Abstract
The Balbiani body or mitochondrial cloud is a large distinctive organelle aggregate found in developing oocytes of many species, but its presence in the mouse has been controversial. Using confocal and electron microscopy, we report that a Balbiani body does arise in mouse neonatal germline cysts and oocytes of primordial follicles but disperses as follicles begin to grow. The mouse Balbiani body contains a core of Golgi elements surrounded by mitochondria and associated endoplasmic reticulum. Because of their stage specificity and perinuclear rather than spherical distribution, these clustered Balbiani body mitochondria may have been missed previously. The Balbiani body also contains Trailer hitch, a widely conserved member of a protein complex that associates with endoplasmic reticulum/Golgi-like vesicles and transports specific RNAs during Drosophila oogenesis. Our results provide evidence that mouse oocytes develop using molecular and developmental mechanisms widely conserved throughout the animal kingdom.
Keywords: germ plasm, gamete biology, oocyte development, ovary, oogenesis
Whether eggs and embryos of diverse species initiate development by highly divergent or basically similar pathways remains a key unresolved issue. One of the strongest arguments for divergent pathways has been the differing origins of germ cells in organisms such as flies, nematodes, and frogs that contain a definitive germ plasm and organisms such as mice and newts that do not (1). In Xenopus and Drosophila, germinal granule components are produced early in oogenesis, associate with a distinctive structure known as the Balbiani body, and are subsequently transported and localized within the egg (2). Shortly after fertilization, germinal granules are taken up by primordial germ cells where they persist (at least in part) as electron-dense particles called nuage. In contrast, none of these processes are thought to take place in mouse oocytes or germ cells. However, several nuage-containing structures have been described in male mouse germ cells, including the chromatoid body, chromatoid satellites, RNF17 granules, sponge bodies, and spherical particles (3–7). The function of these nuage-containing structures in male germ cells is unclear, in part because of the lack of cytological markers to distinguish one nuage body from another.
Molecular studies of germinal granules strongly implicate them in the transport, storage, localization, stability, and regulated utilization of mRNA (8). The Vasa DEAD box RNA helicase is found in germ cells and nuage of diverse organisms (9). Drosophila nuage contains Vasa, as well as Maelstrom, Aubergine, and SpindleE, proteins implicated in RNAi (10). Recently, transport particles used to assemble germinal granule components have been characterized in Drosophila and shown to contain conserved protein components, including me31B/Deadsouth (a DEAD box RNA helicase and translational repressor), Cup/4E-Transporter (4E-T, an eIF4E-binding protein), and Trailer hitch (11–13). These components also associate with the endoplasmic reticulum (ER) (14). Before localization within the egg, germinal granules associate with mitochondria in the Balbiani body (15, 16). The chromatoid body arises in spermatocytes also in conjunction with mitochondria (4) and Golgi (17). The mouse chromatoid body contains the me31B homolog, Ddx25, which is essential for male fertility and has been implicated in translational regulation (18). Other components of the chromatoid body include Vasa and Tudor proteins (19, 20)
The Balbiani body or mitochondrial cloud is a collection of organelles asymmetrically located adjacent to the nucleus in very young oocytes of diverse species (21–23). Balbiani bodies contain mitochondria, ER, and granulofibrillar material (GFM) organized in a characteristic manner. Although Balbiani body mitochondria often aggregate in a cloud around the Golgi, in some species, they extend throughout the perinuclear cytoplasm. The Balbiani body GFM morphologically resembles germinal granule precursors. A connection to germinal granules has been proven in Xenopus, where GFM has been shown to contain germinal granule proteins and RNAs that are incorporated into germ cells (15, 16, 24, 25). Likewise in Drosophila, a key component of the germinal granules, oskar RNA, associates with the newly formed Balbiani body (26).
Mouse oocytes have been thought to be exceptional in lacking a Balbiani body (1, 2). Early researchers reported that fetal and neonatal mouse oocytes contain a perinuclear Golgi body consisting of small vesicles similar to a Balbiani body (27, 28). However, subsequent studies failed to confirm these observations, and it is now widely accepted that mouse oocytes lack such mitochondrial aggregates (1, 2). This situation is surprising, because Balbiani bodies have been found in marsupials (29) and many mammalian species including goat (21), rat (30, 31), hamster (32), and humans (33–35), although the function of mammalian Balbiani bodies is unknown.
The lack of a Balbiani body in mouse oocytes is also hard to understand in light of similarities in the cellular events that precede Balbiani body formation in Drosophila, Xenopus, and mouse oogenesis. In both Xenopus and Drosophila, the Balbiani body arises within interconnected germline cysts by the controlled assembly of organelles and germinal granule components at the time of follicle formation (26, 36). Female mouse germ cells also generate cysts after reaching the genital ridge (37). As the cysts break down around the time of birth, approximately one-third become surrounded by somatic pregranulosa cells to form primordial follicles (38). At this time, mitochondria organize into aggregates that resemble the mitochondrial preclouds that form the Xenopus Balbiani body (38).
Consequently, we have reinvestigated the question of whether early mouse oocytes contain a Balbiani body by using cytological and molecular tools. We now report that female mouse germ cells do contain a Balbiani body similar to those in other mammalian and nonmammalian species. It consists of mitochondria and ER surrounding a distinctive Golgi aggregate. The mouse Balbiani body forms from previously described mitochondrial aggregates just before primordial follicle formation and persists briefly in young primordial follicles. In growing follicles, mitochondria and ER disperse, and a well defined Balbiani body is no longer found. We also present evidence that the molecular composition of the mouse Balbiani body resembles that of other species. In particular, it contains the mouse Trailer hitch protein, a conserved component of the Balbiani body and oocyte transport complexes in Drosophila and other organisms. Our studies support the view that mouse oocytes are not exceptional and develop using mechanisms that have been widely conserved in invertebrates and other vertebrate groups.
Results
Identification of a Murine Balbiani Body.
To look for a mouse Balbiani body, we focused on cyst-stage germ cells and on newly forming primordial follicles, the stages where Balbiani bodies can first be observed in Drosophila and Xenopus ovaries (23). Using electron microscopy, we observed that Golgi stacks are arranged in a distinctive circular manner in young mouse oocytes (Fig. 1A–D). Masses of mitochondria and closely associated ER either surround these Golgi stacks (Fig. 1A) or extend from it around the perinuclear region (Fig. 1B). Despite the limited amount of cytoplasm present in these oocytes, mitochondrial organization was nonrandom, with a strong preference for a perinuclear vs. a periplasma membrane position. Moreover, these mitochondria showed additional evidence of organization as many associate distinctively with thin sheets of ER during this time (38).
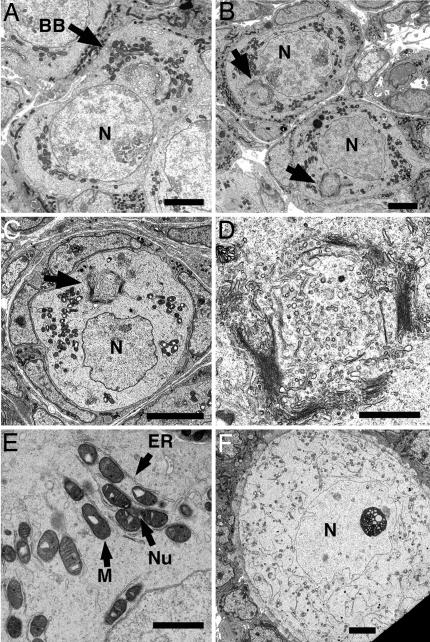
Fig. 1.
Electron micrographs of oocytes in neonatal ovaries. (A) Micrograph of an oocyte within a germline cyst from PND1 showing a well defined Balbiani body (arrow) with Golgi surrounded by mitochondria. (B) Micrograph of two oocytes from PND3 (top oocyte in a newly formed primordial follicle and bottom oocyte still within a germline cyst) with Balbiani bodies (arrows). (C) Micrograph of an oocyte within a primordial follicle at PND7 again showing a Balbiani body (arrow). (D) Higher-power view of Golgi from oocyte in C showing stacks of vesicles arranged in a circular structure. (E) Highly magnified image of mitochondria and associated ER with darkly stained potential nuage. (F) Oocyte with in primary follicle with mitochondria and ER evenly distributed throughout the cytoplasm. BB, Balbiani body; M, mitochondria; n, nucleus; Nu, nuage. (Scale bars: A–C, F, 5 μm; D, 1 μm; E, 0.5 μm.)
Thus, a Balbiani body similar in general structure to those described in other species is present in young mouse oocytes. Oocyte organelles first become organized into a discrete Balbiani body on about postnatal day (PND)1, both in oocytes that still remain in cysts and in oocytes that have already assembled into primordial follicles (Fig. 1 B and C). Darkly staining material is sometimes observed associated with mitochondria and resembles descriptions of nuage (Fig. 1E, arrow). This classic arrangement does not persist indefinitely, however. As follicles begin to grow, the mitochondria and ER disperse and become evenly distributed throughout the cytoplasm (Fig. 1F).
Golgi and Mitochondria Associate with the Balbiani Body.
We used antibodies specific for its component organelles to further characterize the mouse Balbiani body. Because Golgi cisternae lie near its center, we used an antibody against the Golgi localized protein, GM130, to analyze the distribution of Golgi both within the Balbiani body and throughout the oocyte cytoplasm at the level of confocal microscopy. These experiments strongly supported our conclusions based on electron microscopy (Fig. 2). GM130 staining was largely confined to a semicircular perinuclear structure in the oocytes of newly formed follicles (Fig. 2A) whose size, distinctive shape, and intracellular location corresponded to the Golgi portion of the Balbiani body (compare Fig. 2 C and D). Little or no GM130 reactivity was observed elsewhere in the cytoplasm at these stages. Antibodies to the mouse Vasa protein (39) stained the Golgi region at slightly lower levels than throughout the remainder of the cytoplasm (Fig. 2B)
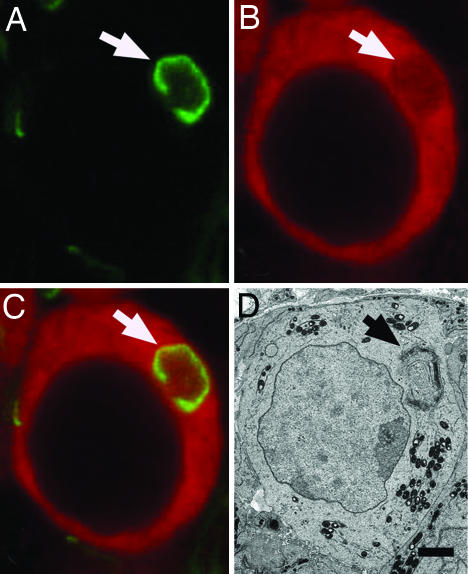
Fig. 2.
Balbiani body-associated Golgi vesicles in neonatal ovaries. (A–C) Confocal section of a PND1 oocyte labeled with a Golgi marker, GM130 (green) (A); Vasa, an oocyte marker (red) (B); and overlay (C) showing ring arrangement of Golgi (arrow). (D) Electron micrograph of Golgi at PND2 showing similar ring arrangement of Golgi (arrow). (Scale bar: 5 μm.)
In electron micrographs, mitochondria lie at the periphery of the Balbiani body, surrounding the Golgi core or extending from it around the nucleus. To further study mitochondrial organization, we labeled neonatal mouse ovaries for the mitochondrial protein cytochrome c and examined them using confocal microscopy. Labeled mitochondria were distributed in clusters in the cytoplasm of oocytes still in cysts and in some newly formed primordial follicles (Fig. 3A–C). Usually, the Golgi stack would lie at the center, with sheet-like aggregates of mitochondria extending away from it within the perinuclear cytoplasm. Strikingly, the mitochondria are excluded from the Golgi-rich region of the Balbiani body, so this region appears as a gap in the mitochondria aggregate. As expected from electron micrographs, in growing follicles mitochondria are evenly distributed throughout the cytoplasm (Fig. 3 D–F).
Fig. 3.
Mitochondria in neonatal oocytes. (A–C) Confocal section of PND3 oocytes becoming assembled into primordial follicles labeled with an antibody against cytochrome c (green) to visualize mitochondria (A), Vasa (red) to visualize oocytes (B), and overlay (C) showing Balbiani body-associated mitochondria (arrow). (D–F) Confocal section of a PND4 oocyte within a growing primary follicle labeled with an antibody against cytochrome c (green) to visualize mitochondria (D), Vasa (red) to visualize oocyte (E), and overlay (F) showing mitochondria evenly distributed in the oocyte cytoplasm. (Scale bars: A–C, 5 μm; D–F, 10 μm.)
Identification of an Antibody That Recognizes Trailer Hitch.
The presence of a Balbiani body in diverse species of young oocytes suggests that it is associated with a conserved function. The strongest candidate for such a role is in the transport and localization of organelles and RNAs within oocytes (23). Consequently, we examined whether conserved proteins known to function in these processes within oocytes of other species are present in the mouse Balbiani body. Recently, specific ribonucleoprotein (RNP) complexes have been characterized that are required to transport and localize oskar RNA, the key germinal granule component in Drosophila (11–13, 40). One component of these complexes is Trailer hitch (11, 14, 40), which is thought to directly interact with other components of the RNP complex, including Me31B and Cup. The trailer hitch (tral) gene is highly conserved in eukaryotes with the highest homology in two regions, the Sm and FDF domains (Fig. 4A) (41, 42). Sm domains are found in proteins involved in RNA metabolism such as splicing (43). FDF domains are found in a family of proteins involved in regulation of mRNA decay (42).
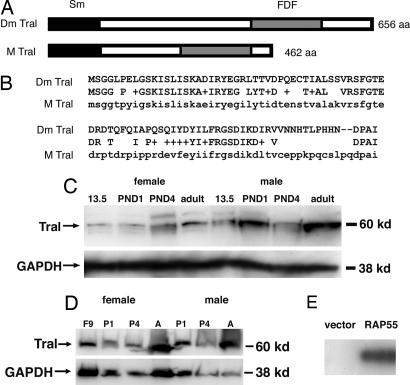
Fig. 4.
Conservation of the Balbiani body-associated protein, Tral. (A) Schematic representation of Drosophila and mouse Trailer hitch proteins showing the conserved Sm-like and FDF domains. (B) Comparison of amino acid sequence of the Sm-like domain from Drosophila and mouse. (C) Western blotting analysis using an antibody raised against Drosophila Trailer hitch. Mouse tissue extracts from 13.5 dpc, PND1, PND4, and PND42 ovaries and testes were probed with the Drosophila Tral antibody. An ≈60-kDa band was detected in all samples. Blots were reprobed with GAPDH (38 kDa) as a loading control. (D) Western blotting analysis of mouse tissue extracts from 9 dpc (F9), PND1 (P1), PND4 (P4), and adult (A) animals using an antibody raised against human RAP55. A band of ≈60 kDa was detected in all samples. Blots were reprobed with GAPDH (38 kDa) as a loading control. (E) Western blotting analysis of bacterially expressed RAP55. Extracts from bacteria with control pGEX4 plasmid (vector) or a plasmid expressing RAP55 were probed with Drosophila Trailer hitch antibody.
In addition to its role in oocytes, Trailer hitch is likely to be involved in RNA localization in other cells types and for general cellular functions such as ER exit-site formation, which is postulated to involve RNA localization (14, 44, 45). The mouse genome contains a single trailer hitch gene, but its function has not yet been characterized (46). However, the human Trailer hitch protein, RAP55, localizes to processing bodies (P bodies) (47). P bodies are cytoplasmic structures involved in mRNA degradation that have been described in both yeast and human cells (48–50). siRNA knockdown of RAP55 results in the loss of P bodies and suggests that RAP55 plays a role in mRNA degradation by promoting assembly of P bodies or by delivering mRNAs to P bodies (47). The mammalian homologue of cup, 4E-T, also localizes to P bodies and siRNA knockdown of 4E-T results in loss of P bodies and decreases mRNA stability (51). In addition, the mouse 4E-T, Clast4, is localized to the cytoplasm of developing oocytes and may play a role in mRNA degradation during female germ-cell development (52).
Drosophila and Mouse Tral proteins are 59% identical and 74% similar within in their N-terminal Sm-like domain (Fig. 4 A and B). To develop a specific antibody that recognizes mouse Trailer hitch, we investigated whether an antibody generated against the Drosophila Tral Sm domain would recognize mouse Tral (14). By Western blot, a band with a nominal molecular weight of ≈60 kDa was detected in extracts prepared from mouse ovaries and testes (Fig. 4C). This is slightly larger than the predicted size of 50 kDa suggesting posttranslational modification (46). We also tested an antibody generated against the human Trailer hitch protein, RAP55 (47) and detected a band of ≈60 kDa (Fig. 4D). We also expressed RAP55 in bacteria and found that it was detected by using the Drosophila Tral antibody (Fig. 4E). Thus, the Drosophila Tral antibody recognizes mammalian Tral protein.
Trailer Hitch Protein Is Associated with the Drosophila Balbiani Body.
We first examined whether Tral protein is enriched in the Drosophila Balbiani body. Using immunofluorescence and confocal microscopy, we observed that the anti-Tral antibody labels organelle clusters in late Drosophila cysts and the large anterior Balbiani body that is present in newly forming follicles [supporting information (SI) Fig. 6 A–C]. Trailer hitch protein distribution becomes perinuclear and on the nuclear envelope in the nurse cells throughout oogenesis and is localized within the oocyte to the posterior pole (SI Fig. 6 D–F).
Trailer Hitch Protein Is Associated with the Mouse Balbiani Body but Not the Chromatoid Body.
The expression and localization of Tral during the early stages of mouse oogenesis were very similar to its expression in Drosophila ovaries. Using whole-mount immunocytochemistry in developing embryonic and neonatal gonads, Tral was not detected at 13.5 days postcoitum (dpc) (data not shown). However, it is detected at 14.5 dpc in developing ovaries (Fig. 5A). At this time, there is a low level of Tral in all cells of ovaries, but expression appears stronger in the germ cells. Tral becomes progressively stronger in the cytoplasm of oocytes over the next several days, whereas expression in somatic cells becomes weaker (Fig. 5B). In addition, Tral is highly enriched in a circular structure in the cytoplasm reminiscent of the Golgi. To verify that Tral is localized in mouse oocytes within the Balbiani body-associated Golgi, we double-labeled ovaries with antibodies specific for GM130 and Tral. PND1 ovaries were exposed to both GM130 and Tral antibodies, and GM130 and Tral were detected in the same circular structure (Fig. 5C). To confirm that the antibody generated against Drosophila Trailer hitch recognized the mouse Tral protein, PND1 ovaries were labeled with the Drosophila Trailer hitch antibody and an antibody generated against the human Trailer hitch protein (RAP55). As shown in Fig. 5D, these two antibodies colocalize to the Balbiani body. Thus, the mouse Balbiani body contains Trailer hitch, a component of a conserved complex that is involved in regulating RNAs in multiple species.
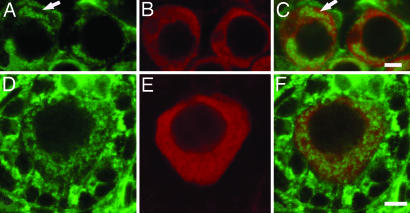
Fig. 5.
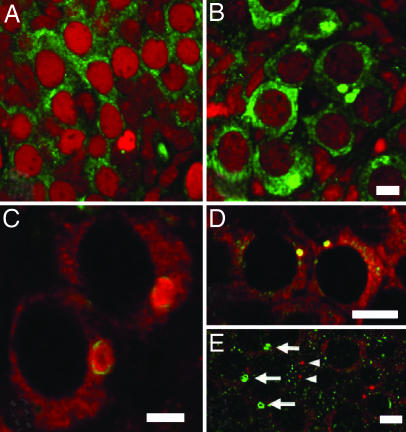
Expression of Tral protein in the Balbiani body of developing mouse ovaries and in adult testes. Immunofluorescence of single confocal ovary sections at 14.5 dpc (A) and at PND3 (B) labeled with an antibody against the Drosophila Tral protein (green) and propidium iodide (red) to visualize nuclei. (C) Immunofluorescence of a single confocal section showing coexpression of Trailer hitch protein (red) and Golgi marker GM130 (green) in developing ovaries at PND1. (D) Codetection of mouse Tral protein by antibodies generated against Drosophila Tral (red) and human RAP55 (green) in the mouse ovary at PND1. (E) Single confocal section through an adult mouse seminiferous tubule labeled with an antibody against the Drosophila Tral protein (green) and Vasa (red) showing that the Tral-rich body (arrows) is distinct from the chromatoid body (arrowheads). (Scale bars: A–C, E, 5 μm; D, 10 μm.)
We previously observed nuage-like structures within the Balbiani body of young mouse oocytes in electron micrographs. Nuage has been best characterized during mouse development in spermatocytes and developing spermatids, where it is found in the chromatoid body (6). Consequently, we stained mouse seminiferous tubules with anti-Tral antibodies and examined them using confocal microscopy. Strong specific labeling of a perinuclear body morphologically similar to the chromatoid body was observed in pachytene spermatocytes and round spermatids (Fig. 5E). This labeling appeared similar to labeling with an antibody to Vasa and Tudor, mouse proteins previously found to be localized to the mouse chromatoid body (19, 20). However, double labeling of seminiferous tubules with antibodies against these proteins showed they do not overlap (Fig. 5E). Several other nuage-containing structures have been described in male germ cells, but a cytological marker exists for only one of these, the RNF17 granule (5). Therefore, we also tested for localization of Tral to the nuage-containing RNF17 granule, but Tral protein did not label this granule either (data not shown). Thus, the mouse Tral protein is not a component of the nuage-containing chromatoid body or the RNF17 granule. Tral protein may be a component of another nuage-containing body in male germ cells, but lack of cytological markers for these structures makes addressing this difficult.
Discussion
Oocytes from Diverse Species Contain a Balbiani Body.
The Balbiani body was first found in spider eggs by von Wittich in 1845 and was subsequently studied in depth by Balbiani in spiders and centipedes (53, 54). The Balbiani body has been described in oocytes of many invertebrate and vertebrate species as a collection of organelles located near the nucleus in germ cells (22). In Xenopus, a special region of the Balbiani body called the message transport organizer (METRO) contains germinal plasm (55). This METRO region functions as a vehicle for transport of germinal plasm and RNAs to the vegetal pole of the oocyte (25). Organelles such as mitochondria also move from the Balbiani body to the vegetal pole, where some will enter germ cells.
Studies in Drosophila demonstrated a connection between germ-line cysts and Balbiani body formation (26). The Balbiani body forms at least in part by the concerted movement of organelles into the oocyte from adjacent germ cells. Germ-line cysts are conserved throughout the animal kingdom, and frequently only a fraction of their germ cells become primordial follicles (56). This might allow the oocyte to initiate oogenesis with more or better-quality organelles. In mice, ≈33% of cyst cells form follicles, with the remainder undergoing programmed cell death at the time of primordial follicle formation (38). It has been postulated that a major function of cysts could be to separate mitochondria with functional and damaged genomes into cells programmed to survive or die, respectively (38). All 16 cells within Xenopus female germ-line cysts are believed to form follicles (23), but some may undergo atresia before maturation.
Young Mouse Oocytes Contain a Balbiani Body.
Our experiments show that young mouse oocytes contain a Balbiani body that is generally similar in its morphology and molecular structure to those of Drosophila, Xenopus, and many other species. As in other organisms, the mouse Balbiani body arises in newly formed primordial follicles but disperses in later oocytes. It temporarily houses most oocyte organelles, with Golgi elements occupying a central position, whereas ER and mitochondrial aggregates reside in a more peripheral location. In Drosophila and Xenopus, mitochondrial clouds that act as precursors to the Balbiani body form earlier, when germ cells reside in cysts of cells interconnected by ring canals. Mitochondrial aggregates were observed in mouse germ-cell cysts before their breakdown (38). Our studies suggest that these aggregates act as precursors to the mouse Balbiani body, as they do in Xenopus. Thus, not only Balbiani body structure, but also its process of formation is at least generally similar in Drosophila, Xenopus, and mouse.
Differences in developmental timing, accessibility, persistence, and relative size probably explain why some investigators previously did not detect a Balbiani body in mouse oocytes. In many species, including Drosophila, the aggregated mitochondria that make up the most visible component of the Balbiani body do not remain for long in a spherical mass but instead spread around the perinuclear cytoplasm (26). Other species, including humans, show similar behavior (35, 57). Once they have spread in this manner, it is more difficult to see that they are specifically clustered, because of the relatively small amount of free cytoplasm in young oocytes. There is also considerable variation in the amount of time that oocytes in different species spend at the early stages when Balbiani bodies are present. They spend less time in these stages in Drosophila and mice than in Xenopus, where many studies of the Balbiani body have been reported. Nonetheless, the presence of true mitochondrial aggregates and their characteristic association with the central Golgi region were clear in our electron and confocal analyses.
We observed some GFM-like structures in electron micrographs of mouse Balbiani bodies, but they were not highly abundant. Because mouse oocytes lack detectable germinal granules (1), our studies show there is not a simple correspondence between the presence of Balbiani bodies in an organism's young oocytes and the localized germinal granules in mature eggs. This is not surprising in light of the fact that oocytes from many other mammals have been shown previously to form Balbiani bodies. Rather, it points to a general and possibly universal role for Balbiani bodies in the young oocyte.
Balbiani Bodies Are Associated with Trailer Hitch, a Protein Involved in RNA Metabolism.
Our identification of Trailer hitch as a Balbiani body constituent strongly supports the view that this structure is related to universal molecular mechanisms of RNA metabolism that may be present in most or all cells. The yeast homologue of mTral, Scd6, was identified as a high-copy suppressor of a deletion of the clathrin heavy-chain locus, suggesting it may play a role in the secretory pathway (58). RNAi of the Caenorhabditis elegans Trailer hitch homologue, CAR-1, results in increased germ cell death in hermaphrodites as well as cytokinesis defects and lethality of embryos (41). In Drosophila, P element insertions in tral result in female sterility (14). These mutants are defective in the secretion of Gurken, which is required for proper dorsal ventral patterning of the embryo. Null alleles of tral have not yet been described in Drosophila.
Previously, the Drosophila Tral protein was shown to be part of an RNP complex involved in mRNA localization and translational regulation in Drosophila oogenesis (40). This complex consists of at least six other proteins, including Me31B (DEAD box helicase), Orb [Cytoplasmic Polyadenylation Element Binding Protein (CPEB)], Yps (Y-box), eIF4E, cup (eIF4E binding), and Exuperentia (11, 40). Complexes containing at least a subset of these proteins exist in C. elegans (41) and Xenopus (59). In C. elegans, CAR-1 localizes to the P granules along with CHG-1, the Me31B homologue (41).
P bodies are cytoplasmic structures involved in mRNA degradation that have been described in both yeast and human cells (48–50). In human cells, these P bodies, also called dcp1 bodies, contain dcp1 and dcp2, proteins involved in decapping RNAs as well as Sm domain-containing proteins (48). The P bodies also have several components in common with the Drosophila RNP complex, including Rck (Me31B homologue), CPEB, 4E-T (Cup homologue), and RAP55 (Trailer hitch homologue) (47, 48, 50, 51). Knockdown of 4E-T or RAP55 causes loss of P bodies, suggesting a role for these proteins in P body assembly and in regulating mRNA decay. In the Drosophila ovary, cup mutants also affect RNP particle assembly of the mRNA localization complex (12). The similarity of components in P bodies and the Drosophila mRNA localization complex suggests these are related structures. The human Trailer hitch protein, RAP55, is localized to P bodies (47). In addition, chromatoid bodies and P bodies also exhibit similarity in their molecular nature (51, 52, 60).
Balbiani Bodies, Oocyte Polarity, and Differences in Early Developmental Regulation Between Invertebrate and Mammalian Embryos.
Drosophila and Xenopus oocytes are highly polar and contain localized RNAs and other components that mediate the patterning of the early embryo (61–63). Mammalian oocytes, in contrast, are often viewed as completely symmetrical and nonpolar. Embryonic polarity is not thought to be established until implantation (64), although this view has been challenged (65, 66). Our experiments show there is not a simple relationship between the presence of a Balbiani body and egg polarity. As proposed above, all oocytes that grow to a larger size than normal cells may require large amounts of the machinery used normally to move and store cytoplasmic constituents. Whether this activity actually leads to the localization of patterning RNAs or germ-cell determinants late in oogenesis may be determined simply later and may vary from species to species. Thus, both patterned and unpatterned eggs may be built using largely conserved processes of organelle and RNA metabolism.
Materials and Methods
Mice.
Ovaries from embryos and neonates and testes from adults were obtained from wild-type CD-1 mice (Charles River Breeding Laboratories, Wilmington, MA). The presence of a vaginal plug the morning after mating was designated 0.5 dpc. Birth occurred at 19.5 dpc and was designated PND1. All animal experimentation was reviewed and approved by the Syracuse University Institutional Animal Care and Use Committee.
Antibodies.
To detect Vasa, an antibody raised against the mouse Vasa protein was used at 1:500 (20) or an antibody raised against the human Vasa protein was used at 1:100 (R&D Systems, Minneapolis, MN). The Drosophila Trailer hitch antibody was used at a dilution of 1:500 for whole-mount immunocytochemistry or 1:1,000 for Western blotting (14). GM130 antibody (BD Biosciences, Franklin Lakes, NJ) was used to label Golgi at a dilution of 1:100. Cytochrome c antibody (BD Biosciences) was used to label mitochondria at a dilution of 1:250. GAPDH antibody (EnCor Biotechnology, Gainesville, FL) was used at 1:2,000. The RAP55 antibody was used at 1:500 for whole-mount immunocytochemistry or 1:1,000 for Western blotting and was a gift of Donald Bloch (Massachusetts General Hospital, Boston, MA) (47).
Immunohistochemistry.
Whole ovaries from embryos and pups and testes from adults of the wild-type CD-1 strain were harvested, fixed, and stained as described (37, 67), except that neonatal and adult ovaries were fixed overnight at 4°C. After adult testes were fixed, they were sliced into small pieces before incubation with antibody. Propidium iodide or Toto-3 (Molecular Probes, Eugene, OR) was used to label nuclei. Drosophila ovaries were dissected and stained as described (14). Samples were imaged on a Zeiss (Oberkochen, Germany) Pascal Confocal microscope.
Western Blot Hybridization.
Mouse gonads were homogenized in Sample Buffer (2% SDS/10% glycerol/25 mM Tris, pH 6.8/0.00005% bromophenol blue/0.025% mercaptoethanol) plus mini complete protease inhibitor (Sigma, St. Louis, MO). PND1 and -4 ovaries were homogenized in 10 μl of sample buffer per ovary and PND1 and four testes in 20 μl per testis. Adult ovaries were homogenized in 200 μl per ovary and adult testes in 800 μl per testis. For each sample, 20 μl of solubilized protein extract was mixed with 1/10 vol of mercaptoethanol, heated to 95°C for 3 min, separated on 10% SDS–polyacrylamide gels, and electroblotted onto Immobilon P membranes (Millipore, Billerica, MA). The blots were incubated with a blocking solution containing 5% nonfat milk powder in PBST (PBS/0.05% Tween 20) overnight at 4°C and incubated with primary antibody in blocking solution for 1 h at room temperature. After three washes in blocking solution, membranes were incubated with horseradish peroxidase-conjugated secondary IgG (1:10,000) in blocking solution at room temperature for 1 h, washed in PBST three times, and signal-visualized by using the Supersignal kit (Pierce, Rockford, IL) on films. Blots were reprobed for GAPDH as a loading control.
A pGEX plasmid (Amersham Pharmacia, Uppsala, Sweden) encoding amino acids 1–266 of RAP55 protein was obtained (47), and RAP55 protein was produced in bacteria as a GST-tagged fusion. Fusion proteins were prepared under denaturing conditions according to the manufacturer's instructions.
Electron Microscopy.
Appropriately aged ovaries were dissected and processed for transmission electron microscopy as described (68). The samples were analyzed by using a Phillips (Eindhoven, The Netherlands) Tecnai 12 microscope, and images were recorded with a GATAN (Pleasanton, CA) multiscan CCD camera in the digital micrograph program.
Supplementary Material
Acknowledgments
We thank Toshiaki Noce (Mitsubish-Kasei Institute of Life Sciences, Tokyo, Japan), Donald Bloch (Massachusetts General Hospital, Boston MA), and Jeremy Wang (University of Pennsylvania, Philadelphia, PA) for generous gifts of antibodies and plasmids. We also thank Eleanor Maine and Scott Erdman for critical reading of the manuscript and Mike Sepanski for assistance with electron microscopy.
Abbreviations
- ER
endoplasmic reticulum
- GFM
granulofibrillar material
- PNDn
postnatal day n
- RNP
ribonucleoprotein
- P body
processing body
- 4E-T
4E-Transporter
- dpc
days postcoitum.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609923104/DC1.
References
- 1.McLaren A. Dev Biol. 2003;262:1–15. doi: 10.1016/s0012-1606(03)00214-8. [DOI] [PubMed] [Google Scholar]
- 2.Kloc M, Etkin LD. J Cell Sci. 2005;118:269–282. doi: 10.1242/jcs.01637. [DOI] [PubMed] [Google Scholar]
- 3.Eddy EM. Biol Reprod. 1970;2:114–128. doi: 10.1095/biolreprod2.1.114. [DOI] [PubMed] [Google Scholar]
- 4.Fawcett DW, Eddy EM, Phillips DM. Biol Reprod. 1970;2:129–153. doi: 10.1095/biolreprod2.1.129. [DOI] [PubMed] [Google Scholar]
- 5.Pan J, Goodheart M, Chuma S, Nakatsuji N, Page DC, Wang PJ. Development (Cambridge, UK) 2005;132:4029–4039. doi: 10.1242/dev.02003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parvinen M. Int J Androl. 2005;28:189–201. doi: 10.1111/j.1365-2605.2005.00542.x. [DOI] [PubMed] [Google Scholar]
- 7.Russell L, Frank B. Anat Rec. 1978;190:79–97. doi: 10.1002/ar.1091900108. [DOI] [PubMed] [Google Scholar]
- 8.Houston DW, King ML. Curr Top Dev Biol. 2000;50:155–181. doi: 10.1016/s0070-2153(00)50008-8. [DOI] [PubMed] [Google Scholar]
- 9.Noce T, Okamoto-Ito S, Tsunekawa N. Cell Struct Funct. 2001;26:131–136. doi: 10.1247/csf.26.131. [DOI] [PubMed] [Google Scholar]
- 10.Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. Development (Cambridge, UK) 2003;130:859–871. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura A, Amikura R, Hanyu K, Kobayashi S. Development (Cambridge, UK) 2001;128:3233–3242. doi: 10.1242/dev.128.17.3233. [DOI] [PubMed] [Google Scholar]
- 12.Wilhelm JE, Hilton M, Amos Q, Henzel WJ. J Cell Biol. 2003;163:1197–1204. doi: 10.1083/jcb.200309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura A, Sato K, Hanyu-Nakamura K. Dev Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- 14.Wilhelm JE, Buszczak M, Sayles S. Dev Cell. 2005;9:675–685. doi: 10.1016/j.devcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 15.Forristall C, Pondel M, Chen L, King ML. Development (Cambridge, UK) 1995;121:201–208. doi: 10.1242/dev.121.1.201. [DOI] [PubMed] [Google Scholar]
- 16.Heasman J, Quarmby J, Wylie CC. Dev Biol. 1984;105:458–469. doi: 10.1016/0012-1606(84)90303-8. [DOI] [PubMed] [Google Scholar]
- 17.Soderstrom KO, Parvinen M. Cell Tissue Res. 1976;168:335–342. doi: 10.1007/BF00215311. [DOI] [PubMed] [Google Scholar]
- 18.Tsai-Morris CH, Sheng Y, Lee E, Lei KJ, Dufau ML. Proc Natl Acad Sci USA. 2004;101:6373–6378. doi: 10.1073/pnas.0401855101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuma S, Hiyoshi M, Yamamoto A, Hosokawa M, Takamune K, Nakatsuji N. Mech Dev. 2003;120:979–990. doi: 10.1016/s0925-4773(03)00181-3. [DOI] [PubMed] [Google Scholar]
- 20.Toyooka Y, Tsunekawa N, Takahashi Y, Matsui Y, Satoh M, Noce T. Mech Dev. 2000;93:139–149. doi: 10.1016/s0925-4773(00)00283-5. [DOI] [PubMed] [Google Scholar]
- 21.de Smedt V, Szollosi D, Kloc M. Genesis. 2000;26:208–212. doi: 10.1002/(sici)1526-968x(200003)26:3<208::aid-gene6>3.3.co;2-e. [DOI] [PubMed] [Google Scholar]
- 22.Guraya SS. Int Rev Cytol. 1979;59:249–321. doi: 10.1016/s0074-7696(08)61664-2. [DOI] [PubMed] [Google Scholar]
- 23.Kloc M, Bilinski S, Etkin LD. Curr Top Dev Biol. 2004;59:1–36. doi: 10.1016/S0070-2153(04)59001-4. [DOI] [PubMed] [Google Scholar]
- 24.Kloc M, Dougherty MT, Bilinski S, Chan AP, Brey E, King ML, Patrick CW, Jr, Etkin LD. Dev Biol. 2002;241:79–93. doi: 10.1006/dbio.2001.0488. [DOI] [PubMed] [Google Scholar]
- 25.Kloc M, Larabell C, Chan AP, Etkin LD. Mech Dev. 1998;75:81–93. doi: 10.1016/s0925-4773(98)00086-0. [DOI] [PubMed] [Google Scholar]
- 26.Cox RT, Spradling AC. Development (Cambridge, UK) 2003;130:1579–1590. doi: 10.1242/dev.00365. [DOI] [PubMed] [Google Scholar]
- 27.Odor DL, Blandau RJ. Am J Anat. 1969;125:177–215. doi: 10.1002/aja.1001250205. [DOI] [PubMed] [Google Scholar]
- 28.Tsuda H. Arch Histol Jpn. 1965;25:533–555. doi: 10.1679/aohc1950.25.533. [DOI] [PubMed] [Google Scholar]
- 29.Ullmann SL, Butcher L. Reprod Fertil Dev. 1996;8:491–508. doi: 10.1071/rd9960491. [DOI] [PubMed] [Google Scholar]
- 30.Eddy EM. Anat Rec. 1974;178:731–757. doi: 10.1002/ar.1091780406. [DOI] [PubMed] [Google Scholar]
- 31.Young JK, Allworth AE, Baker JH. Anat Embryol (Berlin) 1999;200:43–48. doi: 10.1007/s004290050257. [DOI] [PubMed] [Google Scholar]
- 32.Takeuchi IK, Sonta S, Takeuchi YK. J Electron Microsc (Tokyo) 1984;33:388–394. [PubMed] [Google Scholar]
- 33.Hertig AT. Am J Anat. 1968;122:107–137. doi: 10.1002/aja.1001220107. [DOI] [PubMed] [Google Scholar]
- 34.Hertig AT, Adams EC. J Cell Biol. 1967;34:647–675. doi: 10.1083/jcb.34.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Motta PM, Nottola SA, Makabe S, Heyn R. Hum Reprod. 2000;15(Suppl 2):129–147. doi: 10.1093/humrep/15.suppl_2.129. [DOI] [PubMed] [Google Scholar]
- 36.Kloc M, Bilinski S, Dougherty MT, Brey EM, Etkin LD. Dev Biol. 2004;266:43–61. doi: 10.1016/j.ydbio.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Pepling ME, Spradling AC. Development (Cambridge, UK) 1998;125:3323–3328. doi: 10.1242/dev.125.17.3323. [DOI] [PubMed] [Google Scholar]
- 38.Pepling ME, Spradling AC. Dev Biol. 2001;234:339–351. doi: 10.1006/dbio.2001.0269. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T. Genes Dev. 2000;14:841–853. [PMC free article] [PubMed] [Google Scholar]
- 40.Wilhelm JE, Mansfield J, Hom-Booher N, Wang S, Turck CW, Hazelrigg T, Vale RD. J Cell Biol. 2000;148:427–440. doi: 10.1083/jcb.148.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boag PR, Nakamura A, Blackwell TK. Development (Cambridge, UK) 2005;132:4975–4986. doi: 10.1242/dev.02060. [DOI] [PubMed] [Google Scholar]
- 42.Anantharaman V, Aravind L. BMC Genom. 2004;5:45. doi: 10.1186/1471-2164-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Birney E, Kumar S, Krainer AR. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi SB, Wang C, Muench DG, Ozawa K, Franceschi VR, Wu Y, Okita TW. Nature. 2000;407:765–767. doi: 10.1038/35037633. [DOI] [PubMed] [Google Scholar]
- 45.Juschke C, Ferring D, Jansen RP, Seedorf M. Curr Biol. 2004;14:406–411. doi: 10.1016/j.cub.2004.02.034. [DOI] [PubMed] [Google Scholar]
- 46.Ko MS, Kitchen JR, Wang X, Threat TA, Wang X, Hasegawa A, Sun T, Grahovac MJ, Kargul GJ, Lim MK, et al. Development (Cambridge, UK) 2000;127:1737–1749. doi: 10.1242/dev.127.8.1737. [DOI] [PubMed] [Google Scholar]
- 47.Yang WH, Yu JH, Gulick T, Bloch KD, Bloch DB. RNA. 2006;12:547–554. doi: 10.1261/rna.2302706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cougot N, Babajko S, Seraphin B. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheth U, Parker R. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilczynska A, Aigueperse C, Kress M, Dautry F, Weil D. J Cell Sci. 2005;118:981–992. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- 51.Ferraiuolo MA, Basak S, Dostie J, Murray EL, Schoenberg DR, Sonenberg N. J Cell Biol. 2005;170:913–924. doi: 10.1083/jcb.200504039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Villaescusa JC, Allard P, Carminati E, Kontogiannea M, Talarico D, Blasi F, Farookhi R, Verrotti AC. Gene. 2006;367:101–109. doi: 10.1016/j.gene.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 53.Balbiani EG. Compt Rend Hebd Seances Acad Sci. 1864;58:584–588. [Google Scholar]
- 54.von Wittich WH. Dissertatio Sistens Observations Quadem de Sistens Aranearum ex Ovo Evolutione. Halle, Germany: Halis Saxonum; 1845. [Google Scholar]
- 55.Kloc M, Etkin LD. Development (Cambridge, UK) 1995;121:287–297. doi: 10.1242/dev.121.2.287. [DOI] [PubMed] [Google Scholar]
- 56.de Cuevas M, Lilly MA, Spradling AC. Annu Rev Genet. 1997;31:405–428. doi: 10.1146/annurev.genet.31.1.405. [DOI] [PubMed] [Google Scholar]
- 57.Raven C. Oogenesis: The Storage of Developmental Information. New York: Pergamon; 1961. [Google Scholar]
- 58.Nelson KK, Lemmon SK. Mol Cell Biol. 1993;13:521–532. doi: 10.1128/mcb.13.1.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Minshall N, Standart N. Nucleic Acids Res. 2004;32:1325–1334. doi: 10.1093/nar/gkh303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, Sassone-Corsi P. Proc Natl Acad Sci USA. 2006;103:2647–2652. doi: 10.1073/pnas.0509333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anderson O, Heasman J, Wylie C. Int Rev Cytol. 2001;203:215–230. doi: 10.1016/s0074-7696(01)03008-x. [DOI] [PubMed] [Google Scholar]
- 62.Starz-Gaiano M, Lehmann R. Mech Dev. 2001;105:5–18. doi: 10.1016/s0925-4773(01)00392-6. [DOI] [PubMed] [Google Scholar]
- 63.Wylie C. Germ cells. Curr Opin Genet Dev. 2000;10:410–413. doi: 10.1016/s0959-437x(00)00105-2. [DOI] [PubMed] [Google Scholar]
- 64.Gurdon JB. Cell. 1992;68:185–199. doi: 10.1016/0092-8674(92)90465-o. [DOI] [PubMed] [Google Scholar]
- 65.Gardner RL, Davies TJ. Philos Trans R Soc Lond B. 2003;358:1331–1338. doi: 10.1098/rstb.2003.1322. discussion 1338–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zernicka-Goetz M. Development (Cambridge, UK) 2002;129:815–829. doi: 10.1242/dev.129.4.815. [DOI] [PubMed] [Google Scholar]
- 67.Murphy K, Carvajal L, Medico L, Pepling ME. Gene Express Patt. 2005;5:475–482. doi: 10.1016/j.modgep.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 68.Yue L, Spradling AC. Genes Dev. 1992;6:2443–2454. doi: 10.1101/gad.6.12b.2443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.