Abstract
We have found that MHC class II (MHC II) molecules exhibit a distinctive organization on the dendritic cell (DC) plasma membrane. Both in DC lysates and on the surface of living cells, I-A and I-E molecules engaged in lateral interactions not observed on other antigen-presenting cells such as B blasts. Because DCs and B blasts express MHC II at comparable surface densities, the interaction was not due to simple mass action. Instead, it reflected the selective expression of the tetraspanin CD9 at the DC surface. I-A and I-E molecules coprecipitated with each other and with CD9. The association of heterologous MHC II molecules was abrogated in DCs from CD9−/− mice. Conversely, expression of exogenous CD9 in B cells induced MHC II interactions. CD9 is thus necessary for the association of heterologous MHC II, a specialization that would facilitate the formation of MHC II multimers expected to enhance T cell receptor stimulation by DCs.
Keywords: antigen presentation, major histocompatibility complex class II, B cell, CD81, costimulatory molecules
The initiation of adaptive immunity requires presentation of antigens by dendritic cells (DCs) to naïve T lymphocytes (1). Why DCs are more efficient at stimulating naïve T cells than other antigen-presenting cells (APCs) remains incompletely understood. DC specializations that may contribute include the expression of high surface levels of MHC class II (MHC II) and costimulatory molecules, their regulated patterns of maturation and migration that coordinate their abilities to present antigens acquired in the periphery after arrival in lymphoid organs, their elaboration of long membrane projections that facilitate T cell contacts, and their secretion of an array of immunostimulatory cytokines (2). It seems likely, however, that other features remain to be discovered.
Upon antigen-specific contact between APC and CD4+ T cells, molecular reorganizations take place at the surfaces of both cells, including polarization of T cell receptor (TCR) and MHC II to the center of the contact site (3). Recently, MHC II enrichment at T cell contact site was shown to result from LFA-1–ligand interactions and to require the APC actin cytoskeleton (4), suggesting that the APC may not be a passive partner in T cell interactions. Further evidence that T cell-dependent, TCR- or adhesion molecule-driven MHC II recruitment may not fully explain T cell responses is suggested by the fact that T cells can respond to single ligands (5, 6), whereas T cell activation requires TCR cross-linking, possibly induced by MHC II multimers (7, 8). Previous work suggested that MHC II on DCs exhibits a nonrandom distribution at least at the level of immunofluorescence (IF) (9, 10). Such preexisting clusters could facilitate TCR ligation and cross-linking. Although DCs are well known to regulate the intracellular transport of their MHC II molecules (11), little is known about MHC II dynamics at the DC plasma membrane (PM).
MHC II molecules have been reported to cluster with each other and with MHC class I (12, 13); “superdimers” were even observed in the original three-dimensional crystal structure of human MHC II (14). A few reports documenting surface MHC II dimer of dimers have followed, (15–17) but neither the significance nor specificity of these interactions has been established nor has their formation in physiological settings been demonstrated.
MHC II molecules have also been reported to interact with tetraspanins, “master organizers” of the cell surface (18). Several groups have reported interactions of MHC II with numerous tetraspanin family members in whole cell lysates of APCs (19–24). Coordinated interactions with specific tetraspanins at intracellular or PM locations have been proposed to be involved in MHC II distribution and function (25). An Ab to clustered MHC II (26) was found to coprecipitate several tetraspanins in APCs (27).
Conceivably, surface features that differentiate DCs from other potent APCs such as B cells may contribute to the DC's enhanced ability for naïve T cell stimulation. CD9 is one such differentiating feature, a tetraspanin that is selectively expressed in DCs and facilitates the association of heterologous MHC II molecules.
Results
Expression and Distribution of MHC II and Costimulatory Molecules on DCs and B Blasts.
The enhanced efficiency of antigen presentation by DCs may in part simply reflect a higher level of expression of MHC and costimulatory molecules as compared with other APCs. To test this, we quantified the expression of MHC II and costimulatory molecules on mature bone marrow (BM)-derived DCs relative to B blasts, which also express high numbers of MHC II molecules. Quantitative flow cytometry was used, calibrated with fluorescence standards, to measure the numbers of MHC II (I-A or I-E), CD80, and CD86 molecules present on both cell types isolated from C57/B6 (I-Ab) or B10.BR (I-Ek) mice. Although the exact number of copies of a surface molecule cannot reliably be determined with the use of bivalent Ab, a reasonable and internally consistent approximation is possible.
Despite considerable cell to cell variation, it was clear that mature DCs expressed more of all three surface proteins (Table 1). In the case of MHC II, DCs had four to five times more than B blasts. Far greater differences were seen when expression of costimulatory molecules was compared. DCs expressed >10-fold more CD86 and CD80 than B blasts. Although earlier studies reported the numbers of MHC II molecules on the surface of human DCs severalfold higher (28, 29), the fact that our numbers are lower might reflect our having used a different species and Ab that were specific for only a single MHC II allele. In addition, we used directly conjugated primary Ab (molar ratio of fluor to IgG of 1:1), permitting stoichiometric comparisons with fluorescent beads and thus avoiding possible signal amplification by using fluorescent second Ab, as in previous studies.
Table 1.
Expression and surface density of MHC II products, CD80, and CD86 by DCs and B blasts
| Antibody | Mouse strain | Molecules per cell* |
Molecules per μm2 |
||
|---|---|---|---|---|---|
| DC† | B† | DC | B | ||
| I-Ab AF6 120.1 | C57/B6 | 111 ± 36 | 28 ± 9.0 | 77.9 | 115.7 |
| I-Ab TIB120‡ | C57/B6 | 191 ± 47 | 71 ± 43 | 134.1 | 290.5 |
| I-Ek TIB120‡ | B10.BR | 80 ± 21 | 16 ± 1.5 | 56.4 | 63.8 |
| I-Ek 14.4.4S | B10.BR | 124 ± 21 | 30 ± 4.2 | 87.3 | 124.6 |
| I-Ak 11-5.2 | B10.BR | 188 ± 77 | 39 ± 3.4 | 132.1 | 159.7 |
| CD86 GL1 | C57/B6, B10.BR | 208 ± 43 | 16 ± 1.6 | 146.3 | 67.3 |
| CD80 16-10A1 | C57/B6, B10.BR | 132 ± 20 | 1.6 ± 0.7 | 93.1 | 6.5 |
| CD40 3/23 | C57/B6, B10.BR | 17 ± 1.8 | 2.1 ± 1.0 | 11.9 | 8.5 |
| CD11a | C57/B6, B10.BR | 27 ± 7.6 | 9.7 ± 4.8 | 19.2 | 39.7 |
| Isotype control | C57/B6, B10.BR | 3.0 ± 1.6 | 0.9 ± 0.3 | 2.1 | 3.8 |
| Cell volume | 965 μm3 | 254 μm3 | |||
| Cell surface area | 1,421 μm2 | 243 μm2 | |||
Mature DCs or 24-hr B blasts were labeled at saturating concentrations with the indicated PE-conjugated Ab and processed for flow cytometry. Fluorescence standards were used to estimate the number of molecules per cell on mature DCs or fully blasted B cells by using beads with known quantities of fluors to perform a linear regression analysis. Numbers of molecules ± standard deviation (× 103) are listed for each Ab and mouse strain (columns 3 and 4). Pellets of mature DC or 24-hr B blasts were prepared for electron microscopy, serial sections were cut, and cell volume and surface area were calculated using the disector method. Numbers of molecules per square micron were obtained by dividing total number of molecules per cell by surface area in square microns (columns 5 and 6).
*Values are stated as numbers of molecules × 103.
†Gated on CD11c + or B220 + populations, respectively.
‡The mAb TIB 120 (M5/114.15.2) is specific for I-Ab, I-Aq, I-Ad, I-Ed, and I-Ek.
We normalized these numbers to account for differences in cell size. Mean surface area and volume for mature DCs and B blasts were determined from serial sections by electron microscopy and stereology. The average volume and surface area of DCs were four and six times greater than B blasts, respectively (Table 1), so that the effective density of MHC II molecules on the DC surface was ≈1.3 times lower than on B cell blasts. The density of costimulatory molecules, however, remained higher on DCs (2- to 14-fold), with the exception of CD40 present at comparable levels.
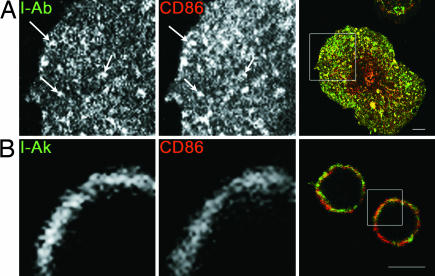
Although the density of MHC II was higher in B blasts, previous work suggested that DCs might organize their MHC II and CD86 in small patches on the PM (10). To determine whether this suggestion was unique to DCs, we visualized the distribution of these molecules in both cell types by IF. As before, the heterogeneous distribution of MHC II and CD86 was best visualized in DCs at an “intermediate” stage of maturation, i.e., before a fully mature “dendritic” phenotype was established (Fig. 1A). In contrast, in B blasts, MHC II and CD86 molecules always appeared homogeneously distributed (Fig. 1B). Morphological differences in these images reflect the fact that DCs spread when they attach to coverslips, whereas the B blasts remain spherical.
Fig. 1.
DCs, but not B cells, cluster surface MHC II and CD86. DC (A) were matured for 6–12 h, after which they or 24-h B blasts (B) were prepared for IF microscopy. Confocal images of a single plane are shown. (Scale bar, 5 μm.) Areas boxed in the right, merged panels are shown at higher magnification in individual colors in the left (MHC II, green) and middle (CD86, red) panels. Arrows show examples of PM regions with clustering of both molecules.
Lateral Interaction of Heterologous MHC II Molecules in Live DCs.
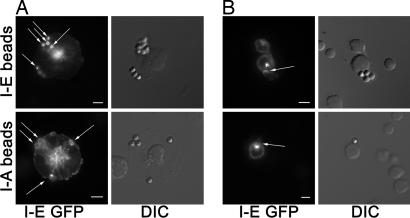
To confirm that the observed patchiness reflected the existence of lateral interactions that did not occur in B blasts, we developed an assay to determine whether cross-linking one species of MHC II resulted in the coclustering of a second MHC II species in live cells, avoiding possible artifacts involving Ab-induced clustering or cell fixation and permeabilization. Polystyrene latex beads were coated with anti-MHC II Ab specific for either I-A or I-E, allowed to interact with DCs or B cells expressing GFP-tagged I-E α chain, and then assayed by confocal microscopy to determine whether I-E-GFP clustered at sites of bead attachment. Under the conditions used, the beads largely remained at the surface of both DCs and B cells. The beads' regular geometry and 1:1 stoichiometry of GFP fluorescence to MHC II facilitated quantitation.
After a short incubation with beads at 37°C, anti-I-E-coated beads (I-E beads), as expected, efficiently clustered I-E-GFP on both cell types (Fig. 2A and B Upper), with brighter GFP fluorescence at sites of bead attachment than elsewhere on the cell. Unattached beads exhibited negligible autofluorescence. On DCs, the fluorescence was typically circumferential around the bead, not because the beads had been internalized but rather because they had adhered to the upper surface of flattened cells, with confocal images taken in a single X-Y plane. On B blasts, beads were generally bound to the cell margins, thus GFP fluorescence was limited in distribution to the point of attachment.
Fig. 2.
DCs, but not B cells, recruit irrelevant MHC II to site of bead contact. After adhering I-E-GFP-expressing DCs (A) or B blasts (B) to coverslips, Ab-coated beads were added and incubated with cells at 37°C for 10 min, followed by fixation and fluorescence microscopy. (Upper) I-E beads. (Lower) I-A beads. (Scale bars, 5 μm.) Confocal images of a single plane are shown (Left), and DIC images show bead position (Right). Arrows show sites of bead contact and/or I-E enrichment.
More interesting were the results obtained by using I-A beads. Despite weaker GFP fluorescence, I-A beads clustered I-E-GFP on DCs, but no such clustering was observed for I-A beads on B blasts (Fig. 2 A and B Lower, arrow). These results are reminiscent of studies reporting glycolipid raft-mediated clustering of irrelevant MHC II molecules at the synapse of T cells with a B cell line (30).
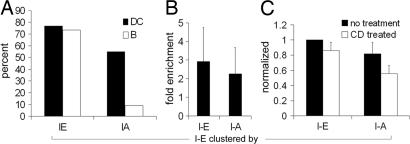
To ensure that the results did not simply reflect a nonspecific accumulation of membrane at the contact sites between beads and DCs or B blasts, we performed control experiments using uncoated beads, beads coated with irrelevant Ab (data not shown), or I-E beads bound to cells expressing CD86-cyan fluorescent protein (in addition to I-E-yellow fluorescent protein) as an unrelated PM protein. I-E beads failed to cluster CD86-cyan fluorescent protein on either DCs or B cell blasts, even at sites where there was abundant clustering of I-E-yellow fluorescent protein [see supporting information (SI) Fig. 7, arrows]. In this image, I-E-yellow fluorescent protein has been recruited to the beads to such an extent that the I-E-yellow fluorescent protein on the remainder of the cell has been depleted, making the overall fluorescence comparatively low. We quantified these data by counting beads in contact with a GFP-expressing cell, scoring the frequency at which the bound beads induced morphologically identifiable clustering (Fig. 3A). Clustering of I-E by I-E-beads (specific clustering) occurred to similar extents in both DCs and B blasts. The frequency of I-E-GFP clustering by I-A beads (nonspecific clustering), however, was 77% that of specific clustering in DCs, whereas in B blasts it was <10%. In those rare instances in which I-E-GFP clustering occurred in B cells, it was barely detectable. Indeed, what appeared as clustering in 2 out of 22 cells scored as positive might have reflected I-E-GFP fluorescence in the Golgi/recycling endosome region of the B cell cytoplasm, which was occasionally close to the PM (Fig. 2B).
Fig. 3.
Quantitation of bead-associated fluorescence. (A) DC vs. B blast bead-induced clustering. The percentage of beads in contact with I-E-GFP-expressing APCs with enrichment of I-E upon incubation with I-A or I-E beads was quantified. DCs, filled bars; B blasts, open bars. (B) DC-specific vs. nonspecific clustering. Enrichment of I-E fluorescence at sites of bead contact vs. elsewhere on the same cell were quantified by fluorescence intensity (see Materials and Methods); fold enrichment of I-E clustering by I-E or I-A beads is shown. (C) Effect of methyl-β-cyclodextrin treatment. After adhering DCs to coverslips, they were treated with 30 mM methyl-β-cyclodextrin, beads were added and processed as above, and fold enrichment of I-E by I-E and I-A beads was measured. Fold enrichment without (filled bars) and with (open bars) cyclodextrin treatment was normalized to without treatment.
The clustering results were further analyzed by measuring relative fluorescence at sites of bead contact vs. nonbead regions of the PM of DCs. Quantitation revealed a 3-fold enrichment of I-E-GFP fluorescence by I-E beads and 2.25-fold by the I-A beads; these were not statistically different (Fig. 3B). Thus, there appeared to be a significant degree of lateral association of I-A and I-E molecules on the PM of DCs. Because a fraction of MHC II has been observed to associate with glycolipid-cholesterol-enriched lipid microdomains (rafts) in B cells and in DCs (31–33), and because complexes of MHC II with other molecules including the tetraspan CD9 (see below) have been reported to be slightly enriched in lipid rafts (24), we wondered whether these microdomains might be involved in corecruitment of I-A and I-E molecules using I-E beads. To test this possibility, I-E-GFP-expressing DCs and B blasts were treated with methyl-β-cyclodextrin to deplete cholesterol and then challenged with I-E or I-A beads. Although the amount of I-A corecruitment was decreased in the treated cells, it decreased in direct proportion to a decrease in I-E clustering (Fig. 3C). These results, while not excluding a role for rafts, provided no evidence in favor of cholesterol-dependent lipid microdomains as responsible for the recruitment of irrelevant MHC II.
I-E and I-A Molecules Can Be Coimmunoprecipitated from DC but Not B Cell Lysates.
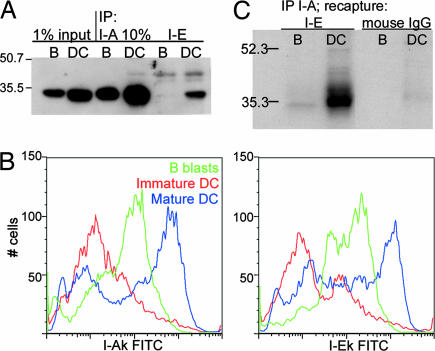
Because glycolipid rafts did not appear to mediate the association between I-A and I-E molecules, we asked whether protein–protein interactions might be responsible. Mature DCs and B blasts were lysed in CHAPS and subjected to immunoprecipitation (IP) by using I-A or I-E Ab. Twice as much total protein was used in B blast as in DC lanes to adjust for higher levels of expression of MHC II in the DCs (Table 1). The precipitates were then probed by Western blotting by using an Ab to I-A (available reagents for I-E were not useful for Western blotting). Input lanes (Fig. 4A, left lanes) and I-A IPs (center lanes) confirmed that comparable amounts of I-A were present in normalized B and DC lysates. I-E IPs, however, contained considerably more I-A in DCs than in B blasts (right lanes in Fig. 4A). This difference did not reflect a disparity in relative levels of I-A and I-E expression because both cell types express both alleles at similar ratios (Fig. 4B). As a control, we used Ab to CD86, another molecule expressed almost exclusively at the DC or B cell surface; only negligible amounts of I-A were detected as coprecipitating with CD86 in either cell type (data not shown).
Fig. 4.
MHC II I-E associates with I-A in DC but not in B blasts. (A) Immunoprecipitations of 1% CHAPS lysates of B blasts and DCs were followed by SDS/PAGE and Western blotting for I-A. B blast lanes contain 2-fold the protein concentration of DC lanes to correct for amounts of MHC II. Lanes 1 and 2 illustrate 1% input of lysates used for IP. I-A lanes, IP with mAb 10-2.16; I-E lanes, IP with mAb 14.4.4S. (B) I-A (Left) and I-E (Right) expression on B blasts (green lines), immature DC (red lines), and mature DC (blue lines) by FACS. (C) DCs (lanes 2 and 4) or B blasts (lanes 1 and 3) were metabolically labeled with 35S, and after an overnight chase, I-A molecules were immunoprecipitated with mAb 10-2.16. Material was released from the beads by treating with Triton X-100, and I-E was recaptured from the IP with mAb 14.4.4S (lanes 1 and 2) or control IgGs (lanes 3 and 4), followed by SDS/PAGE and fluorography.
The biochemical association of I-E with I-A could also be shown via sequential IP following metabolic labeling. Mature DCs and B blasts were labeled with [35S]Met/Cys and, after an overnight chase, I-A molecules were immunoprecipitated from CHAPS lysates. The presence of I-E was determined by recapture of I-E from the I-A IP, followed by SDS/PAGE and fluorography. A band of the correct molecular weight was present in the DC lysate (Fig. 4C, lane 2), with only trace amounts in the case of B blasts (lane 1). No band was recaptured by using a control Ig (lanes 3 and 4).
Because CHAPs is well known to preserve protein–protein interactions, we repeated these experiments using Triton X-100 lysates, another nonionic detergent that more often disrupts such interactions. I-A could not be detected by Western blotting as having coprecipitated with I-E in Triton X-100 lysates from either B cells or DCs (data not shown), consistent with the possibility that protein–protein interactions were responsible.
The DC-Enriched Tetraspanin CD9 Forms a Complex with MHC II Molecules.
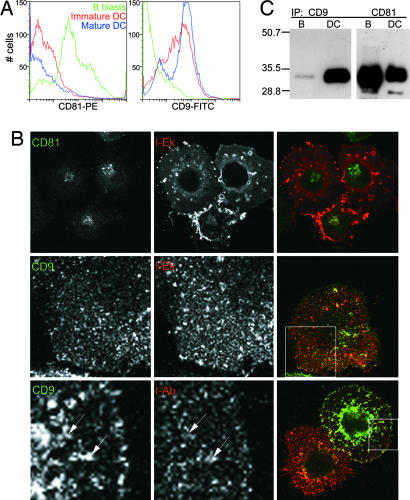
An association between I-A and I-E in DCs might reflect a direct or indirect interaction among dissimilar MHC II molecules. We sought to test whether adaptor proteins were involved, considering members of the tetraspanin family and given the many previous reports of their interactions with MHC II products. Most previous work, however, focused on potential interactions in B cell lines, concentrating on tetraspanins that localized predominantly to MHC II-positive lysosomal compartments as opposed to the DC PM (19–22). In both human B lymphoblastoid cell lines and monocyte-derived DCs, cell fractionation has shown that the MHC II-associated tetraspanins CD63 and CD82 are predominantly localized to lysosomes (23, 25, 34). In contrast, in human monocyte-derived DCs, CD9, CD53, and CD81 are largely nonlysosomal and probably mostly at the PM (25, 34). We screened mouse BM-derived DCs and B blasts for their expression and localization of two predominantly surface tetraspanins, CD9 and CD81. Although CD81 was highly expressed on the surface of B blasts, it was barely detectable at the DC surface by FACS (Fig. 5A). In contrast, CD9 was found on the PM of DCs, enhanced slightly during maturation, at levels >10-fold higher than B blasts, (Fig. 5A). Although the roles of some tetraspanins may be overlapping or redundant (18), we reasoned that because CD9 was expressed at such high levels on DCs, and because at least one other tetraspanin (CD81) was present at much lower levels, CD9 represented the best candidate for facilitating the observed MHC II interactions.
Fig. 5.
Selective association of CD9 with MHC II in DCs as compared with B blasts. (A) Expression of selected tetraspanin molecules on APCs. Green lines, B blasts; red lines, immature DC; blue lines, mature DC. (Left) CD81. (Right) CD9. (B) IF of DCs labeled with I-Ek (red) and the following: (Top) CD81 (green, permeabilized); (Middle) CD9 (green, permeabilized); and (Bottom) CD9 (green, live, nonpermeabilized labeling on ice). Boxed areas on merged right panels are shown at higher magnification in individual colors. Arrows indicate examples of partial CD9 and MHC II surface colocalization. Confocal images of a single plane are shown. (C) Immunoprecipitations and Western blotting for I-A were carried out as in Fig. 4A. CD9 IPs (lanes 1 and 2) as compared with CD81 IPs (lanes 3 and 4) from B blasts and DCs, respectively.
By IF microscopy of fixed and permeabilized cells, CD81 expression on the surface of DCs was found to be low and moreover localized largely to intracellular compartments (Fig. 5B Top). In contrast, CD9 was observed predominately at the PM where it exhibited a patchy distribution, similar to MHC II (Fig. 5B Middle). In fact, it appeared that the distributions of surface CD9 and MHC II at least partially overlapped when live DCs were labeled with Ab to both markers (Fig. 5B Bottom).
We next asked whether either or both CD9 and CD81 would co-IP MHC II molecules. After IPs of CHAPS lysates using anti-CD9 or anti-CD81 Ab, the precipitates were probed for the presence of I-A molecules by Western blot analysis (Fig. 5C). As shown in Fig. 5C Left, far more I-A was present in CD9 IPs from DCs than from B blasts. In the case of CD81, however, similar amounts of I-A were coprecipitated from DCs and B blasts (Right).
CD9 Is Necessary for I-A/I-E Associations.
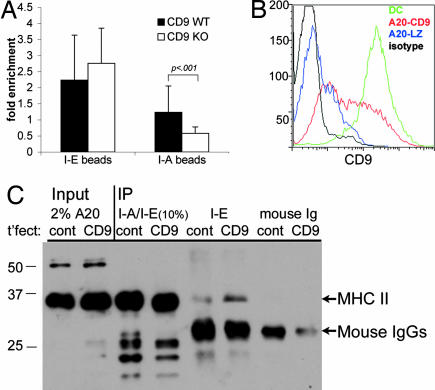
To test whether the I-A/I-E association requires CD9, BM-derived DCs were cultured from CD9−/− mice (35) and tested for I-A/I-E association by using I-A or I-E beads to cluster retrovirally expressed I-E-GFP. In these experiments, the fold enrichment of both specific and nonspecific clustering at sites of bead contact was slightly lower (e.g., Fig. 3) because of a lower level of I-E-GFP expression in the cells used. Nevertheless, after quantitation of fluorescence at sites of bead attachment (performed as in Fig. 3B), it was evident that, in the absence of CD9, no enrichment of heterologous MHC II occurred (Fig. 6A). Thus, CD9 was necessary for heterologous MHC II association in this assay.
Fig. 6.
CD9 is necessary for I-A/I-E clustering. (A) DCs raised from the BM of CD9 WT vs. knockout mice and retrovirally transduced with I-Eα-GFP were adhered to coverslips and allowed to interact with I-E- or I-A-coated beads as in Fig. 2. Fluorescence at sites of bead adherence was quantified as described in Fig. 3. (B) A20 B lymphoma cells were transfected with CD9 cDNA (red line) or lacZ as a control (blue line). DCs are shown for comparison (green line). Black line, isotype control. (C) CHAPS lysates (1%) of stable cells lines (A20 cells transfected with CD9, lane 6, or a control plasmid, cont., lane 5) were immunoprecipitated with Ab to I-A or I-E as in Fig. 4, and I-A was visualized by Western blotting. Total MHC II was precipitated with TIB120, specific for both I-A and I-E in these cells (lanes 3 and 4). IP with an irrelevant Ab is shown in lanes 7 and 8. Arrows indicate MHC II and mouse IgG bands.
To confirm independently that CD9 was responsible for mediating the interaction between I-A and I-E, we performed a gain-of-function experiment, using B cells as naturally occurring CD9-negative cells, and asked whether I-A/I-E interactions could be induced by expression of exogenous CD9. A20 B lymphoma cells, in which, as in primary blasts, I-E association with I-A was found to be negligible, were transfected with the CD9 cDNA (36), and stable lines were created (Fig. 6B). Transfectants expressed highly variable levels of CD9, and only a small minority of cells expressed levels of CD9 comparable to DCs (Fig. 6B). As shown in Fig. 6C, upon exogenous expression of CD9, I-E IPs were observed to contain I-A by Western blotting (Fig. 6C, I-E lanes; bands corresponding to MHC II and IgG are indicated with arrows). Relatively little I-A was detected by blotting I-E IPs from the CD9-negative control. The exogenous expression of CD9 is thus sufficient, given the presence of other tetraspanins, for inducing I-A/I-E association in an otherwise CD9-negative B cell line.
Discussion
We compared DCs to B blasts to seek differences possibly responsible for DCs' superior antigen presentation ability. As expected, DCs express higher numbers of MHC II and costimulatory molecules than B blasts. It is surprising, however, that B blasts, known to be less efficient at T cell stimulation, actually achieve a higher density of MHC II molecules per unit of PM. These results are consistent with earlier findings that B cells require 10-fold more MHC-peptide complexes than DCs to effect the same level of T cell stimulation (37). Thus, although DCs are able to deliver a more potent stimulus to T cells, they do so while expressing a lower surface density of TCR ligand. Although this may in part be explained by DCs' higher density of CD86 and CD80, we propose that surface organization of MHC II may also play a role in facilitating antigen presentation. The patchy distribution of DC surface molecules presents the T cell with preclustered “packets” of MHC II I-A and I-E, potentiating T cell activation by facilitating TCR clustering and signaling (7, 38). The DC is thus able to distribute TCR ligands over a larger surface area, allowing for simultaneous interaction with multiple T cells.
Even with clustered TCR ligand, the next challenge is finding the agonist TCR ligands in a large excess of self-peptide complexes. The unexpected finding of association between heterologous MHC II molecules in DCs provided us with a possible mechanism for TCR clustering. The pseudodimer model proposes interaction of two TCRs with adjacent MHC II-peptide complexes, one loaded with an endogenous, the other with an agonist, peptide, the two receptor-ligand pairs bridged by a CD4 molecule (5, 39). With homogeneously distributed, freely diffusing MHC II molecules, there would be one MHC II molecule per ≈9,200 nm2 of surface area, placing each an average of 96 nm away from its nearest neighbor, such that the probability of forming the pseudodimer would appear to be extremely low. Although lateral diffusion may compensate, tethering similar or dissimilar MHC II molecules together increases the likelihood that a CD4 molecule can bind a neighboring MHC II molecule. Although we did not measure the association of homologous MHC II molecules, we surmise that I-E/I-E pairs are just as likely as I-E/I-A ones. From the standpoint of a role in TCR stimulation, the existence of homodimers would be just as important.
Our results further indicate that the discontinuous distribution of MHC II on the DC surface and the lateral interaction of heterologous MHC II products are mediated by association with members of the tetraspanin family. Although association of MHC II with tetraspanins has been observed, previous work has yet to assign a physiologically relevant function to a specific tetraspanin in APCs. We have provided direct evidence that a major surface tetraspanin on DCs, CD9, plays an essential role in physically tethering dissimilar MHC II molecules together in a fashion that distinguishes DCs from other APCs. We also documented the MHC II/CD9 association in both gain-of-function and loss-of-function experiments, focusing on CD9 as the tetraspanin present in our compartment of interest, the DC PM.
Our results are consistent with but also quite distinct from recent work of Kropshofer et al. (27). In human DCs, an Ab (CDw78) thought to detect “clustered” MHC II was found to coprecipitate the tetraspanins CD81 and CD9, data interpreted to suggest the existence of a tetraspanin domain either at the DC PM and/or intracellularly (27). This pool of MHC II molecules also contained some that were loaded with the invariant chain-derived peptide CLIP, which could be found at T cell interaction sites where it may act to modulate the T cell response to produce Th2 lymphocytes (40). Possibly any tetraspanin localized to the PM could mediate the observed MHC II interactions, but in DCs it is CD9 that is present at high levels and carries out this role. Thus, at a minimum, “tetraspanin domains” on the DC PM would reflect the ability of CD9 to facilitate the lateral interactions of individual MHC II molecules.
Although our data provide no evidence for a role for glycolipid rafts in mediating the observed heterologous MHC II associations, they do not exclude the possibility that these microdomains are involved. The presence of irrelevant MHC II molecules in the immunological synapse (30, 41) is intriguing and may or may not be related to the findings presented here. The MHC II interactions we observe are present before T cell contact and the extensive membrane reorganizations that happen upon antigen-specific encounter between APC and T cell.
Although the question of the function of CD9 in antigen presentation is beyond the scope of this paper, it is interesting to note that ligation of CD9 (by pregnancy-specific glycoprotein 17) inhibits the proliferation of naïve T cells, suggesting a role for CD9 in T cell responses (42).
DCs are not the only cells known to express CD9: defined subsets of B cells have been reported to express CD9 (43), and interestingly it is the same CD9+ subset which has been shown to be more efficient at stimulating naive CD4+ T cells (44). It is possible that the improved T cell stimulatory capacity of CD9-expressing B cells is due to their ability to phenocopy the DC PM, at least with respect to the lateral association of MHC II. With the finding that CD9 is capable of inducing interactions of MHC II in living cells, we have defined one of the very few discrete biochemical roles yet described for this or any other member of the tetraspanin family. It will now be possible to interrogate this interaction by using a variety of approaches to define the larger functional significance of this property of CD9 and MHC II.
Methods
For additional details, see SI Methods.
Mice and Cells.
DC were grown as described (45) from BM progenitors of B10/BR, C57/Black6, (Jackson Labs, Bar Harbor, ME), or CD9 WT vs. knockout mice (35). B blasts were prepared from B10.BR or C57/Black6 spleens as described (46). A20 B lymphoma cells were transfected with the CD9 cDNA in pEF6/V5-His [(36); gift of G. Dveksler, Uniformed Services University of the Health Sciences, Bethesda, MD] or a control plasmid, lacZ-pEF6/V5-His (Invitrogen, Carlsbad, CA), and stable lines created by selection with blasticidin (Sigma, St. Louis, MO), at 2 μg/ml.
Antibodies and IF.
If was done as described (9) by using the following monoclonal Ab: KH74 (I-Ab), GL1 (CD86), AMS 32.1 (I-Ad), and CD81 (TAPA-1) from BD-PharMingen (San Diego, CA); TIB120 (M5/114.15.2, I-A/I-E) and 14.4.4S (I-E) from ATCC (Manassas, VA); CD9 (KMC8.8; BD-PharMingen and ATCC); and 10-2.16 (I-Ak) and Y3P (I-A) (gifts of the Bottomly/Janeway laboratory, Yale University, New Haven, CT). Polyclonal rabbit anti-I-Aβ were also used (Rivoli and Thorax) (10, 47).
For cyclodextrin treatment, DCs were plated as usual on coverslips, washed in serum-free RPMI medium 1640, and then incubated with 30 mM methyl-β-cyclodextrin (Sigma) for 10 min at 20°C, followed by three RPMI medium 1640 washes, after which they were used in the bead assay.
Cell Size Calculations and Stereology.
B cells and DCs were fixed, dehydrated, and embedded in Epon, and serial sections were cut and examined by electron microscopy. Stereology was used to estimate the cell size by using the dissector method.
Bead Assays.
Ab were bound to Dynabeads protein A (Dynal, Great Neck, NY) per manufacturer instructions. Cells were plated as for IF, then 1–5 × 105 beads were added to cells in complete medium and centrifuged for 2 min at 70 × g, incubated at 37°C for 10 min, fixed with 4% paraformaldehyde, and visualized as above. Quantitation of conflated confocal x-z sections was done by using the following formula: fold enrichment = (bead fluorescence − autofluorescence of bead not on GFP-expressing cell − background)/[bead area cell fluorescence (next to bead) − background].
FACS Analysis.
Flow cytometry was done with a FACSCalibur and CellQuest software (Becton Dickinson, Franklin Lakes, NJ) for acquisition and FloJo (TreeStar, Ashland, OR) was used for analysis. Numbers of surface molecules on CD11c+ or B220+ populations were quantified by using QuantiBrite phycoerythrin beads (BD Biosciences, San Jose, CA) as per manufacturer instructions. Events (104) were collected in each of three experiments.
Statistical Analysis.
The two-tailed Student t test was used to determine statistical significance.
Biochemical Assays.
CHAPS lysates (1%) of mature DC or B blasts after 24 h of culture were precleared with protein G Sepharose beads (Amersham Pharmacia, Piscataway, NJ), followed by 4–15 h IP of 1 mg lysate with 10 μg mAb bound to protein G Sepharose, and analyzed by 12 or 15% SDS/PAGE (Bio-Rad, Hercules, CA), transferred to nitrocellulose membrane (Schleicher and Schuell, Florham Park, NJ), Western blotted, and visualized by SuperSignal Femto chemiluminescence reagent (Pierce, Rockford, IL).
Metabolic Labeling and Immunoprecipitation/Recapture.
Cells (DC at day 5, 6 h after maturation stimulus and B blasts after 3 h LPS stimulation) were washed in Cys/Met-free media and then pulsed with [35S]Cys/Met (Amersham Biosciences) for 1 h at 37°C, 5% CO2. After two PBS washes, cells were cultured overnight in complete media. After first IP, material was eluted from beads with 1% Triton X-100 in 100 mM Tris·HCl (pH 7.4). After SDS/PAGE, gel was soaked in 1 M salicylic acid (Sigma) for 1 h before processing for fluorography.
Supplementary Material
Acknowledgments
We thank Gabriela Dveksler and Carolyn Zalepa (Uniformed Services University of Health Sciences, Bethesda, MD) for the gift of CD9 knockout mice; Rachael Couture for animal husbandry and superb technical assistance; Ona Bloom, Nathan Sherer, and Walther Mothes for providing valuable feedback on the manuscript; and members of the I.M. laboratory group for helpful discussions and support. This work was supported by National Institutes of Health Grant R37-AI34098; I.M. is an Affiliate Member of the Ludwig Institute for Cancer Research.
Abbreviations
- DC
dendritic cell
- MHC II
MHC class II
- APC
antigen-presenting cell
- TCR
T cell receptor
- IP
immunoprecipitation
- PM
plasma membrane
- IF
immunofluorescence
- BM
bone marrow
- I-E beads
anti-I-E-coated beads.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609665104/DC1.
References
- 1.Banchereau J, Steinman RM. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 2.Mellman I, Steinman RM. Cell. 2001;106:255–258. doi: 10.1016/s0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 3.Lin J, Miller MJ, Shaw AS. J Cell Biol. 2005;170:177–182. doi: 10.1083/jcb.200503032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de la Fuente H, Mittelbrunn M, Sanchez-Martin L, Vicente-Manzanares M, Lamana A, Pardi R, Cabanas C, Sanchez-Madrid F. Mol Biol Cell. 2005;16:3314–3322. doi: 10.1091/mbc.E05-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irvine DJ, Purbhoo MA, Krogsgaard M, Davis MM. Nature. 2002;419:845–849. doi: 10.1038/nature01076. [DOI] [PubMed] [Google Scholar]
- 6.Purbhoo MA, Irvine DJ, Huppa JB, Davis MM. Nat Immunol. 2004;5:524–530. doi: 10.1038/ni1058. [DOI] [PubMed] [Google Scholar]
- 7.Boniface JJ, Rabinowitz JD, Wulfing C, Hampl J, Reich Z, Altman JD, Kantor RM, Beeson C, McConnell HM, Davis MM. Immunity. 1998;9:459–466. doi: 10.1016/s1074-7613(00)80629-9. [DOI] [PubMed] [Google Scholar]
- 8.Cochran JR, Cameron TO, Stern LJ. Immunity. 2000;12:241–250. doi: 10.1016/s1074-7613(00)80177-6. [DOI] [PubMed] [Google Scholar]
- 9.Pierre P, Turley SJ, Gatti E, Hull M, Meltzer J, Mirza A, Inaba K, Steinman RM, Mellman I. Nature. 1997;388:787–792. doi: 10.1038/42039. [DOI] [PubMed] [Google Scholar]
- 10.Turley SJ, Inaba K, Garrett WS, Ebersold M, Unternaehrer J, Steinman RM, Mellman I. Science. 2000;288:522–527. doi: 10.1126/science.288.5465.522. [DOI] [PubMed] [Google Scholar]
- 11.Trombetta ES, Mellman I. Annu Rev Immunol. 2005;23:975–1028. doi: 10.1146/annurev.immunol.22.012703.104538. [DOI] [PubMed] [Google Scholar]
- 12.Setum CM, Serie JR, Hegre OD. Anat Rec. 1993;235:285–295. doi: 10.1002/ar.1092350212. [DOI] [PubMed] [Google Scholar]
- 13.Jenei A, Varga S, Bene L, Matyus L, Bodnar A, Bacso Z, Pieri C, Gaspar R, Jr, Farkas T, Damjanovich S. Proc Natl Acad Sci USA. 1997;94:7269–7274. doi: 10.1073/pnas.94.14.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown JH, Jardetzky TS, Gorga JC, Stern LJ, Urban RG, Strominger JL, Wiley DC. Nature. 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 15.Schafer PH, Pierce SK. Immunity. 1994;1:699–707. doi: 10.1016/1074-7613(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 16.Roucard C, Garban F, Mooney NA, Charron DJ, Ericson ML. J Biol Chem. 1996;271:13993–14000. doi: 10.1074/jbc.271.24.13993. [DOI] [PubMed] [Google Scholar]
- 17.Cherry RJ, Wilson KM, Triantafilou K, O'Toole P, Morrison IE, Smith PR, Fernandez N. J Cell Biol. 1998;140:71–79. doi: 10.1083/jcb.140.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hemler ME. Nat Rev Mol Cell Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 19.Angelisova P, Hilgert I, Horejsi V. Immunogenetics. 1994;39:249–256. doi: 10.1007/BF00188787. [DOI] [PubMed] [Google Scholar]
- 20.Schick MR, Levy S. J Immunol. 1993;151:4090–4097. [PubMed] [Google Scholar]
- 21.Rubinstein E, Le Naour F, Lagaudriere-Gesbert C, Billard M, Conjeaud H, Boucheix C. Eur J Immunol. 1996;26:2657–2665. doi: 10.1002/eji.1830261117. [DOI] [PubMed] [Google Scholar]
- 22.Szollosi J, Horejsi V, Bene L, Angelisova P, Damjanovich S. J Immunol. 1996;157:2939–2946. [PubMed] [Google Scholar]
- 23.Hammond C, Denzin LK, Pan M, Griffith JM, Geuze HJ, Cresswell P. J Immunol. 1998;161:3282–3291. [PubMed] [Google Scholar]
- 24.Zilber MT, Setterblad N, Vasselon T, Doliger C, Charron D, Mooney N, Gelin C. Blood. 2005;106:3074–3081. doi: 10.1182/blood-2004-10-4094. [DOI] [PubMed] [Google Scholar]
- 25.Engering A, Pieters J. Int Immunol. 2001;13:127–134. doi: 10.1093/intimm/13.2.127. [DOI] [PubMed] [Google Scholar]
- 26.Drbal K, Angelisova P, Rasmussen AM, Hilgert I, Funderud S, Horejsi V. Int Immunol. 1999;11:491–498. doi: 10.1093/intimm/11.4.491. [DOI] [PubMed] [Google Scholar]
- 27.Kropshofer H, Spindeldreher S, Rohn TA, Platania N, Grygar C, Daniel N, Wolpl A, Langen H, Horejsi V, Vogt AB. Nat Immunol. 2002;3:61–68. doi: 10.1038/ni750. [DOI] [PubMed] [Google Scholar]
- 28.Cella M, Engering A, Pinet V, Pieters J, Lanzavecchia A. Nature. 1997;388:782–787. doi: 10.1038/42030. [DOI] [PubMed] [Google Scholar]
- 29.Bhardwaj N, Young JW, Nisanian AJ, Baggers J, Steinman RM. J Exp Med. 1993;178:633–642. doi: 10.1084/jem.178.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hiltbold EM, Poloso NJ, Roche PA. J Immunol. 2003;170:1329–1338. doi: 10.4049/jimmunol.170.3.1329. [DOI] [PubMed] [Google Scholar]
- 31.Meyer zum Bueschenfelde CO, Unternaehrer J, Mellman I, Bottomly K. J Immunol. 2004;173:6119–6124. doi: 10.4049/jimmunol.173.10.6119. [DOI] [PubMed] [Google Scholar]
- 32.Setterblad N, Roucard C, Bocaccio C, Abastado JP, Charron D, Mooney N. J Leukoc Biol. 2003;74:40–48. doi: 10.1189/jlb.0103045. [DOI] [PubMed] [Google Scholar]
- 33.Anderson H, Hiltbold E, Roche P. Nat Immunol. 2000;1:156–162. doi: 10.1038/77842. [DOI] [PubMed] [Google Scholar]
- 34.Mantegazza AR, Barrio MM, Moutel S, Bover L, Weck M, Brossart P, Teillaud JL, Mordoh J. Blood. 2004;104:1183–1190. doi: 10.1182/blood-2004-01-0104. [DOI] [PubMed] [Google Scholar]
- 35.Le Naour F, Rubinstein E, Jasmin C, Prenant M, Boucheix C. Science. 2000;287:319–321. doi: 10.1126/science.287.5451.319. [DOI] [PubMed] [Google Scholar]
- 36.Waterhouse R, Ha C, Dveksler GS. J Exp Med. 2002;195:277–282. doi: 10.1084/jem.20011741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inaba K, Steinman RM. J Exp Med. 1984;160:1717–1735. doi: 10.1084/jem.160.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kupfer A, Singer SJ. Proc Natl Acad Sci USA. 1988;85:8216–8220. doi: 10.1073/pnas.85.21.8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krogsgaard M, Li QJ, Sumen C, Huppa JB, Huse M, Davis MM. Nature. 2005;434:238–243. doi: 10.1038/nature03391. [DOI] [PubMed] [Google Scholar]
- 40.Rohn TA, Boes M, Wolters D, Spindeldreher S, Muller B, Langen H, Ploegh H, Vogt AB, Kropshofer H. Nat Immunol. 2004;5:909–918. doi: 10.1038/ni1108. [DOI] [PubMed] [Google Scholar]
- 41.Wulfing C, Sumen C, Sjaastad MD, Wu LC, Dustin ML, Davis MM. Nat Immunol. 2002;3:42–47. doi: 10.1038/ni741. [DOI] [PubMed] [Google Scholar]
- 42.Bebo BF, Jr, Dveksler GS. Curr Drug Targets Inflamm Allergy. 2005;4:231–237. doi: 10.2174/1568010053586255. [DOI] [PubMed] [Google Scholar]
- 43.Won WJ, Kearney JF. J Immunol. 2002;168:5605–5611. doi: 10.4049/jimmunol.168.11.5605. [DOI] [PubMed] [Google Scholar]
- 44.Attanavanich K, Kearney JF. J Immunol. 2004;172:803–811. doi: 10.4049/jimmunol.172.2.803. [DOI] [PubMed] [Google Scholar]
- 45.Inaba K, Inaba M, Romani N, Aya H, Deguchi M, Ikehara S, Muramatsu S, Steinman RM. J Exp Med. 1992;176:1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hodgkin PD, Yamashita LC, Coffman RL, Kehry MR. J Immunol. 1990;145:2025–2034. [PubMed] [Google Scholar]
- 47.Shin J-S, Ebersold M, Pypaert M, Delamarre L, Hartley A, Mellman I. Nature. 2006;444:115–118. doi: 10.1038/nature05261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.