome is an evolutionarily highly conserved 3′–5′ exoribonucleases complex existing in both the nucleus and the cytoplasm (36–40). The nuclear exosome is required for the 3′ processing of many RNA substrates, including prerRNA, snoRNA, snRNA, and premRNA (41–43). The nuclear exosome also functions in the surveillance system, in which the transcripts with defects generated in the RNA processing and exporting pathways are degraded (44–46). The cytoplasmic exosome plays a key role in the degradation of aberrant or unused intermediate mRNAs and ARE-containing mRNAs (1, 35, 47–53). The effects on the target RNA depend on interactions among the exosome, exosome cofactors, the target RNA, and specific RNA-binding proteins (35, 52–55).
The human exosome is known to contain at least six RNase-PH domain subunits (hRrp41p, hRrp42p, hRrp43p, hRrp46p, PM/Scl-75, and Mtr3), three S1/KH domain subunits (hRrp4p, hRrp40p, and hCsl4), an RNase-D-like domain subunit (PM/Scl-100), a putative RNA helicase (Kiaa0052), and a protein that is specifically phosphorylated in the M phase of the cell cycle (Mpp6) (36, 38). The RNase-PH domain subunits and the S1/KH RNA-binding domain subunits are considered to be the core components of the exosome, whereas Kiaa0052 and Mpp6 are considered to be accessory factors (36, 38). Yeast PM/Scl-100 is found only in the nuclear exosome (36, 46).
The structure of the exosome is not yet determined. Based on the results of mammalian-two-hybrid and yeast-two-hybrid experiments (38, 56–59), the six RNase-PH domain-containing subunits are thought to assemble into a doughnut-shaped ring. The S1/KH RNA-binding domain-containing subunits associate with the RNase-PH domain hexamer and may serve to aid substrate RNA binding and delivery to the exoribonucleaes ring. The accessory factors may modulate the activity and stability of the exosome.
Zinc-finger antiviral protein (ZAP) is a recently isolated host antiviral factor that specifically inhibits the infection of cells by Moloney murine leukemia virus (MLV) (60) and multiple members of the alphavirus family, including Sindbis virus (SIN) (61). Overexpression of ZAP prevents accumulation of the viral RNA in the cytoplasm (60, 61). The N terminus of ZAP contains four CCCH-type zinc-finger motifs (60). ZAP binds directly to specific viral RNA sequences through these zinc-finger motifs (62). The target sequence of ZAP in MLV was mapped to the 3′-LTR, and the target sequences in SIN were mapped to multiple fragments, but no obvious common motifs were found in these sequences yet (62). Particularly, ZAP does not target ARE-containing mRNAs (62).
Despite the lack of primary sequence homology, ZAP shares considerable similarities with TTP. Both ZAP and TTP have the uncommon CCCH-type zinc-finger motifs (12, 60) and directly bind to their cognate target RNAs, and the zinc-finger motifs are required for the binding (30, 62–64). Most importantly, they both mediate specific RNA destabilization (30, 31, 60, 61, 65). These similarities suggested that ZAP and TTP might share a common mechanism for destabilizing specific mRNAs. In the present study, we provide evidence indicating that ZAP directly interacts with the exosome, and we suggest that ZAP destabilizes RNA by directly binding to the target RNA and recruiting the exosome to degrade the target RNA.
Results
ZAP Promotes Target mRNA Degradation in the Cytoplasm.
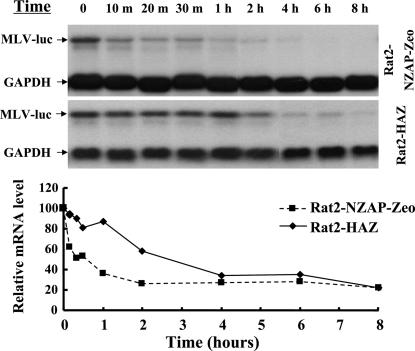
The low levels of MLV mRNA and SIN RNA in the cytoplasm of ZAP-expressing cells suggested that the RNAs were degraded in the cytoplasm (60, 61). To potentiate this notion, we measured the decay rate of the target mRNA in the cytoplasm of ZAP-expressing cells. The cells were transduced with an MLV vector carrying the firefly luciferase reporter gene (MLV-luc). Transcription was blocked by addition of actinomycin D, and the reporter mRNA level in the cytoplasm was measured at different time points (Fig. 1). Consistent with our previous results, the reporter mRNA level was, even before actinomycin D treatment, already much lower in the ZAP-expressing cells than in the control cells. Nonetheless, detectable amounts of the reporter mRNA existed in the cytoplasm of ZAP expressing cells. The amounts of the reporter mRNA before actinomycin D treatment were used as the starting points to measure the decay rates. The half-life of the reporter mRNA in the cytoplasm of ZAP-expressing cells was estimated to be ≈0.5 h, whereas the half-life of the reporter mRNA in the cytoplasm of the control cells was estimated to be ≈2.5 h (Fig. 1). These results demonstrated that ZAP mediated the degradation of the reporter mRNA in the cytoplasm.
Fig. 1.
ZAP promotes target viral mRNA degradation in the cytoplasm. Rat2-HAZ and Rat2-NZAP-Zeo cells were infected with MLV-luc virus. Forty-eight hours after infection, actinomycin D was added to the media at a final concentration of 5 μg/ml to stop transcription. (Upper) At the indicated time points, the cells were lysed and the cytoplasmic RNA was extracted and subjected to Northern blotting. (Lower) The relative levels of the viral mRNA were measured by using a phosphorimager and were normalized by the GAPDH mRNA level.
ZAP Cosediments with the Exosome.
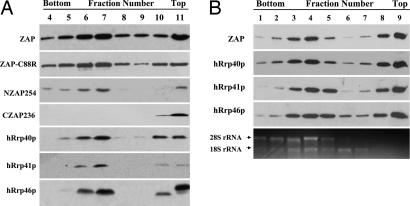
Based on the similarities between ZAP and TTP, we hypothesized that ZAP might recruit the RNA-processing exosome to degrade the target mRNA as TTP does (35). To test this hypothesis, we first analyzed whether ZAP cosediments with the exosome. Myc-tagged full-length ZAP, a mutant ZAP lacking RNA-binding activity (ZAP-C88R), an N-terminal fragment of ZAP (NZAP254, amino acids 1–254), or a C-terminal fragment of ZAP (CZAP236, amino acids 236–776) was expressed in 293TRex cells. The cytoplasmic extracts were fractionated by sucrose velocity sedimentation centrifugation, and fractions were immunoblotted with anti-myc antibody to detect the ZAP proteins or with antibodies against three components of the exosome: hRrp40p, hRrp41p, and hRrp46p. These proteins, except for CZAP236, displayed very similar sedimentation profiles; they were detected in the high mobility fractions, with the peak level in fraction 7 (Fig. 2A). It should be noted that the protein level of NZAP254 in the cytoplasm was so low that it could be detected only with overexposure. The low protein level in the cytoplasm is likely accounted for by the nuclear localization of NZAP254 (66). That NZAP254 and ZAP-C88R displayed the same sedimentation pattern as full-length ZAP and the exosome components suggested that the N-terminal portion of ZAP contains the exosome-interacting domains and that RNA-binding activity is dispensable for ZAP to interact with the exosome. Consistently, CZAP236 was detected only in the low mobility fractions (Fig. 2A).
Fig. 2.
ZAP cosediments with the exosome components. 293TRex cells expressing various versions of ZAP were lysed in the hypotonic buffer. The lysates were fractionated by sucrose (A) or glycerol (B) velocity gradient centrifugation. Equal aliquots of each fraction were subjected to SDS/PAGE and were Western blotted with the 9E10 anti-myc antibody or antibodies against the exosome components hRrp40p, hRrp41p, or hRrp46p. Total RNA was extracted from equal aliquots of each fraction, and the 28S and 18S rRNAs were detected by ethidium bromide staining
The cytoplasmic extracts were also fractionated by using glycerol velocity sedimentation centrifugation. The distribution patterns were very similar to the patterns in the sucrose gradients (Fig. 2B). Under this condition, the peak levels of ZAP and the exosome components in the high mobility fractions were detected in fraction 4 (Fig. 2B). It has been reported that the high mobility fractions are polysome-rich and that the exosome is associated with the polysome (67–69). Indeed, the 28S and 18S rRNAs were detected in these fractions with a distribution pattern similar to those of ZAP and the exosome components (Fig. 2B). In addition, treatment with 40 mM EDTA, which causes dissociation of ribosomes from mRNAs without disrupting the majority of nonribosomal RNA–proteins complexes (70), resulted in a up-shift of the ZAP and exosome peaks (data not shown). The exosome components and ZAP detected in the low mobility fractions could be either free proteins resulting from disassembly of the complex during sample preparation or the ZAP–exosome complex that is not associated with the polysome.
ZAP Interacts with the Exosome.
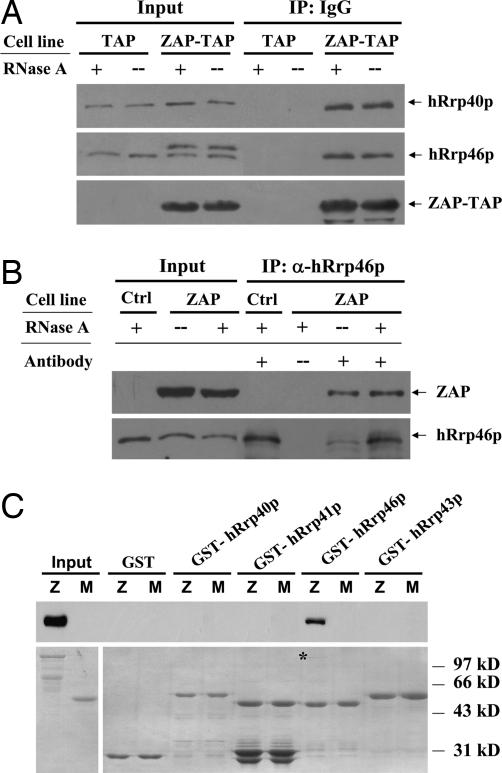
The results in Fig. 2 suggested that ZAP forms a complex with the exosome. To further prove the interaction between ZAP and the exosome, coimmunoprecipitation assays were performed. A 332-aa N-terminal fragment of ZAP was expressed as a fusion protein with the tandem affinity purification (TAP) tag (71, 72) at the C terminus [ZAP (1–332)-TAP] for use in the coimmunoprecipitation assays. ZAP (1–332) includes a nuclear export sequence (NES) and thereby is localized predominantly in the cytoplasm (data not shown). ZAP (1–332)-TAP expressed in HEK293 cells was precipitated by using IgG resin, which binds to the Protein A portion in the TAP tag. To prevent possible RNA tethering, RNase A was added to the lysates. Endogenous hRrp40p and hRrp46p were coprecipitated with or without RNase treatment from the ZAP-expressing cells but not from the control cells (Fig. 3A). In the reverse experiment, immunoprecipitation using the anti-hRrp46p antibody coprecipitated myc-tagged ZAP in the presence or absence of RNase A (Fig. 3B), further confirming the interaction between ZAP and the exosome.
Fig. 3.
Interaction between ZAP and the exosome components. (A) The 293 cells expressing ZAP (1–332)-TAP or TAP alone were lysed in the lysis buffer in the presence (+) or absence (−) of 100 μg/ml RNase A. The proteins were precipitated with IgG Sepharose and were Western blotted with anti-protein A antibody to detect the TAP-tag or with the anti-hRrp40p or anti-hRrp46p antibody to detect the exosome components. Input, total cell lysates; TAP, TAP-expressing control cells; ZAP-TAP, ZAP (1–332)-TAP-expressing cells. (B) 293TRex-ZAP cells were treated with tetracycline to induce ZAP expression and lysed in the presence (+) or absence (−) of 100 μg/ml RNase A. The proteins were immunoprecipitated with preimmune (−) or anti-hRrp46p antisera and were Western blotted with the 9E10 anti-myc antibody. Input, total cell lysates; Ctrl, 293TRex control cells; ZAP, 293TRex-ZAP cells. (C) GST, GST-hRrp40p, GST-hRrp41p, GST-hRrp43p, and GST-hRrp46p expressed in E. coli were treated with RNase A at a final concentration of 100 μg/ml, immobilized onto glutathione-Sepharose 4B resin, and incubated with MBP-ZAP-myc in the presence of RNase A for 2 h at 4°C. The resins were washed, and the proteins were resolved by SDS/PAGE and detected by Western blotting by using the 9E10 anti-myc antibody (Upper) or Coomassie blue staining (Lower). Z, MBP-ZAP; M, MBP.
To test whether ZAP directly interacts with any of the separate exosome components, hRrp40p, hRrp41p, hRrp43p, and hRrp46 were expressed as GST fusion proteins in Escherichia coli and analyzed for their abilities to bind to partially purified ZAP expressed in E. coli with the maltose binding protein (MBP) fused at the C terminus. To prevent possible RNA tethering, these proteins were treated with RNase A. ZAP bound to GST-hRrp46p but not to other exosome components (Fig. 3C).
Mapping the Binding Domains of ZAP and hRrp46p.
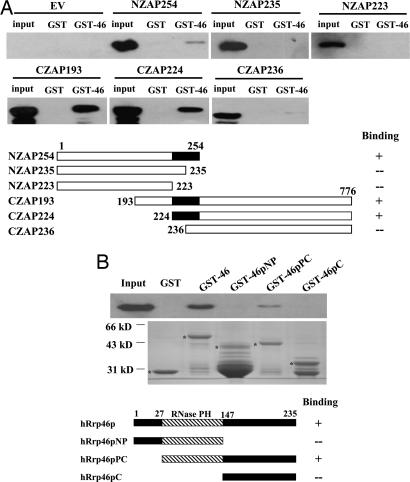
To map the binding domain of ZAP, a series of ZAP truncation mutants were expressed in HEK293T cells and analyzed for their abilities to bind to GST-hRrp46p. In consistence with the results that NZAP254 comigrated with the exosome components (Fig. 1) and that NZAP-Zeo displayed the same antiviral activity as full-length ZAP (60), NZAP254 displayed detectable binding to hRrp46. In contrast, NZAP223 and NZAP235 displayed no detectable binding to hRrp46p (Fig. 4A), suggesting that the region spanning amino acids 224–254 is required for ZAP to bind to hRrp46p. In the complementary experiment, a series of N-terminal deletion mutants of ZAP were tested for their abilities to bind to hRrp46p. The two mutants that contain the fragment of amino acids 224–254, CZAP193 (amino acids 193–776) and CZAP224 (amino acids 224–776), both bound to hRrp46p (Fig. 4A). In contrast, binding of CZAP236 (amino acids 236–776) to hRrp46p was dramatically reduced to a barely detectable level (Fig. 4A). Collectively, these results indicated that the region of amino acids 224–254 of ZAP is necessary and sufficient for the binding to hRrp46p.
Fig. 4.
Mapping the interaction region for ZAP and hRrp46p. (A) Recombinant GST or GST-hRrp46p was immobilized onto glutathione-Sepharose 4B resin and incubated with the lysates of the indicated ZAP truncation mutants in the presence of RNase A (final concentration, 100 μg/ml) for 2 h at 4°C. The resins were washed, and the proteins were resolved by SDS/PAGE and detected by Western blotting by using the 9E10 anti-myc antibody. Input, total cells lysates. (B) The indicated recombinant GST proteins were immobilized onto glutathione-Sepharose 4B resin. 293TRex-ZAP cells were treated with tetracycline to induce ZAP expression, and the lysate was treated with RNase A (final concentration, 100 μg/ml) at 37°C for 30 min and then incubated with the resins for 2 h at 4°C. The resins were washed, and the proteins were resolved by SDS/PAGE and detected by Western blotting by using the 9E10 anti-myc antibody (Upper) or Coomassie blue staining (Lower). The positions of the GST proteins are indicated by asterisks.
To map the binding region of hRrp46p, a series of hRrp46p truncation mutants were expressed as GST-fusion proteins and analyzed for their abilities to bind to ZAP expressed in 293TRex-ZAP cells. The hRrp46p-PC fragment, which contains the central RNase-PH domain and the C-terminal domain, bound to ZAP at a reduced level compared with the full-length hRrp46p (Fig. 4B). In contrast, the hRrp46p-NP fragment, which contains the central RNase-PH domain and the N-terminal domain, failed to display detectable binding to ZAP (Fig. 4B). Because both hRrp46p-NP and hRrp46p-PC share the RNase-PH domain, the binding region of hRrp46p appeared to be located in the C-terminal domain. However, the C-terminal fragment alone did not display detectable binding (Fig. 4B). Collectively, these results suggested that the C-terminal fragment of hRrp46p is necessary but not sufficient for binding to ZAP and that the whole protein of hRrp46p is required for efficient binding to ZAP.
Depletion of hRrp41p or hRrp46p Reduces the Activity of ZAP.
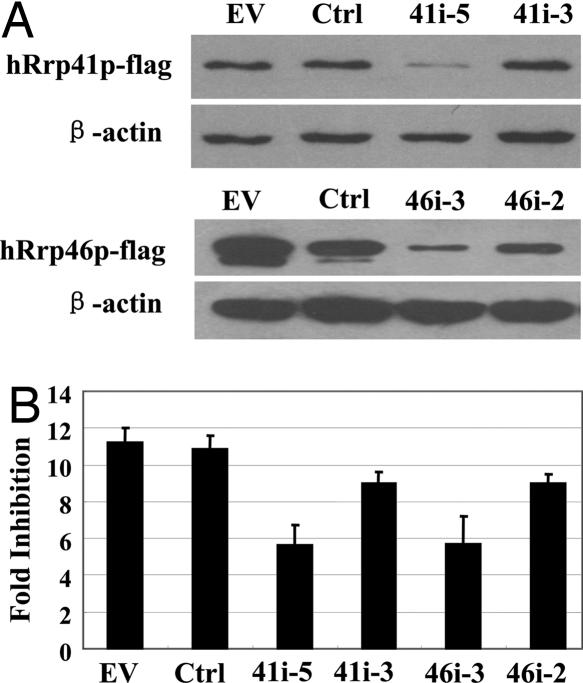
To further determine whether the exosome is required for ZAP's activity, we used RNAi to down-regulate the expression of endogenous exosome components and analyzed the effects on the destabilizing activity of ZAP. We first designed shRNAs to down-regulate hRrp46p, the exosome component that directly bound to ZAP. Plasmids expressing hRrp46p-RNAi directed against two different sites of hRrp46p were constructed. Because the transfection efficiency was not high enough to accurately evaluate the inhibitory effect of these RNAi on the expression of the endogenous hRrp46p, the ability of the RNAi to inhibit the expression of hRrp46p was tested by cotransfection into HEK 293 cells with a construct expressing FLAG-tagged hRrp46p. Compared with the empty vector or control RNAi, hRrp46p-RNAi-3 significantly reduced the expression level of FLAG-tagged hRrp46p, whereas hRrp46p-RNAi-2 displayed only a modest effect (Fig. 5A Lower). To test the effect of the depletion of hRrp46p on the activity of ZAP, 293TRex-ZAP cells were cotransfected with the MLV-luc reporter and the hRrp46p-RNAi expressing plasmid and assayed for inhibition of the reporter expression by ZAP. Consistent with the inhibitory effects of the RNAi constructs on the expression levels of hRrp46p, hRrp46p-RNAi-2 displayed only a marginal effect, whereas hRrp46p-RNAi-3 reduced the fold inhibition significantly (Fig. 5B). Using the same method, we also tested the effect on ZAP's activity of the down-regulation of hRrp41p, an exosome component that did not directly bind to ZAP. hRrp41p-RNAi-5, which efficiently inhibits the expression of FLAG-tagged hRrp41p (Fig. 5A, Upper), also reduced ZAP's activity (Fig. 5B). It is worth noting that down-regulating either exosome component reduced ZAP's activity by only ≈50%. This could be accounted for by either the incomplete depletion of the exosome components by the RNAi method, the existence of other mechanisms not involving the exosome in ZAP-mediated RNA degradeation, or both. Nonetheless, the results in Fig. 5 indicated that the integrity of the exosome is important for ZAP's activity.
Fig. 5.
Down-regulation of hRrp41p and hRrp46p with the interfering RNA reduces ZAP's activity. (A) The plasmids expressing the indicated FLAG-tagged exosome components were cotransfected into HEK293 cells with empty vector, control RNAi, or plasmids expressing the RNAi directed against the indicated exosome components. The relative expression levels of the FLAG-tagged exosome components were analyzed by Western blot analysis. (B) pMLV-luc, pRL-TK, and the indicated RNAi expressing plasmid were cotransfected into 293TRex-ZAP cells. Immediately after transfection, tetracycline was added to induce ZAP expression. Forty-eight hours after transfection, the luciferase activities were measured. The expression level of pRL-TK, a plasmid expressing Renilla luciferase and not responsive to ZAP, was used to normalize transfection efficiency. Fold inhibition is calculated as the normalized luciferase activity in the untreated cells divided by the normalized luciferase activity in the tetracycline-treated cells. The data are means + SD of three independent experiments.
Discussion
mRNA decay is an important mechanism that regulates gene expression (1, 2). The rate of mRNA decay is largely determined by the cis-acting elements. ARE is the most common cis element so far identified that mediates mRNA decay (19, 23–25). In principle, other cis-acting elements that regulate the stability of non-ARE-containing mRNAs should also exist. However, such elements and the trans-acting factors binding to them remain largely unidentified. ZAP has been shown to target non-ARE mRNAs, although the features of the ZAP responsive element are not yet well defined (62). In this article, we showed that ZAP directly binds to the exosome and that the integrity of the exosome is required for ZAP's activity. These results suggest that ZAP represents a trans-acting factor that modulates the stability of ZAP responsive element-containing mRNAs.
ZAP was originally isolated as an antiviral factor based on the antiviral activity of the protein when overexpressed. The function of ZAP under physiological conditions remains to be determined. Ryman et al. (73) recently reported that ZAP was expressed at an undetectable level in murine bone marrow-derived dendritic cells. Upon IFN-α treatment or infection of the cells by SIN virus, the expression level of ZAP was significantly up-regulated. These results suggest that ZAP might be involved in host antiviral mechanisms under physiological conditions. In addition, our preliminary results demonstrated that, in ZAP-expressing cells, the expression of a cellular gene was down-regulated (Z. Huang and G.G., unpublished work), suggesting that ZAP might also be involved in regulating the stability of cellular mRNAs. Further investigation is needed to fully understand the physiological functions of ZAP.
Exosome-mediated mRNA degradation is a complex process involving deadenylation, decapping, and degradation of the body of the RNA (40, 48, 74). It has been reported (23, 30, 65) for the degradation of ARE-containing RNA mediated by AUPBs that the process is initiated by removal of the poly(A) tail. The body of the RNA is degraded by the exosome in the 3′–5′ direction (35, 52). Because ZAP degrades the target RNA by recruiting the exosome, we speculate that the degradation should be similar. Further investigation is necessary to fully understand the mode of ZAP-mediated RNA degradation and the relationship among ZAP and the deadenylation and decapping machineries.
Materials and Methods
Plasmids.
The plasmids pcDNA4TO/myc-NZAP, pcDNA4TO/myc-ZAP, and pcDNA4TO/myc-ZAP-C88R, which express myc-tagged NZAP254, ZAP, and ZAP-C88R, respectively, have been described in ref. 60. pcDNA4TO/myc-CZAP236 expresses the C-terminal fragment of ZAP (amino acids 236–776). To generate CZAP236, amino acids 4–235 were deleted from ZAP. First, the BamHI site in the multiple cloning sites of pcDNA4TO/myc-ZAP was first destroyed. The AflII–NotI fragment, which spans from the 5′ UTR to the end of the coding sequence of ZAP, was replaced with AflII–BamHI and BamHI–NotI PCR-derived fragments. The AflII–BamHI fragment, which covers the 5′ UTR and the first four amino acids of ZAP, was generated by using forward primer CZAP236-5′-FP upstream of the AflII site and reverse primer CZAP236-5′-RP bearing a silent mutation to create a BamHI site. The BamHI–NotI fragment, which covers the sequence encoding amino acids 236–776 of ZAP, was generated by using forward primer CZAP236-3′-FP bearing a silent mutation to create a BamHI site and reverse primer CZAP236-3′-RP downstream of the NotI site. pcDNA4TO/myc-CZAP193 and pcDNA4TO/myc-CZAP224 express myc-tagged CZAP193 (amino acids 193–776) and CZAP224 (amino acids 224–776), respectively. To generate pcDNA4TO/myc-CZAP193, a ZAP fragment was amplified by using forward primer CZAP193-FP bearing a BamHI site and reverse primer CZAP193-RP bearing a NotI site and was used to replace the BamHI–NotI fragment of pcDNA4TO/myc-CZAP236. pcDNA4TO/myc-CZAP224 was generated by using the same strategy with primers CZAP224-FP and CZAP224-RP. pcDNA4TO/myc-NZAP223 and pcDNA4TO/myc-NZAP235 express myc-tagged NZAP223 (amino acids 1–223) and NZAP235 (amino acids 1–235), respectively. The plasmids were generated by replacing the BamHI–NotI fragment of pcDNA4TO/myc-ZAP with PCR fragments generated from pcDNA4TO/myc-ZAP by using a forward primer NZAP-FP upstream of the BamHI site and reverse primers NZAP223-RP or NZAP235-RP.
pZome-1-C, an expression vector of the TAP tag, was provided by the European Molecular Biology Laboratory. pZAP (1–332)-Zome expresses the N-terminal fragment of ZAP (amino acids 1–332) fused with a TAP tag at the C terminus. The ZAP (1–332) fragment was amplified from pcDNA4To/myc-ZAP by using primers ZAP (1–332)-FP and ZAP (1–332)-RP and was cloned into pZome-1-C by using the BamHI site to generate pZAP (1–332)-Zome.
pCMV-HA-Flag-hRrp46p and pCMV-HA-Flag-hRrp41p express FLAG-tagged hRrp46p and hRrp41p, respectively. The expression vector pCMV-HA-Flag was modified from pCMV-HA (Clontech, Mountain View, CA) by inserting a FLAG-tag coding sequence between the HA coding sequence and the multiple cloning site. The coding sequences of hRrp46p and hRrp41p were amplified from human cDNA libraries by using primers hRrp46p-FP and hRrp46p-RP for hRrp46p. hRrp41p-FP and hRrp41p-RP were used for hRrp41p.
The coding sequence of hRrp40p was amplified from a human fetal liver cDNA library by using primers hRrp40p-FP and hRrp40p-RP and was cloned into pCMV-HA-Flag. pGEX-5M-hRrp40p, pGEX-5M-hRrp41p, pGEX-5M-hRrp43p, and pGEX-5M- hRrp46p express GST-hRrp40p, GST-hRrp41p, GST-hRrp43p, and GST-hRrp46p fusion proteins, respectively. The GST expression vector pGEX-5x-3 (Amersham Pharmacia, Piscataway, NJ) was first modified to introduce a KpnI site and a BglII site in the multiple cloning sites to generate pGEX-5M. The coding sequences of hRrp40p, hRrp41p, and hRrp46p were cloned into pGEX-5M by using EcoRI and KpnI sites. The coding sequence of hRrp43p (kindly provided by Sydney Altman, Yale University, CT) was PCR amplified and cloned into pGEX-5M. The GST-hRrp46p truncation mutants were constructed by cloning the PCR fragments amplified from hRrp46p into pGEX-5M.
pBabe-SuperhRrp41pRNAi and pBabe-SuperhRrp46pRNAi express the siRNAs directed against hRrp41p and hRrp46p, respectively. To construct the RNAi expression vector, pBabe-puro was first modified by sequentially destroying the HindIII and EcoRI sites. The siRNA expression cassette was PCR-amplified from pSuper (OligoEngine, Seattle, WA) and cloned into the modified pBabe-puro. Oligonucleotides hRrp41p-RNAi-3-FP and hRrp41p-RNAi-3-RP were annealed to generate hRrp41p-RNAi-3; oligonucleotides hRrp41p-RNAi-5-FP and hRrp41p-RNAi-5-RP were annealed to generate hRrp46p-RNAi-5. Oligonucleotides hRrp46p-RNAi-2-FP and hRrp46p-RNAi-2-RP were annealed to generate hRrp46p-RNAi-2; oligonucleotides hRrp46p-RNAi-3-FP and hRrp46p-RNAi-3-RP were annealed to generate hRrp46p-RNAi-3. The annealed fragments were cloned into pBabe-super to generate pBabe-SuperhRrp41pRNAi-3, pBabe-SuperhRrp41pRNAi-5, pBabe-SuperhRrp46pRNAi-2, and pBabe-SuperhRrp46pRNAi-3.
The sequences of the oligonucleotides are as follows, with the restriction sites in italics: CZAP236-5′-FP, 5′-GCTCTCCCTATCAGTG-3′; CZAP236-5′-RP, 5′-ATATAGGGATCCGCCATGGCGCGCTATGG-3′; CZAP236-3′-FP, 5′-ATATAGGGATCCACACCGCAGAGGCGG-3′; CZAP236-3′-RP, 5′-GAGGGGCAAACAACAG-3′; CZAP224-FP, 5′-CGGGATCCAGCCAGGAGGAACCCGCC-3′; CZAP224-RP, 5′-CTTAGCCACATTCTTCCAAG-3′; CZAP193-FP, 5′-ATATAGGGATCCCTGATGGACAGAAAGGTG-3′; CZAP193-RP, 5′-GAGGGGCAAACAACAG-3′; NZAP-FP, 5′-GACCTCCATAGAAGACAC-3′; NZAP223-RP, 5′-AGACTCGAGCGGCCGCCGTGTTTGTTGTTGCAGATG-3′; NZAP235-RP, 5′-AGACTCGAGCGGCCGCCTGGATGGGCAGCTCTCGTG-3′; ZAP (1–332)-FP, 5′-ATATAGGGATCCAAAGCCGAGTCCCGCGC-3′; ZAP (1–332)-RP, 5′-ATATAGGGATCCCCCATCCTCTGAAAACTC-3′; hRrp40p-FP, 5′-GCGAATTCGCCGAACCTGCGTCTGTC-3′; hRrp40p-RP, 5′-GCGGTACCTCAACTTTCTGCCAATCTGGAGA-3′; hRrp41p-FP, 5′-GCGAATTCGCGGGGCTGGAGCTCTTG-3′; hRrp41p-RP, 5′-GCGGTACCTCAGTCCCCCAGCAAGATAGA-3′; hRrp43p-FP, 5′-GAAGATCTGCGGCTGGGTTCAAAAC-3′; hRrp43p-RP, 5′-ATGGTACCTTATTTGGGTTTCATACTC-3′; hRrp46p-FP, 5′-GCGAATTCGAGGAGGAGACGCATACTGA-3′; hRrp46p-RP, 5′-GCGGTACCTCAGCTCTTGGAGTAACGCC-3′; hRrp46pNP-RP, 5′-GCGGTACCTCAGGGCACACCTGCATCCAC-3′; hRrp46pPC-FP, 5′-GCGAATTCAGCCTCCGGCACTTTGCC-3′; hRrp46pC-FP, 5′-GCGAATTCATGCGGGCTCTCTTCTG-3′; hRp41p-RNAi3-FP, 5′-GATCCCCCTGTCAATATAGTTCAGCTTTCAAGAGACGCTGAACTATATTGACAGTTTTTA-3′; hRp41p-RNAi-3-RP, 5′-AGCTTAAAAACTGTCAATATAGTTCAGCGTCTCTTGAAAGCTGAACTATATTGACAGGGG- 3′; hRp41p-RNAi-5-FP, 5′-GATCCCCTATAGTTCAGCGACCTTCCTTCAAGAGATGAAGGTCGCTGAACTATATTTTTA-3′; hRp41p-RNAi-5-RP, 5′- AGCTTAAAAATATAGTTCAGCGACCTTCATCTCTTGAAGGAAGGTCGCTGAACTATAGGG- 3′; hRrp46p-RNAi-2-FP, 5′-GATCCCCCGGAAGCTGCTGATGTCCATTCAAGAGATGGACATCAGCAGCTTCCGTTTTTGGAAA-3′; hRrp46p-RNAi-2-RP, 5′-AGCTTTTCCAAAAACGGAAGCTGCTGATGTCCATCTCTTGAATGGACATCAGCAGCTTCCGGGG-3′; hRrp46p-RNAi-3-FP, 5′-GATCCCCGGATCCTACATCCAAGCAATTCAAGAGATTGCTTGGATGTAGGATCCTTTTTGGAAA; hRrp46p-RNAi-3-RP, 5′-AGCTTTTCCAAAAAGGATCCTACATCCAAGCAATCTCTTGAATTGCTTGGATGTAGGATCCGGG-3′.
Cell Culture.
All of the cells were maintained in DMEM supplemented with 10% FBS. The MLV-luc virus is reported in ref. 60. Infection was conducted for 3 h followed by replacement of the infection media with fresh media. Transfection was performed by using FuGENE 6 (Roche Diagnostics, Basel, Switzerland) according to the manufacturer's instructions. The assay to evaluate the inhibition of ZAP on the reporter is described in ref. 62.
Rat2-HAZ, Rat2-NZAP-Zeo, 293TRex, 293TRex-ZAP, and 293TRex-NZAP cell lines are described in ref. 62. The CZAP236 protein was expressed by transiently transfecting 293TRex cells with pcDNA4TO/myc-CZAP236, followed by induction with tetracycline.
To express ZAP (1–332)-TAP, pZAP (1–332)-Zome was packaged into MLV pseudovirus to transduce 293 cells. The cells were selected in 3 μg/ml puromycin, and the resistant cells were pooled.
Northern Blot Analysis.
Cytoplasmic RNA was isolated from cells with an RNeasy kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The RNA samples were separated by electrophoresis, transferred to nylon membrane, and hybridized for 15–20 h with 32P-labeled probes prepared by a random primer labeling kit (Stratagene, La Jolla, CA). The nylon membrane was washed three times with 0.1× SSC (0.015 M sodium chloride/0.0015 M sodium citrate, pH 7) and 0.1% SDS at 65°C and exposed to either x-ray film or phosphorimaging to quantitate the relative RNA levels.
Velocity Sendimentation Centrifugation.
To prepare a discontinuous sucrose density gradient, sucrose was completely dissolved in hypotonic buffer [10 mM Tris·HCl (pH7.6)/1.5 mM Mg(Ac)2/1 mM KAc/10 μM ZnCl2/2 mM DTT] at concentrations of 50%, 40%, 30%, 20%, and 10%. Two milliliters of the 50% sucrose was first loaded at the bottom of the polyallomer centrifuge tube (Beckman, Fullerton, CA), on top of which was layered 2 ml each of the 40%, 30%, 20%, and 10% sucrose solutions. To prepare a discontinuous glycerol density gradient, glycerol was completely dissolved in hypotonic buffer at concentrations of 30%, 25%, 20%, 15% 10%, and 5%. Thirty percent glycerol (1.6 ml) was first loaded at the bottom of the polyallomer centrifuge tube (Beckman), on top of which was layered 1.6 ml of each of the 25%, 20%, 15%, 10%, and 5% glycerol solutions. To prepare the cytoplasmic extract, 293TRex-ZAP cells were treated with tetracycline (final concentration, 1 μg/ml) for 24–36 h to induce ZAP expression, and the cells were trypsinized, washed two times with ice-cold PBS, and suspended in the hypotonic buffer. For complete lysis, the suspension was homogenized in a Dounce homogenizer by 20 up and down strokes. The cell lysates were clarified by centrifugation for 10 min at 13,000 rpm in a microcentrifuge (Sorvall, Bad Homberg, Germany), and the supernatant was loaded on top of the sucrose or glycerol gradients. Centrifugation was performed in a Beckman SW41 rotor at 4°C for 3 h at 36,000 rpm. The fractions were collected from the bottom of the tube with ≈1 ml per fraction.
Coimmunoprecipitation.
The ZAP-expressing and control cells were lysed in lysis buffer B [30 mM Hepes (pH 7.6)/100 mM NaCl/0.5% Nonidet P-40 and protease inhibitors mixture] with or without RNase A (final concentration, 100 μg/ml) on ice for 10 min, and the lysates were clarified by centrifugation at 4°C for 10 min at 13,000 rpm in a microcentrifuge (Sorvall). The supernatant was mixed with IgG Sepharose (Amersham Pharmacia) to precipitate ZAP (1–332)-TAP or mixed with protein G plus agarose (Santa Cruz Biotechnology, Santa Cruz, CA) and anti-hRrp46p antibody (kindly provided by Ger Prujin, University of Nijmegen, Nijmegen, Netherlands) to precipitate hRrp46p. After incubation at 4°C for 2 h, the resins were washed three times with lysis buffer B and the bound proteins were detected by Western blotting.
Acknowledgments
We thank Drs. Stephen P. Goff and Margaret R. MacDonald for helpful discussion and critical readings of the manuscript and Dr. Ger Prujin for generously providing the exosome antibodies. This work was supported by National Science Foundation Grants 30470092 and 30530020 (to G.G.) and by Ministry of Science and Technology of China 973 Program 2002CB513001.
Abbreviations
- ZAP
zinc-finger antiviral protein
- TTP
tristetraprolin
- MLV
murine leukemia virus
- SIN
Sindbis virus
- ARE
AU-rich element
- AUBP
ARE binding protein
- TAP
tandem affinity purification.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Bevilacqua A, Ceriani MC, Capaccioli S, Nicolin A. J Cell Physiol. 2003;195:356–372. doi: 10.1002/jcp.10272. [DOI] [PubMed] [Google Scholar]
- 2.Guhaniyogi J, Brewer G. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 3.Khodursky AB, Bernstein JA. Trends Genet. 2003;19:113–115. doi: 10.1016/S0168-9525(02)00047-1. [DOI] [PubMed] [Google Scholar]
- 4.Vasudevan S, Peltz SW. Curr Opin Cell Biol. 2003;15:332–337. doi: 10.1016/s0955-0674(03)00051-6. [DOI] [PubMed] [Google Scholar]
- 5.Vasudevan S, Peltz SW, Wilusz CJ. BioEssays. 2002;24:785–788. doi: 10.1002/bies.10153. [DOI] [PubMed] [Google Scholar]
- 6.Wagner E, Lykke-Andersen J. J Cell Sci. 2002;115:3033–3038. doi: 10.1242/jcs.115.15.3033. [DOI] [PubMed] [Google Scholar]
- 7.Anderson JS, Parker RP. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpousis AJ, Vanzo NF, Raynal LC. Trends Genet. 1999;15:24–28. doi: 10.1016/s0168-9525(98)01627-8. [DOI] [PubMed] [Google Scholar]
- 9.Dehlin E, Wormington M, Korner CG, Wahle E. EMBO J. 2000;19:1079–1086. doi: 10.1093/emboj/19.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erlitzki R, Long JC, Theil EC. J Biol Chem. 2002;277:42579–42587. doi: 10.1074/jbc.M207918200. [DOI] [PubMed] [Google Scholar]
- 11.Loflin P, Chen CY, Shyu AB. Genes Dev. 1999;13:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blackshear PJ. Biochem Soc Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 13.Brennan CM, Steitz JA. Cell Mol Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Orso I, Frasch AC. J Biol Chem. 2001;276:15783–15793. doi: 10.1074/jbc.M010959200. [DOI] [PubMed] [Google Scholar]
- 15.Lee JH, Jeon MH, Seo YJ, Lee YJ, Ko JH, Tsujimoto Y, Lee JH. J Biol Chem. 2004;279:42758–42764. doi: 10.1074/jbc.M407357200. [DOI] [PubMed] [Google Scholar]
- 16.Paste M, Huez G, Kruys V. Eur J Biochem. 2003;270:1590–1597. doi: 10.1046/j.1432-1033.2003.03530.x. [DOI] [PubMed] [Google Scholar]
- 17.Sengupta TK, Bandyopadhyay S, Fernandes DJ, Spicer EK. J Biol Chem. 2004;279:10855–10863. doi: 10.1074/jbc.M309111200. [DOI] [PubMed] [Google Scholar]
- 18.Stoecklin G, Lu M, Rattenbacher B, Moroni C. Mol Cell Biol. 2003;23:3506–3515. doi: 10.1128/MCB.23.10.3506-3515.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tourriere H, Chebli K, Tazi J. Biochimie. 2002;84:821–837. doi: 10.1016/s0300-9084(02)01445-1. [DOI] [PubMed] [Google Scholar]
- 20.Wilusz CJ, Wormington M, Peltz SW. Nat Rev Mol Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 21.Bakheet T, Frevel M, Williams BR, Greer W, Khabar KS. Nucleic Acids Res. 2001;29:246–254. doi: 10.1093/nar/29.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakheet T, Williams BR, Khabar KS. Nucleic Acids Res. 2003;31:421–423. doi: 10.1093/nar/gkg023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Hoof A, Parker R. Curr Biol. 2002;12:R285–R287. doi: 10.1016/s0960-9822(02)00802-3. [DOI] [PubMed] [Google Scholar]
- 24.Xu N, Chen CY, Shyu AB. Mol Cell Biol. 1997;17:4611–4621. doi: 10.1128/mcb.17.8.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen CY, Shyu AB. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 26.Chen CY, Xu N, Shyu AB. Mol Cell Biol. 1995;15:5777–5788. doi: 10.1128/mcb.15.10.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng SS, Chen CY, Shyu AB. Mol Cell Biol. 1996;16:1490–1499. doi: 10.1128/mcb.16.4.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen CY, Xu N, Shyu AB. Mol Cell Biol. 2002;22:7268–7278. doi: 10.1128/MCB.22.20.7268-7278.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shim J, Lim H, Yates JR, Karin M. Mol Cell. 2002;10:1331–1344. doi: 10.1016/s1097-2765(02)00730-x. [DOI] [PubMed] [Google Scholar]
- 30.Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. J Biol Chem. 2000;275:17827–17837. doi: 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- 32.Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY. Mol Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 33.DeMaria CT, Brewer G. J Biol Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 34.Sarkar B, Xi Q, He C, Schneider RJ. Mol Cell Biol. 2003;23:6685–6693. doi: 10.1128/MCB.23.18.6685-6693.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 36.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Estevez AM, Kempf T, Clayton C. EMBO J. 2001;20:3831–3839. doi: 10.1093/emboj/20.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehner B, Sanderson CM. Genome Res. 2004;14:1315–1323. doi: 10.1101/gr.2122004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 40.Raijmakers R, Schilders G, Pruijn GJ. Eur J Cell Biol. 2004;83:175–183. doi: 10.1078/0171-9335-00385. [DOI] [PubMed] [Google Scholar]
- 41.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- 43.Torchet C, Bousquet-Antonelli C, Milligan L, Thompson E, Kufel J, Tollervey D. Mol Cell. 2002;9:1285–1296. doi: 10.1016/s1097-2765(02)00544-0. [DOI] [PubMed] [Google Scholar]
- 44.Allmang C, Mitchell P, Petfalski E, Tollervey D. Nucleic Acids Res. 2000;28:1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bousquet-Antonelli C, Presutti C, Tollervey D. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- 46.Phillips S, Butler JS. Rna. 2003;9:1098–1107. doi: 10.1261/rna.5560903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gatfield D, Izaurralde E. Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- 48.Haile S, Estevez AM, Clayton C. RNA. 2003;9:1491–1501. doi: 10.1261/rna.5940703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hilleren PJ, Parker R. Mol Cell. 2003;12:1453–1465. doi: 10.1016/s1097-2765(03)00488-x. [DOI] [PubMed] [Google Scholar]
- 50.Lejeune F, Li X, Maquat LE. Mol Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- 51.Mitchell P, Tollervey D. Mol Cell. 2003;11:1405–1413. doi: 10.1016/s1097-2765(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 52.Mukherjee D, Gao M, O'Connor JP, Raijmakers R, Pruijn G, Lutz CS, Wilusz J. EMBO J. 2002;21:165–174. doi: 10.1093/emboj/21.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi S, Araki Y, Sakuno T, Katada T. EMBO J. 2003;22:3951–3959. doi: 10.1093/emboj/cdg374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tran H, Schilling M, Wirbelauer C, Hess D, Nagamine Y. Mol Cell. 2004;13:101–111. doi: 10.1016/s1097-2765(03)00481-7. [DOI] [PubMed] [Google Scholar]
- 56.Aloy P, Ciccarelli FD, Leutwein C, Gavin AC, Superti-Furga G, Bork P, Bottcher B, Russell RB. EMBO Rep. 2002;3:628–635. doi: 10.1093/embo-reports/kvf135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mitchell P, Tollervey D. Nat Struct Biol. 2000;7:843–846. doi: 10.1038/82817. [DOI] [PubMed] [Google Scholar]
- 58.Raijmakers R, Egberts WV, van Venrooij WJ, Pruijn GJ. J Mol Biol. 2002;323:653–663. doi: 10.1016/s0022-2836(02)00947-6. [DOI] [PubMed] [Google Scholar]
- 59.Symmons MF, Williams MG, Luisi BF, Jones GH, Carpousis AJ. Trends Biochem Sci. 2002;27:11–18. doi: 10.1016/s0968-0004(01)01999-5. [DOI] [PubMed] [Google Scholar]
- 60.Gao G, Guo X, Goff SP. Science. 2002;297:1703–1706. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 61.Bick MJ, Carroll JW, Gao G, Goff SP, Rice CM, MacDonald MR. J Virol. 2003;77:11555–11562. doi: 10.1128/JVI.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo X, Carroll JW, Macdonald MR, Goff SP, Gao G. J Virol. 2004;78:12781–12787. doi: 10.1128/JVI.78.23.12781-12787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Nat Struct Mol Biol. 2004;11:257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 64.Lai WS, Kennington EA, Blackshear PJ. J Biol Chem. 2002;277:9606–9613. doi: 10.1074/jbc.M110395200. [DOI] [PubMed] [Google Scholar]
- 65.Lai WS, Kennington EA, Blackshear PJ. Mol Cell Biol. 2003;23:3798–3812. doi: 10.1128/MCB.23.11.3798-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu L, Chen G, Ji X, Gao G. Biochem Biophys Res Commun. 2004;321:517–523. doi: 10.1016/j.bbrc.2004.06.174. [DOI] [PubMed] [Google Scholar]
- 67.Bharucha AD, Ven Murthy MR. Methods Enzymol. 1992;216:168–179. doi: 10.1016/0076-6879(92)16020-k. [DOI] [PubMed] [Google Scholar]
- 68.Brengues M, Teixeira D, Parker R. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ven Murthy MR, Bharucha AD, Charbonneau R. Nucleic Acids Res. 1986;14:6337. doi: 10.1093/nar/14.15.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Calzone FJ, Angerer RC, Gorovsky MA. Nucleic Acids Res. 1982;10:2145–2161. doi: 10.1093/nar/10.6.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 72.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 73.Ryman KD, Meier KC, Nangle EM, Ragsdale SL, Korneeva NL, Rhoads RE, MacDonald MR, Klimstra WB. J Virol. 2005;79:1487–1499. doi: 10.1128/JVI.79.3.1487-1499.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Z, Kiledjian M. Cell. 2001;107:751–762. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]