Abstract
The Oka vaccine strain is a live attenuated virus that is routinely administered to children in the United States and Europe to prevent chickenpox. It is effective and safe but occasionally produces a rash. The vaccine virus has accumulated mutations during its attenuation, but the rashes are not explained by their reversion, unlike complications reported for other viral vaccines. Indeed, most of the novel mutations distinguishing the Oka vaccine from the more virulent parental virus have not actually become fixed. Because the parental alleles are still present, the vaccine is polymorphic at >30 loci and therefore contains a mixture of related viruses. The inoculation of >40 million patients has consequently created a highly replicated evolutionary experiment that we have used to assess the competitive ability of these different viral genotypes in a human host. Using virus recovered from rash vesicles, we show that two vaccine mutations, causing amino acid substitutions in the major transactivating protein IE62, are outcompeted by the ancestral alleles. Standard interpretations of varicella disease severity concentrate on the undeniably important effects of host genotype and immune status, yet our results allow us to demonstrate that the viral genotype is associated with virulence and to identify the key sites. We propose that these loci have pleiotropic effects on the immunogenic properties of the virus, rash formation, and its epidemiological spread, which mould the evolution of its virulence. These findings are of practical importance for reducing the incidence of vaccine-associated rash and promoting public acceptance of the vaccine.
Keywords: chickenpox, shingles, virulence, epidemiology, IE62
The live attenuated Oka strain varicella vaccine is licensed for immunization of healthy children in the United States and Germany and for healthcare workers in the United Kingdom (1, 2). Vaccine-associated rash after inoculation occurs in 5% of healthy children but is more common, and potentially life-threatening, when the vaccine has inadvertently been administered to immunocompromised subjects (3). Evidence from work in the SCID hu mouse epithelial implant model has firmly linked attenuation of the Oka vaccine strain with reduced skin replication (4). Rash formation is more than an incidental nuisance in varicella infection. It is fundamental to the transmission of the virus, because infectious virions are shed from skin vesicles (5, 6). In line with these findings, the attenuated vaccine strain exhibits limited human-to-human transmission (three cases reported in 15 million doses) and no associated mortality (3). The reduction of free vaccine virus within epithelial vesicles could also explain the lower incidence of herpes zoster in Oka vaccines (3, 7). This decreased free vesicular virus is thought to reduce the infection of sensory nerve endings, the retrograde spread, and, consequently, the viral burden in the dorsal root ganglia (3, 6).
Sequencing by Gomi et al. (8) of the vaccine Oka genome and the parental Oka strain from which it is derived identified 42 single nucleotide polymorphisms (SNPs), 20 nonsynonymous and 22 synonymous, over one-third of which were located in the ORF 62 major transactivating gene. The vaccine preparation is not genetically uniform because only three of the novel vaccine mutations are fixed (9). The remainder of the SNPs are polymorphic, so the vaccine dose comprises a diverse but related viral population (10). The repeated inoculation of many millions of vaccine doses into vaccinees and the recovery of viruses from rashes presents a remarkable opportunity to follow the relative competitive success of different viral genotypes within the human host. It is unlikely that all 42 of the SNPs are implicated in attenuation of virulence. The attenuation of the vOka vaccine strain was achieved by serial passaging of the virus in both animal and human cell lines (11). Some of the novel mutations will have been selected for fitness in this unnatural new environment; some may have become established by genetic drift. Some, but not necessarily all, must have pleiotropic effects that reduce virulence in the human host; however, up to now, there has been no evidence directly linking individual SNPs with loss of virulence. One possibility is that the occurrence of vaccine-associated rash is determined by the host's genotype and immunological status alone. This study was designed to test the less-conventional hypothesis that the viral genotype affects rash formation, in which case, the viral genotypes recovered from rashes would differ from those originally inoculated.
We have shown previously that, unlike the vaccine preparation, viruses from a single vaccine-associated lesion are genetically uniform (12), but viruses sampled from different lesions in the same individual, or from different individuals, are genetically distinct (9, 12). This preliminary result suggested that no single preexisting vaccine genotype was responsible for rash formation (9), in contrast to other live attenuated vaccines, notably poliovirus, in which a single point mutation can cause reversion to virulence (13). In this report, we evaluate the evidence that a subset of the vaccine virus genotypes is disproportionately successful in spreading through the human body and causing rashes. An initial step made use of information from 10 of the viral genotypes obtained by Quinlivan et al. (12) from vaccine-associated rashes collected in the 1980s. We excluded from consideration those viruses derived from a batch of vaccine that was not subsequently forwarded to licensure for public use. Comparison of the remaining genotype frequencies with those in the vaccine identified eight candidate loci (of 37 surveyed, including all of the nonsynonymous SNPs; Table 1) at which alleles had shown the most dramatic increase in frequency from inoculation to rash. A larger study then evaluated the existence of consistent selection at these loci by genotyping virus recovered from 70 additional rashes collected a decade or more later. Significant deviations in the genotype frequencies between the vaccine and the rashes of this second, independent sample would provide evidence of selection acting between the point at which the live virus was inoculated and the production of virions in the rash vesicle.
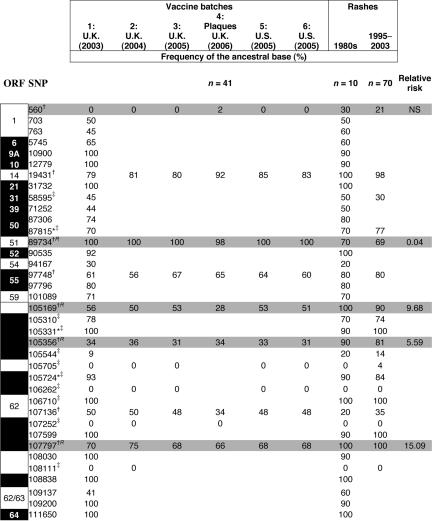
Table 1.
The frequency of ancestral bases in viral samples recovered from rashes and from various vaccine batches
Allele frequencies were estimated in the Merck vaccine by using comparative sequence analysis. The allele frequencies in vaccine batches 2–6 were estimated by pyrosequencing. ∗, SNPs identified by us. The remaining 34 SNPs, including all 20 nonsynonymous SNPs and 14 of 22 nonsynonymous SNPs, were identified by Gomi et al.(8). †, SNPs at which the allele frequency in the original 10 rashes screened differed from the vaccine by ≥20%. ‡, Control SNPs that differed by <20%. Relative risk is a ratio calculated from the parameters of the logistic regression. It is the relative probability of being a vaccine genotype when the mutant allele is present at the locus (compared with a genotype ancestral at all four loci). The numbers in the SNP column indicate the location in the vOka genome at which mutations have been described. They denote the corresponding nucleotide position in the VZV Dumas genome. Black areas indicate SNPs that change the protein sequence; white numbers in black areas denote the VZV ORF. Gray areas indicate loci at which allele frequencies in skin rash viruses differed significantly from those in the original vaccine (P < 0.001: 2 × 2 contingency tables with Bonferroni correction).
Results
Variation in Allele Frequencies Among Vaccine Batches.
The rashes were caused by at least seven different vaccine batches. To exclude vaccine batch-to-batch variation as a reason for the differences in allele frequencies between the vaccine and rashes, we estimated the allele frequencies in five different batches (Table 1) (14, 15). Three batches were from the United Kingdom, including one manufactured during the period over which the 70 rashes were collected (batch 1) and two from the United States (batches 5 and 6), where the rash viruses were collected. Batch-to-batch variation of the vaccines was minimal (SD of allele frequency was 2.3%) and significantly less than the differences seen when allele frequencies in the rashes and vaccines were compared (12–50%; SD, 27%; P < 0.001). This negligible variation between the batches can be explained by the use of a single master-seed for production of the seed stocks produced since licensure. Each seed stock is used for 1–2 years (Alan Shaw, personal communication). A second test for variation between vaccine stocks made use of the vaccine rash genotypes themselves. None of the candidate loci showed significant deviation in allele frequency between vaccine lots produced in different years.
Allele frequencies in 41 virus plaques purified from one vaccine batch (batch 4) were also very close to the pyrosequencing estimates for the other batches (Table 1) but significantly different from those recovered from the rashes (P < 0.001; individually significant SNPs are shaded in Table 1, P < 0.05). Only one locus in the plaque 105169 (ORF 62) differed significantly from the pyrosequencing results.
Comparison of Vaccine and Rash Viruses.
The viral haplotypes recovered from the vaccine preparation and vaccine rashes are extraordinarily diverse, each being unique [see supporting information (SI)]. There was no significant difference between allele frequencies in the zoster rashes (distinguished by a code starting with a Z in the SI) and the varicella rashes (P = 0.89). Using logistic regression, we compared allele frequencies at the 8 candidate and 12 control loci in vaccine preparations and 70 rashes. From this, we identified four of the candidate loci as significantly and individually contributing to rash formation (marked R in Table 1). None of the four could be excluded from the minimal model (P < 0.005), and no pairwise interactions or linkage disequilibria were significant. When the minimal model was used to predict the type of a virus (rash or vaccine), it correctly classified 95% of the rash viruses, whereas only 21% of vaccine viruses were predicted to be rash-forming.
After immunization, vOka virus replicates locally, then infects peripheral blood mononuclear cells and may be carried to distant sites including the skin (3, 5), lungs, brain, and visceral organs (3). The observed bias in rash viral genotypes could, therefore, have arisen before replication in skin. To investigate this possibility, we genotyped virus sampled from the lung of a patient with vOka vaccine viral pneumonia (9). The allele frequencies were, like skin viruses, 100% wild type at position 107797 (446L) but closer to vaccine frequencies at other loci (Table 1). Selection at this locus may therefore have occurred before the virus reached the skin. In vitro observations that the vOka vaccine mixture replicates better in lung than in skin fibroblasts (16) may indicate that the majority of vaccine genotypes are better adapted to lung tissue and hence explain the persistence of a predominantly vaccine-genotypic composition in the lung.
Discussion
It was known previously that there are only 42 sites where the vOka vaccine DNA sequence has accrued novel mutations that distinguish it from the more virulent parental Oka virus (8). However, these differences were not established by direct selection for reduced virulence. Instead, they accumulated during the attenuation procedure, during which there will have been selection for efficient propagation during serial passaging in cell lines and random genetic drift. Some of the novel alleles clearly must have pleiotropic effects that reduce virulence because the clinical effects of the vaccine virus are less severe. However, distinguishing the particular loci responsible has been hindered because no animal model adequately replicates varicella-zoster virus (VZV) propagation and virulence in vivo.
In this study, we have overcome this problem to distinguish a subset of loci that have major effects on propagation in the human body. This was achieved by exploiting the natural experiment occurring within recipients of the Oka varicella vaccine. The approach was possible because the vaccine is extensively polymorphic at the loci in question. In interpreting the results, it was feasible that the occurrence of rash would have proved to be determined by the host genotype and immune status irrespective of the viral genotype. In that case, the virus recovered from the rash would have been a random selection of the genotypes in the vaccine. We observed the converse: four residues were strongly and consistently selected in vivo during viral spread and the formation of skin rashes.
Reversion to a more virulent phenotype is not explained by mutation occurring in the human host. We identified only three previously undescribed alleles in ≈200 kb of sequencing, and each occurred in the rashes at frequencies indistinguishable from the vaccine. Furthermore, the rash alleles identified by the analysis were all detectable in the vOka mixture that was inoculated. The genotypes that carry one or more of the four selected mutations have outcompeted other components of the live vaccine inoculum. Of these four selected mutations, two code for amino acid changes at positions 446 and 1260 in ORF 62, and two are synonymous. More than one-third of vaccine mutations are located in ORF 62, and vaccine IE62 is one of four proteins with significantly reduced mRNA expression in MRC5 cells compared with parental virus (17). ORF 62 is essential for replication of virus in skin (18), and position 446 (107797), present in all rash viruses, lies within a region that binds the immediate early protein IE63 (19). Binding of IE63 has been shown to increase IE62 transactivation of the ORF 67 promotor, which codes for glycoprotein I, another protein essential to VZV replication in skin (19, 20). The ancestral leucine residue may therefore be functionally important for skin replication and also forms part of an HLA A2-restricted T cell epitope (21). Computer algorithms (22) predict that the vaccine substitution (proline) would abolish immune recognition of this epitope. One corollary of this interpretation is the prediction that the vaccine virus will be found to spread more effectively in subjects who are HLA A2-negative or have lost immunity to this epitope.
We cannot speculate on the functional significance, if any, of the synonymous substitutions at positions 89734 and 105169. However, the implication of position 1260 is reinforced by the analysis of the Ellen strain, another multiply passaged, laboratory-adapted VZV. A substitution of valine for the ancestral isoleucine at position 1260 is again observed, as are glycine residues, which are fixed at positions 628 and 928 in the vaccine. It is therefore possible that the ancestral amino acid at these positions confers an advantage in replication of the virus in differentiated epithelium.
Why should the novel vaccine mutations persist in culture but, unlike other live attenuated vaccines, notably poliovirus (23), not be able to circulate in the wild? The explanation appears to be in the central importance of skin rashes in the epidemiology of the virus: infectious virions are shed from the rash vesicles. Our data show that the development of rash after vOka vaccination is due to selection of viral strains that occur infrequently in the vaccine (Table 1) and probably depends on specific interactions by these viruses with host immunity.
Our analysis has identified 4 of 37 sites with vaccine mutations, which are most clearly implicated in rash formation. We suggest that these alleles could be targeted for reduction in the preparation of vaccine batches. Any reduced incidence of rash would benefit the public acceptance of the vaccine. Although the effects of vaccine rash are generally mild, they can be life threatening when immunocompromised patients are inadvertently immunized (3, 24–26). Because these patients are at great risk from natural varicella infection, vaccination would be valuable, and measures to reduce vaccine rash would be particularly beneficial.
Materials and Methods
Samples.
Samples for the pilot study were obtained from subjects immunized in the 1980s. All samples had been passaged two to five times in human embryo lung fibroblast (HEL) cells. Samples for the second study were obtained as part of a postlicensure surveillance program conducted between 1995 and 2003 (3). Five of the 70 samples had been passaged between two and five times. The remainder were uncultured.
DNA Extractions and PCRs.
DNA was extracted from clinical samples (vesicle fluid) and vaccine batches by using a QIAamp DNA Mini Kit (Qiagen, Crawley, U.K.) according to the manufacturer's instructions. Before extraction, lyophilized vaccine preparations were resuspended in the 500 μl of accompanying buffer. DNA from each preparation was extracted from 2 × 200-μl and 1 × 100-μl (mixed with 100 μl of PBS) aliquots. The 3 × 200-μl aliquots of eluted DNA were merged to form a 600-μl DNA pool that was used for subsequent experiments.
All PCRs were performed in a GeneAmp 2700 thermocycler (PerkinElmer, Beaconsfield, U.K.), initiated by hot start activation at 95°C for 10 min and ending with a step of 72°C for 10 min, before being incubated at 4°C. For pyrosequencing, reactions included 45 cycles of amplification (95°C, 15 s; the primer annealing temperature specified in the SI, 30 s; 72°C, 15 s).
Direct Sequencing and Comparative Sequence Analysis (CSA).
DNA from the initial 10 vaccine rash virus samples was amplified and directly sequenced on an ABI Prism 3700 automated sequencer (Applied Biosystems, Warrington, U.K.) according to the manufacturer's instructions. The vaccine allele frequencies in batch 1 were determined by CSA (14). For the CSA, 0.5 μl of LIZ 500 size standard was mixed with each sample before loading onto the 3700 platform. Peak heights were automatically measured by Genescan software (Applied Biosystems) by using the LIZ 500 size standard as a marker and are displayed in arbitrary units. Sequence traces from the known wild type and unknown sample were overlaid as described in the CSA methodology (14). The frequency of the ancestral allele at mixed or substituted loci in the vaccine was calculated. The standard error of the method was estimated to be 7% by using replicates of known mixtures of polymorphic loci (data not shown).
Pyrosequencing.
Pyrosequencing was used to analyze allele frequencies in batches 2, 3, 5, and 6 and the 70 additional rash genotypes. We have shown previously that virus from one lesion is genetically uniform (4), so detection of polymorphic SNPs in 22 of the 70 rashes may indicate cases when more than one lesion was sampled; at the time of sampling, the genetic distinctiveness of individual lesions was not known. Comparison of pyrosequencing and CSA results for batch 1 were within the confidence limits of batch-to-batch variation. Pyrosequencing primers (SI) were generated for each locus that had been identified by the pilot survey. Single-stranded DNA was purified by using 5′ biotin-labeled primers. The biotinylated PCR product was captured with streptavidin Sepharose and purified with a vacuum prep workstation according to the manufacturer's instructions (Biotage, Uppsala, Sweden). Pyrosequencing reactions were performed according to the manufacturer's instructions.
Plaque Purification of Vaccine Virus.
To assess haplotype variation, vaccine virus was plaque-purified, and each virus was genotyped by using pyrosequencing. The viral plaques were grown in human embryo lung fibroblast monolayers, which were cultured in 24-well tissue culture plates (1 ml; Nunc, Roskilde, Denmark) until 70–80% confluent. The growth medium was removed, and each well was inoculated with 100 μl of vaccine virus diluted 1:100 from neat in maintenance medium (98% Earl's medium and 2% FCS). After 2 hours of absorption at 37°C, excess medium was removed from each well, and the cells were overlaid with 1 ml of low-melting-point agarose (5% in water) mixed in a ratio of 1:10 with maintenance medium (98% Earl's medium, 2% FBS, 1% penicillin/streptomycin/nystatin, and 1% l-glutamine) that had been cooled to 37–40°C. After 7–10 days of culture at 37°C in 5% CO2, individual vaccine plaques showing syncytia were picked with a sterile pipette tip, mixed with 1 ml of maintenance medium, and recultured for am additional 7 days in fresh human embryo lung fibroblast monolayers.
Statistical Methods.
Generalized linear modeling, assuming binomial error, was used to assess the difference in allele frequencies at the eight candidate loci for the comparisons of zoster and varicella rashes, the comparisons between year of vaccine-lot manufacture, and the comparisons between rash and plaque genotypes.
The comparison of rash allele frequencies between the years of vaccine-lot manufacture (listed in the SI) was made by using a separate logistic regression for each candidate locus except for SNP 107797, which was monomorphic in the rashes. Adding the year of manufacture did not improve any model: F12/60 values fell in the range of 0.63–1.67 (for SNPs 105169 and 19431, respectively), and none was significant (P > 0.05 in all cases).
For the rash–plaque comparison, the logistic regression was carried out on all plaque and post-1995 rashes for which we obtained complete genotypes. The SNPs were encoded as two-level factors. The dependant variable was the virus' origin (isolated from rash or from vaccine). The model with the minimal Akaike information criterion (AIC) was obtained by sequentially discarding SNPs from the model. None of the two-way interactions had a significant effect. For each candidate locus, the fitted value was estimated for a genotype bearing the ancestral allele at all but that locus (the fitted value is the probability of such a genotype being a vaccine sample). The relative risk given in Table 1 was obtained by dividing this estimate by the fitted value for a genotype consisting entirely of ancestral alleles.
The variation among frequencies in different vaccine batches was compared with the deviation between rash and vaccine frequencies. First, the vaccine allele frequencies were arcsine-transformed. A generalized model, assuming Gaussian error, was fitted to the transformed values. An ANOVA was used to compare the deviation between vaccine and allele frequencies with the residual variance among vaccine frequencies: F6/40 = 25.99, P < 0.001.
Supplementary Material
Acknowledgments
We thank Dr. Robert Sharrar of Merck Sharp Dohme for his helpful comments and advice and Susan Galea and Ann Sweet, research nurses at Merck. This work was funded by Welcome Trust Grant GR075427MA-075427 and National Institutes of Health Grants AI 24021 and AI 127187.
Abbreviations
- CSA
comparative sequence analysis
- VZV
varicella-zoster virus.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
See Commentary on page 7.
This article contains supporting information online at www.pnas.org/cgi/content/full/0605688104/DC1.
References
- 1.Holmes SJ, Reef S, Hadler SC, Williams WW, Wharton M. MMAR. 1996;45:RR–11. [Google Scholar]
- 2.Rentier B, Gershon AA. Pediatr Infect Dis J. 2004;23:379–389. doi: 10.1097/01.inf.0000122606.88429.8f. [DOI] [PubMed] [Google Scholar]
- 3.Sharrar RG, LaRussa P, Galea SA, Steinberg SP, Sweet AR, Keatley RM, Wells ME, Stephenson WP, Gershon AA. Vaccine. 2000;19:916–923. doi: 10.1016/s0264-410x(00)00297-8. [DOI] [PubMed] [Google Scholar]
- 4.Moffat JF, Zerboni L, Kinchington PR, Grose C, Kaneshima H, Arvin AM. J Virol. 1998;72:965–974. doi: 10.1128/jvi.72.2.965-974.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arvin AM. Curr Opin Microbiol. 2001;4:442–449. doi: 10.1016/s1369-5274(00)00233-2. [DOI] [PubMed] [Google Scholar]
- 6.Chen JJ, Gershon AA, Li ZS, Lungu O, Gershon MD. J Med Virol. 2003;70:S71–78. doi: 10.1002/jmv.10325. [DOI] [PubMed] [Google Scholar]
- 7.Hardy I, Gershon AA, Steinberg SP, LaRussa P. N Engl J Med. 1991;325:1545–1550. doi: 10.1056/NEJM199111283252204. [DOI] [PubMed] [Google Scholar]
- 8.Gomi Y, Sunamachi H, Mori Y, Nagaike K, Takahashi M, Yamanishi K. J Virol. 2002;76:11447–11459. doi: 10.1128/JVI.76.22.11447-11459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quinlivan ML, Gershon AA, Nichols RA, LaRussa P, Steinberg SP, Breuer J. J Infect Dis. 2006;193:927–930. doi: 10.1086/500835. [DOI] [PubMed] [Google Scholar]
- 10.Gomi Y, Imagawa T, Takahashi M, Yamanishi K. J Med Virol. 2000;61:497–503. doi: 10.1002/1096-9071(200008)61:4<497::aid-jmv13>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi M, Okuno Y, Otsuka T, Osame J, Takamizawa A. Biken J. 1975;18:25–33. [PubMed] [Google Scholar]
- 12.Quinlivan ML, Gershon AA, Steinberg SP, Breuer J. J Infect Dis. 2004;190:793–796. doi: 10.1086/423210. [DOI] [PubMed] [Google Scholar]
- 13.Westrop GD, Wareham KA, Evans DM, Dunn G, Minor PD, Magrath DI, Taffs F, Marsden S, Skinner MA, Schild GC. J Virol. 1989;63:1338–1344. doi: 10.1128/jvi.63.3.1338-1344.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattocks C, Tarpey P, Bobrow M, Whittaker J. Hum Mutat. 2000;16:437–443. doi: 10.1002/1098-1004(200011)16:5<437::AID-HUMU9>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 15.Qiu P, Soder GJ, Sanfiorenzo VJ, Wang L, Greene JR, Fritz MA, Cai XY. Biochem Biophys Res Commun. 2003;309:331–338. doi: 10.1016/j.bbrc.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Shiraki K, Yoshida Y, Asano Y, Yamanishi K, Takahashi M. J Infect Dis. 2003;188:875–877. doi: 10.1086/379835. [DOI] [PubMed] [Google Scholar]
- 17.Cohrs RJ, Gilden DH, Gomi Y, Yamanishi K, Cohen JI. J Virol. 2006;80:2076–2082. doi: 10.1128/JVI.80.5.2076-2082.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sato B, Ito H, Hinchliffe S, Sommer MH, Zerboni L, Arvin AM. J Virol. 2003;77:5607–5620. doi: 10.1128/JVI.77.10.5607-5620.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sommer MH, Zagha E, Serrano OK, Ku CC, Zerboni L, Baiker A, Santos R, Spengler M, Lynch J, Grose C, et al. J Virol. 2001;75:8224–8239. doi: 10.1128/JVI.75.17.8224-8239.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynch JM, Kenyon TK, Grose C, Hay J, Ruyechan WT. Virology. 2002;302:71–82. doi: 10.1006/viro.2002.1555. [DOI] [PubMed] [Google Scholar]
- 21.Frey CR, Sharp MA, Min AS, Schmid DS, Loparev V, Arvin AM. J Infect Dis. 2003;188:40–52. doi: 10.1086/375828. [DOI] [PubMed] [Google Scholar]
- 22.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 23.Cherkasova EA, Korotkova EA, Yakovenko ML, Ivanova OE, Eremeeva TP, Chumakov KM, Agol VI. J Virol. 2002;76:6791–6799. doi: 10.1128/JVI.76.13.6791-6799.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramer JM, LaRussa P, Tsai WC, Carney P, Leber SM, Gahagan S, Steinberg S, Blackwood RA. Pediatrics. 2001;108:E39. doi: 10.1542/peds.108.2.e39. [DOI] [PubMed] [Google Scholar]
- 25.Levin MJ, Dahl KM, Weinberg A, Giller R, Patel A, Krause PR. J Infect Dis. 2003;188:954–959. doi: 10.1086/378502. [DOI] [PubMed] [Google Scholar]
- 26.Levy O, Orange JS, Hibberd P, Steinberg S, LaRussa P, Weinberg A, Wilson SB, Shaulov A, Fleisher G, Geha RS. J Infect Dis. 2003;188:948–953. doi: 10.1086/378503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.