Abstract
Smooth muscle α-actin gene activity appears in promyocardial cells well before cardiac myocyte differentiation and is down-regulated during the onset of rhythmic contractility and cardiac morphogenesis. The levels of LIM-only CRP2 correlated well with smooth muscle gene activity. Cardiomyocyte-specific expression of CRP2 in transgenic mice showed robust expression of smooth muscle cell-specific transcripts and protein filaments in the adult heart. Protein transduction of a recombinant CRP2 protein, fused to the protein transduction domain of HIV, into neonatal heart cells induced de novo synthesis of smooth muscle cell-specific transcripts and proteins. The LIM zinc fingers in CRP2 were found to collaborate with Brg1 of the SNF/SWI complexes, recruited serum response factor, and remodeled smooth muscle target gene chromatin through histone acetylation. CRP2 may have a cytoskeletal role, but as a nuclear protein, CRP2 acted as a potent transcription coadaptor that remodeled silent cardiac myocyte chromatin and directed serum response factor-dependent smooth muscle gene activity.
Keywords: chromatin remodeling, smooth muscle cell differentiation
Smooth muscle α-actin (SMA), a member of an actin multigene family, is selectively activated in the heart-forming field (HFR) in avian embryos (1). The HFR represents the precontractile developmental stage that marks a primitive mesodermal state, which will generate definitive cardiac cell types. The earliest cell types are the noncontractile or promyocardial cardiomyoblast within the cardiac lineage before cardiomyocyte differentiation (defined by organized myofibrils and rhythmic contractions) identified several years ago in early avian embryos (2). De novo activation of smooth muscle genes in cardiomyoblasts, coincided well with the appearance of LIM-only proteins CRP1 and CRP2 (3). CRP1 and CRP2 act as coadaptor molecules that tether cardiovascular enriched nuclear factors, SRF, and GATA4/5/6 to confer strong activation of smooth muscle cell (SMC)-specific genes (3). Thus, SMA appears within a significant time interval between the stages of precontractile cardiomyoblast and cardiomyocyte differentiation (1) and represented the first demonstration of coexpression of both smooth muscle and striated α-actin transcripts and proteins within the same myogenic cells, well before the events of looping and maturation to the multichambered heart (4, 5). Myocardin, another powerful regulator of smooth muscle gene activity that also appears during early cardiovascular development (6, 7) was shown not to be obligatory for the appearance of smooth muscle-specified genes in embryonic stem cells (8). In addition, during the onset of rhythmic contractility and formation of cardiac chambers, even in the presence of myocardin, smooth muscle gene activity was switched off in the myocardium, correlating well with the down-regulation of both CRP1 and CRP2 in the heart (3–5).
CRP2 may function in the assembly of multiprotein DNA-binding complexes that collectively mediate SMC-specific gene transcription. Conversely, CRPs may also play a role in the assembly and maintenance of a SMC cytoskeleton involved in cell-substrate adhesion and contractile responses to vasoactive stimuli (9). In addition, a cytoarchitectural role of CRP2 may be related to the strong binding of the N-terminal LIM domain to cytoskeleton-associated proteins such as α-actinin and zyxin (10). Because CRP2 cycles in and out of the nucleus, is it possible that CRP2 is sequestered on the cytoskeleton to simply repress its nuclear regulatory role? Or does CRP2 have another regulatory agenda to foster smooth gene activity by signaling through its association with the cytoskeleton? We tested the idea that CRP2 largely controls the expression of smooth muscle gene activity in the heart and that one reason smooth muscle contractile protein gene activity was vastly reduced might be the diminished levels of a potent SMC differentiation cofactor, such as CRP2. If CRP2 protein is reintroduced back to heart cells, will exogenous synthesis of SMC fibers occur? This experiment would test the notion of the reversibility of smooth muscle gene activity in terminally differentiated cardiac myocytes.
Results
Up-Regulation of Smooth Muscle Target Genes in the Adult Heart of α MHC-CRP2 Transgenic Mice.
Does CRP2 expression in adult heart of transgenic mice lead to exogenous reexpression of SMC contractile proteins? FLAG epitope-tagged rat CRP2 cDNA transgene was driven by the α-myosin heavy-chain (α MHC) promoter in adult hearts (Fig. 1A). Three founder lines with the highest copy number of transgenes were chosen to establish subsequent colonies. Mice were screened by Southern blot analysis and confirmed by detection of both mRNA (RT-PCR data not shown) and protein products (Fig. 1B) of the transgene in the hearts of positive animals. FLAG-CRP2 appeared specifically in the transgenic but not in the nontransgenic hearts. SMC-specific contractile filament proteins such as SM-MHC, calponin, SM γ-actin, SMA, SM22α, and caldesmon were up-regulated in the transgenic hearts, whereas cardiac-specific markers (cardiac troponin T and sarcomeric α-actin) and transcription factors (SRF, GATA6, and Nkx2–5) were not affected. CRP1 proteins in the hearts remained at basal level in transgenic mice, even though RT-PCR analysis indicated a slight increase at the mRNA level (data not shown). CRP2 is normally absent in nontransgenic adult myocardium. On the other hand, CRP3/MLP is augmented in transgenic hearts, a possible compensatory response to CRP2 expression. RT-PCR analysis revealed no other alteration in marker genes for cardiac hypertrophy (β-myosin heavy chain, skeletal α-actin, and atrial natriuretic factor), calcium-handling (SERCA2, phospholamban) transcription factors (myocardin, MEF2C, GATA4, and dHAND), or immediate-early gene, c-Fos (data not shown). Thus, an in vivo mouse model demonstrated induction of SMC phenotype when CRP2 was overexpressed in cardiomyocytes and, as shown [see supporting information (SI) Figs. 1–3], caused mild cardiac hypertrophy with no signs of cardiomyopathy.
Fig. 1.
Cardiac-specific overexpression of CRP2 in α MHC-CRP2 transgenic mice results in stable production of SMC filaments. (A) DNA construct used to generate mice expressing FLAG-tagged rat CRP2 protein in the adult cardiomyocytes. Genotyping was performed by Southern blot of mouse tail genomic DNA using probe (≈1 kb) specific to the transgene confirmed by RT-PCR analysis using primers amplifying a 472-bp fragment starting from the FLAG epitope tag. To profile the effects of CRP2 misexpression in the heart, we examined the expression level of several SMC and cardiac marker genes. Protein lysates and total RNA were collected from the ventricular portion of the hearts from three pairs of transgenic and nontransgenic littermates and subsequently analyzed by Western immunoblot analysis. Equal loadings of protein were shown by β-actin staining. (B) Western blot analysis revealed exogenous synthesis of SMC contractile proteins in adult hearts of α MHC-CRP2 mice. Cardiac proteins from three pairs of NTg and Tg littermates were harvested from the ventricular portions of the hearts. Equal loadings of whole-cell lysate (≈100 μg) were subjected to SDS/PAGE and transferred to PVDF membrane. Immunoblots were carried out by using protein-specific antibodies. Small intestine and soleus muscle were used as positive controls for smooth (SM) and striated (SK) muscle, respectively.
In Vivo Evidence of CRP2 Interactions with Chromatin SWI/SNF Cofactors.
Recent reports have indicated that Brg1 and Brm ATPases of the SWI/SNF chromatin remodeling complexes, through selective association with certain classes of transcription factors, play a critical role in cellular differentiation (11, 12). In particular, Brg1 SWI/SNF complexes coactivated β-globin transcription by zinc finger-containing factors such as EKLF and GATA1 (11). We hypothesized that the LIM modules of CRP2 could interact with chromatin remodeling complexes. We found that CRP2, as well as zinc-finger GATA4, were coimmunoprecipitated with endogenous Brg1, but not with Brm, complexes in cardiomyocytes (Fig. 2A). In contrast, MADS-box protein SRF and homeodomain factor Nkx2–5 did not associate with Brg1 or Brm complexes. We explored further to uncover the specificity of Brg1-containing SWI/SNF for CRP2. Various V5-tagged CRP2 deletion constructs were transfected into C2C12 mouse myoblasts. We found that the second zinc finger of the N-terminal LIM domain of CRP2 was required for in vivo interaction with Brg1 (Fig. 2B) and was sufficient to activate smooth muscle genes (3) In addition, the deletion of the entire N-terminal LIM domain in CRP2ΔN effectively shut down vascular smooth muscle differentiation and acted as a powerful dominant-negative mutant (3).
Fig. 2.
CRP2 coassociated with nuclear transcription factors and chromatin remodeling complexes. Nuclear extracts were harvested from adult heart cells of α MHC-CRP2 transgenic mice overexpressing CRP2 in the cardiomyocytes. (A) Pulldown analysis of protein interactions between recombinant CRP2, SRF, GATA4, or NKx2.5 and nuclear extracts from α MHC-CRP2 cardiomyocytes. Bound proteins (lanes 2–7) and 10% of input nuclear extracts (lane 1) were resolved on 4–20% SDS/PAGE gels and immunoblotted with antibodies against Brg1 (Upper) or Brm (Lower) of SWI/SNF remodeling complexes. (B) The second zinc finger of the CRP2 LIM domain is necessary for in vivo coassociation with Brg1. Nuclear lysates of C2C12 cells cotransfected with V5-tagged wild type or deletion mutants of CRP2 were subjected to coimmunoprecipitation assays using either anti-V5 or anti-Brg1 antibody. Immune complexes were separated by 4–20% SDS/PAGE gels and immunoblotted with antibodies against V5 or Brg1. Summary of Brg1 interactions and the schematic representation of CRP2 constructs were shown on the right.
CRP2 Bound to the Endogenous cis-Elements of Up-Regulated SMC Target Genes.
ChIP analyses were performed to examine the protein occupancies of a variety of tissue-specific target genes in heart cells harvested from either the α MHC-CRP2 transgenic mice or from the nontransgenic littermates (Fig. 3). Anti-FLAG-CRP2 (Fig. 3, lane 1) and anti-SRF (Fig. 3, lane 2) antibodies specifically pulled out the 5′ chromatin regions of up-regulated SMC genes (SM-MHC, calponin, and SM γ-actin) embedded with multiple SRF-binding CArG elements in the transgenic, but not the nontransgenic, hearts. SRF also occupied the promoter regions, laden with multiple CArG boxes, of two activated cardiac-specific genes (cardiac α-actin and α MHC) in the hearts of both the transgenic and the nontransgenic mice. Because CRP2 does not bind to DNA directly, its occupancy on the enhancer areas of the SM-MHC, calponin, SM γ-actin, and even cardiac α-actin genes is possibly through SRF. Regions upstream of the cardiac troponin C gene contains GATA but no CArG-binding elements, and was amplified from the ChIP assay by using anti-GATA6 antibody (Fig. 3, lane 3). The CRP2 transgene facilitated SMC genes (SM-MHC, calponin, and SM γ-actin) chromatin remodeling by the binding of Brg1 (Fig. 3, lane 4) plus Ini1 (Fig. 3, lane 6) and the acetylation of histone H3 (Fig. 3, lane 9) and histone H4 (Fig. 3, lane 10), presumably because of the recruitment of p300 (Fig. 3, lane 8) and the loss of HDAC1 (Fig. 3, lane 7). Although the association of CRP2 on the cardiac α-actin promoter appeared to increase bound SWI/SNF complexes (Fig. 3, lanes 4 and 6), the inactive adult β-globin gene was not associated with the cardiovascular transcription factors, the chromatin remodeling subunits, acetylated histones, p300 coactivator/HAT, or even the corepressor HDAC1.
Fig. 3.
Promoter-specific recruitment of CRP2, SWI/SNF complexes, and chromatin modifiers in transgenic cardiomyocytes. ChIP analyses of transcriptional activators (CRP2 and SRF; lanes 1 and 2), SWI/SNF remodeling complexes (Brg1, Brm, and core subunit Ini1; lanes 3–5), chromatin coactivators/corepressors (HDAC1 and p300; lanes 6 and 7), and histone modifications (Ac-H3 and AcH4; lanes 8 and 9) were performed by using a variety of promoters (schematic on the left), which are either exogenously up-regulated (SM-MHC, calponin, and SM γ-actin), constitutively activated (Ca α-actin, and Ca α MHC), or permanently inactivated (β-globin) in the heart cells. Briefly, cardiomyocytes from α MHC-CRP2 transgenic (Tg) or nontransgenic (NTg) littermate mice were harvested and treated with formaldehyde to cross-link DNA. DNA recovered from immunoprecipitated samples by using specific antibodies (lanes 1–9) or no-antibody negative control (10) was subjected to PCR amplification, employing primers directed against various promoters. Amplification products contain the regulatory sequence elements (CArG boxes, filled ovals or GATA-binding sites) were shown schematically in the top box. Genomic DNA input (10%, lane 11, open oval) was included as a positive control.
Cycling of CRP2 to The Nucleus Was Essential for Transcription Activation.
To distinguish between the nuclear or the cytoplasmic roles of CRPs, we modified CRP2 (Fig. 4A) to contain SV40 T antigen nuclear localization signal, amino acid 26-33 PKKKRKVE (13), PKI nuclear export signal, amino acid 37-46 LALKLAGLDI (14), Mxi1 strong repression domain SR, amino acid 1-69 (15), or mutated SRpm domain that is defective in repression (16). In CV1 fibroblasts, overexpressed wild-type CRP2 localized in both the nucleus and the cytoplasmic compartments, similar to the subcellular distribution pattern of endogenous CRP2 (2). CRP2-NLS was present in ≈90% of the fibroblast nuclei, whereas CRP2-NES was localized in the cytoplasm of ≈93% of the transfected cells (Fig. 4A). In cotransfection assays with SRF and GATA6, expressions of CRP2-NLS stimulated SMA promoter reporter activity by 39% (Fig. 4B, lane 11), whereas the presence of CRP2-NES reduced reporter activity by 37% (Fig. 4B, lane 12). To examine whether CRP2's nuclear occupancy alone is directly responsible for gene transactivation, we used SR-CRP2. The addition of the SR domain, which recruits mSin3A corepressor complex that include N-CoR and HDAC1 (16), to CRP2 inhibited CRP2-dependent transcription by 75% in a dominant-negative fashion (Fig. 4B, lane 13). In contrast, the defective inhibitor SRpm-CRP2 served as a control and did not block SMA promoter activity (Fig. 4B, lane 14). Together, these experiments demonstrate unequivocally CRP2's nuclear role in transcriptional regulation.
Fig. 4.
Intracellular localization of CRP2 regulates gene transactivation. (A) A summary showing schematic view of wild-type CRP2 and four fusion protein expression constructs and their cellular localization and transfection efficiency. Indirect immunofluorescent staining of CV1 cells overexpressing V5-tagged proteins (green). NLS, SV40 nuclear localization signal; NES, PKI nuclear export signal; SR, Mxi1 strong repression domain; SRpm, Mxi1 strong repressor domain point mutant. (B) Effects of regulated cellular redistribution of CRP2 on SMA gene activation. CV1 cells were cotransfected with SMA-luciferase reporter and expression vectors encoding SRF, GATA6, and various CRP2 constructs, as indicated. Values are expressed as fold-activation increases in luciferase activity ± SEM compared with the level of activity with empty expression vector alone (lane 1).
CRP2 Directed Production of SMC-Specific Filaments in Heart Cells.
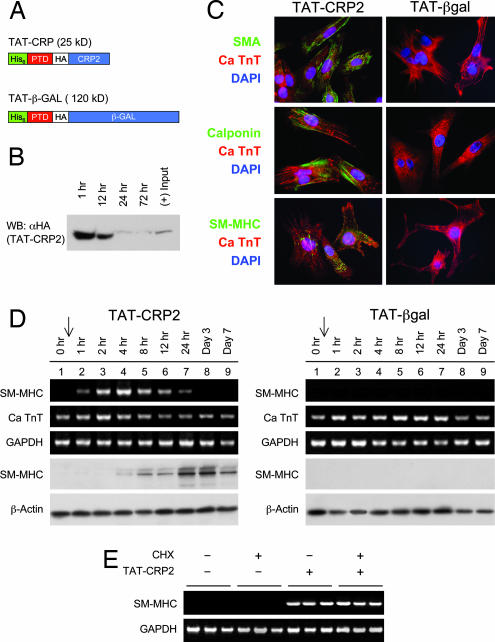
We next turned to tissue culture studies to gain further understanding at the mechanistic level of this differentiation event. Because primary tissue culture cells such as heart cells are impervious to DNA transfection, a recombinant CRP2 protein, fused to the protein transduction domain (PTD) from the HIV TAT protein (Fig. 2A). Immunoprotein blots revealed protein transduction of TAT-CRP2 into cardiomyocytes within 1 h (Fig. 2B). Even though the transduced proteins were degraded within 24 h, TAT-CRP2, but not the control TAT-β gal, induced up-regulation of SMC contractile proteins, including SMA, calponin, and SM-MHC, in neonatal rat cardiac myocytes harvested 3 d after treatment (Fig. 2C). A time course showed the acute and potent induction of SM-MHC in response to TAT-CRP2 treatment (Fig. 2D). Although SM-MHC mRNA level leveled off after 24 h, coincident with TAT-CRP2 depletion in cardiomyocytes, SM-MHC protein was stably maintained up to 7 d, possibly because of slow turnover rate of the thick filament. Experiments performed in the presence of the protein synthesis inhibitor cyclohexamide showed that the rapid effects obtained for SM-MHC transcripts occurred independently of de novo protein synthesis (Fig. 2E). Thus, in cardiac cells that already express SRF and GATA4, CRP2 alone can immediately initiate the SMC gene program.
Discussion
Sequential activation of SMA to α-striated actins in the embryonic heart (4) also occurs during somitogenesis (1) and the transdifferentiation of esophageal cells (17) and may be a general property of primitive muscle mesoderm undergoing a transition from the earliest stages of myogenic commitment in a cardiomyoblast toward terminal differentiation of a cardiac myocyte. Elevated levels of CRP2. GATA4/6 and SRF may be obligatory for activating smooth muscle gene activity during the biogenesis of the cardiovascular system. We learned from knockouts of SMA and cardiac α-actin that there is greater flexibility in genetic reprogramming in smooth and cardiac muscles than previously imagined (18, 19), and here, we showed that this process is reversible with the sole addition of CRP2 in the adult heart. This study may focus attention toward how we define cardiogenic cell types in the future.
How can CRP2, which does not bind to DNA directly, stimulate the smooth muscle differentiation program, even in adult heart cells? Our results indicate that, when inside the nucleus, CRP2 is found complexed with DNA-binding transcription activators and chromatin-modifying factors and drives immediate and lasting smooth muscle phenotypic switch (Figs. 2, 3, and 5). Several two-LIM-only proteins are also known to act as bridging molecules that bring together DNA-binding factors important in cell lineage specification (3, 20–22). CRP2 physically associates with other cardiovascular lineage regulators, such as SRF and GATA proteins, to synergistically activate the transcription of SMC target genes. Moreover, we discovered a property of CRP2's LIM domain in directing chromatin modification that might generate a DNA template more accessible to the general transcription machinery. Two major classes of complexes, ATP-dependent SWI/SNF remodeling complexes and histone-modifying HAT or HDAC complexes, regulate accessibility of the nucleosomal DNA to binding factors (23). CRP2 interacted with Brg1-containing SWI/SNF chromatin remodeling complexes (Fig. 2 A and B). The SWI/SNF complexes, which use ATP hydrolysis to expose nucleosomal DNA sequences to transfactors, are targeted to activated SMC gene loci in the transgenic cardiac myocytes (Fig. 3). In addition, using specific antibodies to acetylated histones, ChIP studies revealed that the increased acetylation in proximal promoter regions of up-regulated SMC genes correlated with recruitment of histone acetyltransferase p300 and loss of histone deacetylase HDAC1 (Fig. 3). The p300 family of proteins is known to mediate diverse enhancer-binding factors with components of the basal transcriptional apparatus (24, 25). It remains to be proven whether p300 binds to CRP2 directly or indirectly through SRF (26) or GATA4/6 (27) partners of the transcriptosome complexes. In summary, CRP2 exert its potent differentiation role through a combinatorial interaction involving DNA-binding transactivators, chromatin remodeling, and histone modifiers.
Fig. 5.
Protein transduction of recombinant TAT-CRP2 into neonatal cardiomyocytes induces up-regulation of SMC genes. (A) Schematic representation of recombinant TAT fusion protein constructs. His-6, polyhistidine tag; PTD, protein transduction domain of HIV TAT protein; HA, epitope tag of influenza hemagglutinin protein. Recombinant TAT-PTD-CRP2 proteins (50 μg/ml) were added to tissue culture medium incubating freshly isolated neonatal cardiomyocytes. Protein transduction occurs in a rapid (<30 min), efficient (>97%), and concentration-dependent fashion (10–70 μg/ml). (B) Freshly isolated neonatal rat cardiomyocytes were plated overnight and then treated with recombinant TAT-CRP2 fusion protein. Whole-cell lysates were harvested at various time points, and Western blot analysis was performed to verify transduction of HA epitope-tagged TAT-CRP2. (C) Immnuofluorescent staining was carried out to detect coexpression of SMC marker genes (SMA, calponin, and SM-MHC) and cardiac-specific troponin T (TnT) in neonatal cardiomyocytes 3 d after TAT-CRP2 or TAT-β gal treatment. SMC contractile proteins were present in cardiomyocytes treated with TAT-CRP2 but not in the TAT-β gal control group. Indirect immunofluorescent staining was performed after 72 h of TAT fusion protein treatment. (D) The effect of recombinant TAT-CRP2 (Left) or TAT-β gal (Right) treatment on SM-MHC induction in cardiomyocyte assessed over time by semiquantitative RT-PCR (top three blots) and Western immunoblot analysis (bottom two blots). Arrow, time at which TAT-fusion proteins were added to the media. (E) The effect of cycloheximide (CHX) on TAT-CRP2-induced SM-MHC mRNA levels assessed by semiquantitative RT-PCR analysis. Cardiac myocytes were preincubated with cycloheximide (25 μg/ml) for 2 h before exposure to TAT proteins.
The subcellular location of CRP2 has been reported to be exclusively cytoplasmic, exclusively nuclear, or both cytoplasmic and nuclear, depending on the cell type and the experimental conditions. We propose a model suggesting that CRPs function as molecular sensors on the contractile filament. This function could be a feedback mechanism whereby the cytoskeletal status regulates CRPs' subcellular distribution and, hence, controls when cells proliferate or differentiate. For example, during the initial transition of proepicardial cells to coronary SMCs and in early skeletal myoblast differentiation, endogenous CRP2 and CRP3/MLP, respectively, concentrate in the nuclei of these progenitor cells (2, 28). Later, as the cells undergo maturation, these CRPs colocalize with actin filaments. In contrast, during the pathological progression of right ventricular hypertrophy, CRP3/MLP, which is normally anchored along the Z discs of contractile sarcomeres, accumulates in the nucleus of dedifferentiated cardiomyocytes associated with ultrastructural disorganization of the myofibrils (29). Because CRP2 does not contain an obvious motif resembling the leucine-rich nuclear export signal (NES) of other CRM1 cargo, CRP proteins might be exported through their association with other cycling proteins that contain an NES, such as unpolymerized actin. A highly related LIM-only protein, CRIP2/HLP, has been shown to be a substrate for a Ser/Thr kinase, the cGMP-dependent protein kinase I (30). The subcellular distribution of CRIP2/HLP is altered after its phosphorylation. It is possible, however, that the shuttling response of CRP2 to Rho-mediated signals may be a secondary response and due to the “piggy-back” of CRP2 on SRF or GATA factors that are targets for Rho-initiated nuclear translocation (31, 32).
Experimental Procedures
Cells and Reagents.
Neonatal ventricular myocytes from 1-d-old Sprague–Dawley rats were isolated and cultured as described (33) by using a Percoll step gradient followed by preplating for 1 h to enrich for cardiac myocytes and deplete nonmyocytes. Primary cardiac myocytes were maintained in DMEM/Ham's nutrient mixture F12 (1:1) containing 5% horse serum, 2 mM l-glutamine and 50 μg/ml gentamycin. Rabbit polyclonal anti-FLAG antibody and monoclonal antibodies for calponin (hCP), smooth muscle α-actin (SMA, 1A4), h-caldesmon (hHCD), sarcomeric α-actin (5C5), and β-actin (AC-15) were from Sigma (St. Louis, MO). Monoclonal anti-smooth muscle γ-actin (B4) antibody was from ICN (Aurora, OH). Monoclonal anti-smooth muscle MHC (G-4) and anti-HA-epitope (F-7) antibodies, goat polyclonal antibodies against cardiac troponin T (C-19), GATA6 (C-20), Nkx2–5 (A-16), Brg1 (N-15), Brm (C-20), Ini1 (N-20), HDAC1 (N-19), rabbit polyclonal antibodies specific for SRF (G-20), ID1 (C-20), α-actinin (H-300), α-tubulin (H-300), and p300 (N-15) were from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal V5-epitope antibody was from Invitrogen (Carlsbad, CA). Rabbit polyclonal anti-acetyl-histone H3 and anti-acetyl-histone H4 antibodies were from Upstate Biotechnology (Lake Placid, NY). Monoclonal anti-SM22α (1-B8) was a generous gift from Saverio Sartore (Padua, Italy). Rabbit polyclonal antibodies against CRP1, CRP2, and CRP3/MLP were generously provided by Mary Beckerle (Salt Lake City, UT). The horseradish peroxidase-conjugated secondary antibodies were from Santa Cruz Biotechnology. SuperSignal West Pico Chemiluminescent Substrate for Western blotting was from Pierce (Rockland, IL). The fluorescent Alexa Fluor-conjugated secondary antibodies, rhodamine-phalloidin, and DAPI nuclear acid stain were from Molecular Probes (Eugene, OR). Leptomycin B, cytochalasin D, and latrunculan B were from Sigma.
Construction of Plasmids and Transfection of Cells.
Luciferase reporter plasmids SMA-Luc and expression vectors pCGN-SRF, pCG-GATA6, pcDNA-CRP2-V5, pGEX-CRP2, pGEX-SRF, pMAL-Nkx2–5, and pMAL-GATA4 were reported in refs. 3, 34, and 35. Expression vectors pcDNA3.1-V5 (Invitrogen), pGEX-4T2 (Amersham, San Francisco, CA), and pMAL (New England Biolabs, Beverly, MA) were purchased commercially. CRP2 deletion mutants were constructed by PCR subcloning using pcDNA-CRP2-V5 as the template. Synthetic double-strand oligonucleotides specifying the amino acid positions amino acid 26-33 (PKKKRKVE) in SV40 T antigen and amino acid 37-46 (LALKLAGLDI) in PKI, respectively, were fused to the C terminus of pcDNA-CRP2-V5, generating the plasmids pcDNA-CRP2-NLS and pcDNA-CRP2-NES. Dominant-negative pcDNA-SR-CRP2-V5 and the control pcDNA-SRpm-CRP2-V5 mammalian expression vectors were constructed as described (16) by using pcDNA-CRP2-V5 as the backbone. Bacterial expression vectors pTAT-HA and pTAT-HA-β GAL were generous gifts of Steven Dowdy (La Jolla, CA). The TAT-CRP2 expression vector was constructed by subcloning a mouse full-length CRP2 cDNA fragment into the EcoRI and KpnI sites of the pTAT-HA bacterial expression vector (16). All constructs were verified by DNA sequencing. Transfection of plasmids into CV1 or C2C12 cells were performed by using Fugene 6 according to manufacturer's recommendation.
Animal and Gene Expression Profile.
A full-length rat CRP2 cDNA with a N-terminal FLAG-epitope tag was inserted between the 5.5-kb mouse α MHC promoter (36) and 0.7-kb polyadenylation sequence of human growth hormone cDNA. The α MHC-FLAG-rCRP2 cassette was released from the vector backbone by digestion with BamH1, purified, and injected into the pronuclei of FVB/N zygotes. The resulting mice were screened for the presence of the transgene in tail DNA by Southern blot analysis.
Western Blot and Immunofluorescent Staining.
Protein lysates were resolved by SDS/PAGE and transferred to PVD membrane, followed by immunoblotting by using SuperSignal West Pico Chemiluminescent reagent (Pierce). Myocytes were washed with PBS and then fixed with 4% formaldehyde, permeabilized with 0.5% Triton X-100, and blocked with 5% BSA in PBS for 1 h. Cells were incubated with primary antibodies [SMA 1A4, 1:400 (Sigma), calponin hCP, 1:400 (Sigma), SM-MHC, 1:200 (Santa Cruz Biotechnology), and cardiac troponin-T, 1:200 (Santa Cruz Biotechnology) for 2 h, followed by 1-h incubation with Alexa Fluor488-conjugated goat anti-mouse IgG (1:1,000; Molecular Probes) or Alexa Fluor594-conjugated donkey anti-goat IgG (1:1,000; Molecular Probes) and DAPI (1:1,000; Molecular Probes). Transduced cells were analyzed by immunofluorescence microscopy.
Protein Transduction.
Expression and purification of TAT-CRP2 and TAT-β GAL fusion proteins were based on methods reported in ref. 37. TAT-CRP2 fusion protein was expressed in BL21 bacteria by IPTG-induction (1 M), isolated by using a urea-denaturing protein-purification protocol, and affinity purified by using a nickel column. After the removal of the urea denaturant, the purified TAT-CRP2 fusion proteins were stored at −80°C and desalted by using PD-10 Desalting Columns (Amersham Pharmacia, Piscataway, NJ) before usage. Neonatal ventricular myocytes were treated with 50 μg/ml TAT-recombinant proteins in serum free media.
Recombinant Fusion Proteins.
GST-CRP2, GST-SRF, MBP-GATA4, MBP-Nkx2.5, control GST, and maltose-binding protein (MBP) were expressed in BL21 bacteria by 1 M IPTG induction and affinity purified by using either B-Per GST Fusion Protein Spin Purification kit (Pierce) or amylose resin (New England Biolabs). Pull-down assays were performed by incubating 500 μg of nuclear extracts from cardiac myocytes with 10 μg of purified GST-CRP2 or GST-SRF immobilized on glutathione-Sepharose beads (Amersham) or with 10 μg of purified MBP-Nkx2.5 or MBP-GATA4 immobilized on amylose resin (New England Biolabs). After extensive washing, the associated proteins were resolved on 4–20% SDS-polyacrylamide gel, followed by Western immunoblot analysis (anti-Brg1, 1:400; anti-Brm, 1:400).
ChIP Assay.
Cardiac myocytes were enriched as in co-IP experiments. Cells were treated with 1% formaldehyde and incubated for 15 min at room temperature. The fixed cardiomyocytes were harvested and prepared by following the protocol of the ChIP assay kit (Upstate Biotechnology). Cells were lysed and then sonicated to produce ≈400 bp of DNA-protein fragments. Samples were precleared with salmon sperm DNA/protein A agarose (rotating 1 h in the cold room). Ten micrograms of antibodies to the proteins FLAG-CRP2, SRF, Brg1, Brm, Ini1, HDAC1, p300, acetyl-H3, and acetyl-H4 were used to incubate with DNA extracts. No-antibody ChIP was carried out as a negative control. After extensive washes, immune complexes were eluted and reverse cross-linked by heating at 65°C for 4 h. One microliter of each sample was subjected to a PCR amplification. PCR analysis was performed by using primers from different regions 5′ of target genes SM-MHC, calponin, smooth muscle γ-actin, cardiac α-actin, cardiac α-MHC, cardiac troponin C, β-globin and will be provided on request. PCRs were carried out for 30 cycles at 58°C annealing temperature. Genomic DNA isolated from the starting material was used as the positive control PCR template. DNA products were resolved on 2% agarose gels.
Gene Reporter Assay.
A series of luciferase assays was done by transient transfection using Fugene 6 (3). Cotransfection experiments of CMV-driven expression vectors and SMA-Luc gene reporter were performed in duplicate and repeated three times. Luciferase activities were normalized to baseline reporter gene activity as fold-activation, with error bars representing SEM.
Supplementary Material
Acknowledgments
We thank Dr. Mark Majesky for helpful discussions. This work was supported by National Institutes of Health (NIH) Grant R01 HL50422 (to R.J.S.) and Department of Physiology NIH Postdoctoral Training Grant Fellowship HL7676 (to D.F.C.).
Abbreviations
- α MHC
α-myosin heavy chain
- SMA
smooth muscle α-actin
- SMC
smooth muscle cell.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0605635103/DC1.
References
- 1.Colas JF, Lawson A, Schoenwolf GC. Dev Dyn. 2000;218:316–330. doi: 10.1002/(SICI)1097-0177(200006)218:2<316::AID-DVDY6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 2.Gannon M, Bader D. Development (Cambridge, UK) 1995;121:2439–2450. doi: 10.1242/dev.121.8.2439. [DOI] [PubMed] [Google Scholar]
- 3.Chang DF, Belaguli NS, Iyer D, Roberts WB, Wu SP, Dong XR, Marx JG, Moore MS, Beckerle MC, Majesky MW, Schwartz RJ. Dev Cell. 2003;4:107–118. doi: 10.1016/s1534-5807(02)00396-9. [DOI] [PubMed] [Google Scholar]
- 4.Ruzicka DL, Schwartz RJ. J Cell Biol. 1988;107:2575–2586. doi: 10.1083/jcb.107.6.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugi Y, Lough J. Dev Dyn. 1992;193:116–124. doi: 10.1002/aja.1001930203. [DOI] [PubMed] [Google Scholar]
- 6.Li S, Wang DZ, Wang Z, Richardson JA, Olson EN. Proc Natl Acad Sci USA. 2003;100:9366–9370. doi: 10.1073/pnas.1233635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 8.Pipes GC, Sinha S, Qi X, Zhu CH, Gallardo TD, Shelton J, Creemers EE, Sutherland L, Richardson JA, Garry DJ, et al. Dev Biol. 2005;288:502–513. doi: 10.1016/j.ydbio.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 9.Faggin E, Puato M, Zardo L, Franch R, Millino C, Sarinella F, Pauletto P, Sartore S, Chiavegato A. Arterioscler Thromb Vasc Biol. 1999;19:1393–1404. doi: 10.1161/01.atv.19.6.1393. [DOI] [PubMed] [Google Scholar]
- 10.Pomies P, Louis HA, Beckerle MC. J Cell Biol. 1997;139:157–168. doi: 10.1083/jcb.139.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadam S, Emerson BM. Mol Cell. 2003;11:377–389. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 12.Kadam S, McAlpine GS, Phelan ML, Kingston RE, Jones KA, Emerson BM. Genes Dev. 2000;14:2441–2451. doi: 10.1101/gad.828000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodel MR, Corbett AH, Hodel AE. J Biol Chem. 2001;276:1317–1325. doi: 10.1074/jbc.M008522200. [DOI] [PubMed] [Google Scholar]
- 14.Elfgang C, Rosorius O, Hofer L, Jaksche H, Hauber J, Bevec D. Proc Natl Acad Sci USA. 1999;96:6229–6234. doi: 10.1073/pnas.96.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi AI, DePinho RA. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- 16.Jiang Y, Drysdale TA, Evans T. Dev Biol. 1999;216:57–71. doi: 10.1006/dbio.1999.9469. [DOI] [PubMed] [Google Scholar]
- 17.Patapoutian A, Wold BJ, Wagner RA. Science. 1995;270:1818–1821. doi: 10.1126/science.270.5243.1818. [DOI] [PubMed] [Google Scholar]
- 18.Schildmeyer LA, Braun R, Taffet G, Debiasi M, Burns AE, Bradley A, Schwartz RJ. FASEB J. 2000;14:2213–2220. doi: 10.1096/fj.99-0927com. [DOI] [PubMed] [Google Scholar]
- 19.Kumar A, Crawford K, Close L, Madison M, Lorenz J, Doetschman T, Pawlowski S, Duffy J, Neumann J, Robbins J, et al. Proc Natl Acad Sci USA. 1997;194:4406–4411. doi: 10.1073/pnas.94.9.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong Y, Flick MJ, Kudla AJ, Konieczny SF. Mol Cell Biol. 1997;17:4750–4760. doi: 10.1128/mcb.17.8.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mead PE, Deconinck AE, Huber TL, Orkin SH, Zon LI. Development (Cambridge, UK) 2001;128:2301–2308. doi: 10.1242/dev.128.12.2301. [DOI] [PubMed] [Google Scholar]
- 22.Wadman IA, Osada H, Grutz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narlikar GJ, Fan HY, Kingston RE. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 24.Abraham SE, Lobo S, Yaciuk P, Wang HG, Moran E. Oncogene. 1993;8:1639–1647. [PubMed] [Google Scholar]
- 25.Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, Livingston DM. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 26.Ramirez S, Ait-Si-Ali S, Robin P, Trouche D, Harel-Bellan A. J Biol Chem. 1997;272:31016–31021. doi: 10.1074/jbc.272.49.31016. [DOI] [PubMed] [Google Scholar]
- 27.Wada H, Hasegawa K, Morimoto T, Kakita T, Yanazume T, Sasayama S. J Biol Chem. 2000;275:25330–25335. doi: 10.1074/jbc.M000828200. [DOI] [PubMed] [Google Scholar]
- 28.Arber S, Halder G, Caroni P. Cell. 1994;79:221–231. doi: 10.1016/0092-8674(94)90192-9. [DOI] [PubMed] [Google Scholar]
- 29.Ecarnot-Laubriet A, De Luca K, Vandroux D, Moisant M, Bernard C, Assem M, Rochette L, Teyssier JR. J Mol Cell Cardiol. 2000;32:2385–2395. doi: 10.1006/jmcc.2000.1269. [DOI] [PubMed] [Google Scholar]
- 30.Huber A, Neuhuber WL, Klugbauer N, Ruth P, Allescher HD. J Biol Chem. 2000;275:5504–5511. doi: 10.1074/jbc.275.8.5504. [DOI] [PubMed] [Google Scholar]
- 31.Charron F, Tsimiklis G, Arcand M, Robitaille L, Liang Q, Molkentin JD, Meloche S, Nemer M. Genes Dev. 2001;15:2702–2719. doi: 10.1101/gad.915701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hill CS, Wynne J, Treisman R. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 33.Paradis P, MacLellan WR, Belaguli NS, Schwartz RJ, Schneider MD. J Biol Chem. 1996;271:10827–10833. doi: 10.1074/jbc.271.18.10827. [DOI] [PubMed] [Google Scholar]
- 34.Sepulveda JL, Belaguli N, Nigam V, Chen CY, Nemer M, Schwartz RJ. Mol Cell Biol. 1998;18:3405–3415. doi: 10.1128/mcb.18.6.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belaguli NS, Sepulveda JL, Nigam V, Charron F, Nemer M, Schwartz RJ. Mol Cell Biol. 2000;20:7550–7558. doi: 10.1128/mcb.20.20.7550-7558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gulick J, Subramaniam A, Neumann J, Robbins J. J Biol Chem. 1991;266:9180–9185. [PubMed] [Google Scholar]
- 37.Becker-Hapak M, McAllister SS, Dowdy SF. Methods. 2001;24:247–256. doi: 10.1006/meth.2001.1186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.