Abstract
Nuclear functions for IκB kinase (IKK), including phosphorylation of histone H3 and nuclear corepressors, have been recently described. Here, we show that IKK is activated in colorectal tumors concomitant with the presence of phosphorylated SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) corepressor that is aberrantly localized in the cytoplasm. In these tumors, IKKα associates to the chromatin of specific Notch targets, leading to the release of SMRT. Abrogation of IKK activity by BAY11-7082 or by expressing dominant negative IKKα restores the association of SMRT with Notch target genes, resulting in specific gene repression. Finally, BAY11-7082 significantly reduces tumor size in colorectal cancer xenografts (CRC-Xs) implanted in nude mice.
Keywords: hes1, SMRT, corepressor
Solid tumors are the most common type of cancer and their malignancy is mainly associated with increased proliferation capacity, decreased apoptosis and as well as the ability to keep away from the differentiation program. To achieve these characteristics, colorectal cancer cells accumulate multiple mutations in specific sets of genes. Mutation of adenomatous polyposis coli (APC) that leads to the accumulation of nuclear β-catenin, is the most prevalent event in this multistep process (1, 2). However not only β-catenin-dependent transcription is required for colorectal cancer progression, because abrogation of the Notch pathway prevents tumorigenesis in APCmin/+ mice whereas activation of Notch results in the expansion of the progenitor compartment in the intestine (3, 4). A role for Notch signaling in cancer has extensively been reported (reviewed in ref. 5) and increased expression of the Notch-target gene hes1 is a common feature in many neoplasias such as intestinal adenomas (3, 6), meningiomas (7), medulloblastomas (8), and leukemias (9). This observation is consistent with the role of Notch-target genes in maintaining the undifferentiated phenotype in the proliferative compartments of the intestinal epithelium (4) and other tissues (10–14).
NFκB signaling pathway is involved in inflammatory and immune responses as well as in tumor development in breast, liver or intestine (15, 16). In the classical NFκB pathway, a ternary IκB kinase complex (IKK) formed by IKKα, IKKβ, and NFκB essential modulator (NEMO) is responsible for inducing IκB phosphorylation, thus leading to the activation of the NFκB pathway. An alternative pathway involving the activation of IKKα is responsible for phosphorylating p100, inducing its processing to p52 that together with RelB activates specific gene transcription (reviewed in ref. 17). Recently, chromatin-associated functions for IKK have been described (18–23). For example, in response to TNF-α, EGF, or estrogens, IKKα associates with downstream target promoters to phosphorylate components of the transcriptional complex including Ser-10 of histone 3, resulting in gene activation (18, 21–23). Laminin attachment induces the association of IKKα to the cIAP2 and IL-8 promoters to phosphorylate Ser-2410 of the nuclear corepressor SMRT (silencing mediator of retinoic acid and thyroid hormone receptor), promoting its release and permitting transcriptional activation (20). TNF-α also induces recruitment of IKK to the Notch targets hes1 and herp2/hrt1 leading to gene activation (19), however the possibility that IKK regulate Notch target genes in colorectal cancer remains to be elucidated.
In this work, we show that IKK activity plays an NFκB-independent role in colorectal cancer. In these cancer cells, IKKα is aberrantly activated and recruited to the chromatin in a variety of Notch targets, including hes1, hes5, and herp2, leading to the release of SMRT and the transcriptional activation of these genes. In addition, inhibition of IKK activity restores the association of SMRT with the chromatin directly affecting specific gene transcription, apoptosis and tumor cell growth in nude mice.
Results
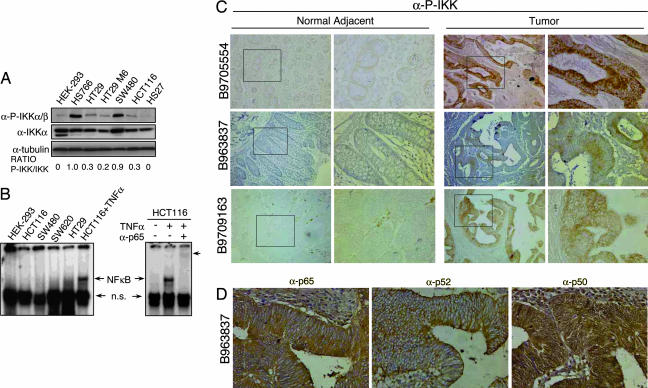
To investigate whether the NFκB pathway plays a role in colorectal cancer, we analyzed the IKK activity with the α-P-IKKα/β antibody (20) in different cancer cell lines. As shown in Fig. 1A, total levels of IKKα protein were comparable among cancer cells and HEK-293 or HS27 controls. In contrast, activated/phosphorylated IKK proteins were predominantly detected in tumor cell lines. To test whether IKK activity resulted in the activation of the classical NFκB pathway, we determined the subcellular distribution of p65 and NFκB DNA binding activity by EMSA. We did not detect nuclear p65 [supporting information (SI) Fig. 5 A and B] or NFκB-binding activity in the nuclear extracts from these cells (Fig. 1B) in basal conditions. As a control, TNF-α treatment resulted in both nuclear translocation of p65 (data not shown) and increased DNA-binding activity in HCT116 colorectal cancer cells (Fig. 1B), indicating that the NFκB pathway is not impaired. These results indicate that the canonical NFκB pathway is not active in colorectal cancer cell lines and are consistent with the presence of comparable levels of IκBα protein in tumor and nontransformed control cells (SI Fig. 5C). In addition, we did not detect the presence of the active/processed p52 protein in these cell lines by Western blot (SI Fig. 5D), indicating that the alternative NFκB pathway was not activated.
Fig. 1.
IKK activation in colorectal and pancreatic cancer cell lines. (A) Western blot analysis of P-IKKα/β and total IKKα from the different cancer cell lines compared with HEK-293 and HS27. α-tubulin is used as a loading control. (B) Different cell lines were tested for NFκB-binding activity by EMSA. p65-containing complexes are indicated as NFκB. Arrowhead indicates the supershift with α-p65 antibody. (C) IHC staining of three representative primary colon tumor sections and normal adjacent tissue with α-P-IKKα/β. Images were obtained at ×100 and ×600. (D) IHC staining of serial sections from a representative primary colorectal tumor with the indicated antibodies. Images were obtained at ×400.
We next tested whether IKK were activated in primary colorectal tumors. By immunohistochemistry (IHC), we detected phosphorylated IKK in all tested tumor tissues at different stages of transformation (n = 9)(Fig. 1C). We also determined the subcellular distribution of p50, p65, and p52 in these samples and observed that both p65 and p52 were exclusively cytoplasmic in all colorectal tumors (n = 4) whereas p50 was homogenously distributed in the cell (Fig. 1D and SI Fig. 6A). We also performed ChIP assays with α-p65 and α-p52 antibodies, and we did not detect a consistent interaction of these proteins with the promoters of different tumor-related NFκB-target genes (SI Fig. 6B). Altogether, these results demonstrate that NFκB pathway is not active in colorectal tumors with phosphorylated/activated IKK.
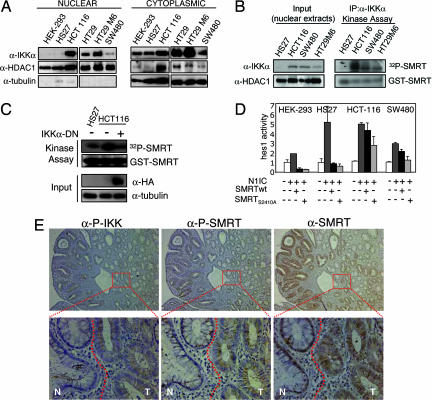
Because it has been shown that IKKα can activate gene transcription by directly phosphorylating nuclear substrates (20–22) we speculated that IKKs may play a nuclear function in colorectal cancer cells. By subcellular fractionation followed by Western blot analysis, we first determined the subcellular distribution of IKKα in different cancer cell lines. In Fig. 2A, we show that IKKα is present in the nucleus of most colorectal cancer cells but not in HS27 or HEK-293 control cells. We next tested the capability of this nuclear IKK to phosphorylate SMRT. To do this with this aim, we precipitated IKKα from normal and tumor nuclear extracts and tested the kinase activity of the precipitates on a GST-SMRT (amino acids 2321–2525). In Fig. 2B, we show that nuclear IKKα, immunoprecipitated from SW480, HCT116, and HT29-M6 colorectal cancer cells, efficiently phosphorylates the GST-SMRT construct containing Ser-2410, but not a mutant GST-SMRT S2410A (SI Fig. 7A). To confirm that the IKKα subunit was responsible for SMRT phosphorylation, we transfected HCT116 cells with a dominant negative IKKα mutant and tested the kinase activity of total cell extracts on the GST-SMRT construct. As seen in Fig. 2C, phosphorylation of GST-SMRT was completely abrogated by the expression of the DN-IKKα. Because IKKα-dependent phosphorylation of Ser-2410 abrogates the SMRT repression function (20), we investigated the ability of WT or the mutant SMRTS2410A to repress the previously reported target hes1 (24) in normal or colorectal tumor cell lines. By cotransfecting a hes1-luc reporter together with activated Notch1-IC we showed that ectopically expressed SMRTwt failed to repress the Notch-dependent hes1 activity in SW480 cells and HCT-116 cells compared with the control HEK-293 or HS27. As expected, the SMRTS2410A mutant was more efficient than the WT in repressing hes1-luc in tumor cells (Fig. 2D).
Fig. 2.
Nuclear IKKα phosphorylates SMRT and associates with its cytoplasmic translocation in colorectal tumors. (A) Western blot analysis of IKKα in cytoplasmic and nuclear extracts from the different cell lines. α-HDAC1 and α-tubulin are used as fractionation and loading controls. (B) Kinase activity assay of precipitated nuclear IKKα tested on GST-SMRT (amino acids 2321–2525) detected by [32P]ATP incorporation. Nuclear levels of IKKα in the different cell lines and HDAC1 as a nuclear input control are shown. (C) Kinase activity assay of total extracts from cells transfected with or without DN-IKKα tested on GST-SMRT (amino acids 2321–2525) detected by [32P]ATP incorporation. Levels of transfected DN-IKKα are detected with α-HA antibody, and α-tubulin is used as a loading control. (D) Indicated cell lines were cotransfected with hes1-luc and the indicated constructs. The graph represents the activity of hes1-luc in the different conditions. The average and standard deviation from duplicates of one representative of three experiments are shown. (E) IHC of sequential sections containing both normal (N) and adenoma (T) tissue (see dotted line) with α-P-IKKα/β, α-P-SMRT, and α-SMRT antibodies. Images were obtained at ×100 and ×600.
To investigate whether IKK-mediated phosphorylation of SMRT is occurring in primary colorectal tumors, we performed IHC on serial sections from colorectal tumor samples compared with adjacent normal tissue. Similar to Fig. 1C, we detected phosphorylated IKK in all tumor tissues but not in normal adjacent mucosa (Fig. 2E). In all tested samples (n = 9), the tumor areas containing active IKK showed SMRT phosphorylation at Ser-2410 (Fig. 2E and SI Fig. 7B). Because SMRT phosphorylation has been associated with its nuclear export (20, 25, 26), we next analyzed the subcellular distribution of SMRT in these colorectal tumor samples. As shown in Fig. 2E, SMRT corepressor is primarily localized in the nucleus in normal colorectal tissues whereas its distribution is mainly citoplasmic or excluded from the nucleus in tumors (n = 7). These results and those obtained from tumor cell lines strongly suggest that in colorectal carcinomas, IKKα is constitutively active and participate in the specific phosphorylation of SMRT.
To further characterize the molecular mechanism by which IKKα participates in colorectal carcinogenesis, we used a set of colorectal cancer xenografts (CRC-X) as an enriched source of tumor tissue. For this study, we grew and maintained six CRC-X during different passages as s.c. tumors in nude mice. All these tumors contained activated IKK and phosphorylated SMRT (data not shown).
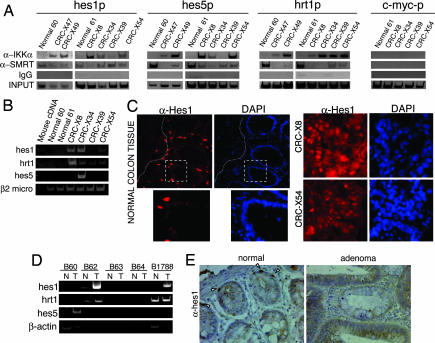
Because phosphorylation of Ser-2410 of SMRT by IKKα occurs on the chromatin (20), we performed ChIP assay to analyze whether IKK from CRC-X was recruited to specific gene promoters previously described as IKKα-target genes (18–21). Our results demonstrate that IKKα associates with the hes1, hes5, and herp2/hrt1 gene promoters in five of six tumor samples whereas we did not detect this interaction in two different normal colon tissues (Fig. 3A). Interestingly, association of IKKα to the different Notch-dependent promoters nicely correlates with a decrease in the levels of chromatin-bound SMRT in most of the samples (Fig. 3A). Comparable results were obtained with the antiapoptotic gene cIAP2 (data not shown) whereas we did not detect IKKα bound to the c-myc (Fig. 3A) or the IL6 (data not shown) promoters. Of note, differences in the amplification efficiency among the different xenografts correlate with differences in the amount of human tissue enrichment as observed by microscopic analysis (data not shown). We also tested whether the presence of IKKα correlated with phosphorylation of H3 in tumor samples. We did not detect a consistent association between IKKα recruitment and H3 phosphorylation in these samples (SI Fig. 8), suggesting that H3 is not the main target for chromatin-bound IKKα in colorectal tumors.
Fig. 3.
IKKα is recruited to different Notch target gene promoters correlating with transcriptional activation in colorectal tumors. (A) ChIPs with α-IKKα or α-SMRT and nonrelevant IgG from CRC-X compared with normal colon. PCR detection of the indicated promoters is shown. (B) Semiquantitative RT-PCR from patient-derived CRC-X. (C) Hes1 staining by immunofluorescence of frozen sections from normal colon tissue (×400 to ×600) and two different CRC-X (×600). (D) Semiquantitative RT-PCR from paired normal adjacent and tumor colon samples. (E) IHC of colorectal sections from normal and adenoma tissues with α-Hes1 antibody. Images were obtained at ×100 and ×600. Arrows indicate the presence of nuclear Hes1 in few cells in the bottom of the normal crypt.
To further demonstrate that recruitment of IKKα to the chromatin and SMRT release correlates with the specific activation of gene transcription, we determined the expression levels of Notch target genes (hes1, hes5, herp2/hrt1) by RT-PCR in both CRC-X samples (Fig. 3B) and five primary CRC paired with normal adjacent mucosa (Fig. 3D). Our results showed that hes1 and herp2 genes are overexpressed in CRC-X coinciding with IKKα recruitment. In contrast we only detected increased expression of hes5 in CRC-X34 suggesting that other mechanisms contribute to regulate hes5 expression. Most important, we detected up-regulation of hes1 and herp2/hrt1 in three of five, and of hes5 in one of five primary CRC tumor samples compared with normal controls (Fig. 3D). By immunostaining we showed that Hes1 protein was restricted to scarce cells in the bottom of the crypt in normal colon mucosa (Fig. 3 C and E) likely representing the proliferating/progenitor compartment as described in adult small intestine (3, 27) whereas high levels of Hes1 protein were detected in virtually all of the tumor tissues (Fig. 3 C and E).
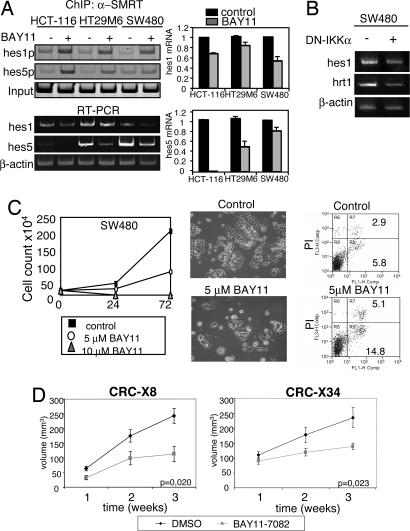
To directly demonstrate that IKK activity is responsible for the release of SMRT from specific promoters in cancer cells, we performed ChIP assays from cells treated with the IKK inhibitor BAY11-7082 (28) that prevents IKKα-mediated SMRT phosphorylation (29). Our results demonstrated that treatment with BAY11-7082 is sufficient to increase the association of SMRT to the hes1 and hes5 promoters in HCT116, HT29M6 and SW480 cells, nicely correlating with their transcriptional repression (Fig. 4A). In contrast, inhibition of the canonical NFκB pathway by IκBα32–36 did not affect hes1 transcriptional activity in HCT116 suggesting that the effect of BAY11-7082 on hes1 gene is IKK-dependent but NFκB-independent (SI Fig. 9). To further confirm the specific involvement of the IKKα subunit in the activation of endogenous Notch target genes in colorectal cancer cells, we transfected SW480 cells with the DN-IKKα and measured mRNA levels by RT-PCR. As shown in Fig. 4B, both hes1 and herp2/hrt1 genes were down-regulated in DN-IKKα expressing cells compared with the control. Consistent with this result, SMRT association to the hes1 promoter is restored in the presence of the DN-IKKα (SI Fig. 10).
Fig. 4.
IKK-target genes are up-regulated in primary tumors and inhibition of IKK reduces tumor volume in vivo. (A) ChIP with α-SMRT antibody and PCR detection of the hes1 and hes5 gene and mRNA expression by semiquantitative RT-PCR from HCT-116, HT29M6, and SW480 treated with DMSO or BAY11-7082 for 48 h. (B) Semiquantitative RT-PCR showing the expression levels of hes1 and hrt1/herp2 in SW480 cells transfected with control vector or DN-IKKα. (C) Representative culture of SW480 cells after 3-day treatment with DMSO or BAY11-7082. (Left) Graphs represent daily cell counts from cultures treated with the indicated concentrations of BAY11-7082. (Center) Photographs show a representative image of the cultures at day 3 of treatment. Images were obtained in an Olympus IX-10 at ×100. (Right) Flow cytometry analysis of AnnexinV staining. (D) CRC-X8 and CRC-X34 tumors were implanted s.c. in nude mice (n = 40). Ten days after transplantation, the animals were treated with BAY11-7082 or DMSO as a control. Tumors were measured weekly, and the results are presented as mean ± SEM. Student's t test for independent analysis was applied to evaluate differences in tumor volume [A(B2)π/6] between treated and control mice.
It has been previously shown that IKK inhibitor BAY11-7085 induces apoptosis and inhibits cell growth of colorectal cancer cell lines (30). To check the effect of the IKK inhibitor BAY11-7082 on colorectal cancer cells, we cultured SW480 (Fig. 4C) and HT29M6 (SI Fig. 11) cell lines with DMSO or BAY11-7082 and analyzed cell growth and apoptosis. In both experimental conditions, we detected a similar number of colonies after three days in culture, however, colony size was greatly reduced in the BAY11-7082 treated cultures, consistent with a lower cell count. The decrease in the number of cells because of the BAY11-7082 treatment correlates with increased apoptosis as detected by AnnexinV staining (Fig. 4C and SI Fig. 11).
To determine the potential effect of BAY11-7082 on an in vivo system, we transplanted equivalent pieces (50–75 mg) of CRC-X8 and CRC-X34 s.c. in nude mice (n = 40), and, 1 week after implantation, we treated the animals for 3 weeks with the drug (5 μg/g) or with the vehicle DMSO. As shown in Fig. 4D in the animals treated with BAY11-7082, tumor growth was significantly reduced at the end of the treatment (P = 0.02) in both CRC-X8 and CRC-X34 compared with the animals treated with DMSO, similar to that observed in cell cultures.
Altogether, our results indicate that nuclear IKKα activity induces phosphorylation of chromatin associated SMRT thus modulating the expression of specific genes such as hes1 and herp2/hrt1 in colorectal cancer cells. Inhibition of IKK activity restores the normal transcriptional level of these genes leading to a decrease in cell proliferation and increased apoptosis in tumor cells.
Discussion
In the present study, we have identified a role for IKKα in colorectal carcinomas, independent of the NFκB pathway. Increased IKK activity in colorectal tumors nicely correlates with SMRT phosphorylation and with its cytoplasmic translocation. At the chromatin level, association of IKKα to specific Notch target promoters results in the release of chromatin-bound SMRT thus activating hes1, hes5, or herp2/hrt1 transcription.
Constitutive activation of NFκB has been found in many types of tumor cells (for review, see refs. 31 and 32). Most of these studies report increased IKK activity that results in phosphorylation of IκBα (33, 34), however minor or no changes in the subcellular localization of p65 have been described in most of the tumor cells (34–36). Because activation of the canonical NFκB pathway requires the nuclear translocation of p65 (for review, see ref. 37), the role of this pathway in tumorigenesis is not clearly understood. We have now demonstrated that colorectal cancer cell lines and primary CRC show increased IKK activity that is concomitant with undetectable levels of nuclear p65 and p52. This result is consistent with the absence of p65 and p52 in different NFκB-target genes as detected by ChIP analysis. Together these results indicate that NFκB activation is not the main consequence of IKK activity in colorectal tumors, maybe reflecting substrate specificity of different IKK complexes; Alternatively, in some conditions IκBα could be Lys-63-linked polyubiquitinated instead of Lys-48 after phosphorylation, protecting the protein from degradation such is the case for TNF receptor-associated factor (TRAF) 2, TRAF6, or NEMO (38).
There is increasing evidence supporting the idea that IKKs are important in directly regulating transcription of NFκB-dependent but also NFκB-independent genes. In this sense, some nuclear proteins such as histone H3, estrogen receptor (ER), and the SMRT corepressor are direct substrates for IKKα kinase activity (18, 20, 21, 23). Nuclear corepressors are crucial for the formation of repression complexes; therefore, changes in their subcellular distribution should have striking effects on the overall regulation of transcription (39). This is the case for cytoplasmic translocation of N-CoR after Akt-dependent phosphorylation that is required for astrocyte differentiation (40), the aberrant recruitment of SMRT by PML-RAR in myeloid leukemias (41), or down-regulation of SMRT in non-Hodgkin lymphomas (42). We show that active IKKs from colon tumor cells are able to specifically phosphorylate Ser-2410 of SMRT that has been previously associated with its cytoplasmic translocation and degradation (20). This observation is consistent with our finding that SMRT is aberrantly distributed in all of the primary tumors tested. Moreover, not only subcellular distribution but also association of SMRT and IKKα with the chromatin is affected in colorectal cancer cells. We detected high levels of IKKα bound to hes1, hes5, and herp2/hrt1 promoters in colon tumor cells compared with normal colon tissue whereas association of SMRT with these promoters was preferentially found in normal tissues. In contrast, we did not detect association of IKKα with the c-myc promoter, although it is possible that other SMRT regulated genes that are important in cancer are also affected by this mechanism.
Chromatin-associated IKKα was shown to be important in the transcriptional activation of NFκB-target genes such as IL-6 and IκBα (18, 21), but also in NFκB-independent genes such as c-fos (22). In all these reports, the mechanism for IKKα regulation relies on their ability to phosphorylate Ser-10 of histone H3. In our study, we did not detect a significant correlation between the presence of chromatin-bound IKKα and histone H3 phosphorylation in tumor samples, suggesting that different kinases phosphorylate histone H3 in specific genes (20, 43). More recently the kinase activity of IKKα has also been implicated on estrogen-mediated gene activation through phosphorylation of estrogen receptor (23), and we previously showed that IKK is important in the TNF-α-dependent regulation of the classical Notch target genes hes1 or herp2/hrt1 (19). Together with our current data, these results suggest that IKKα may operate downstream of different signaling pathways to regulate specific sets of genes.
The fact that IKK activity can regulate gene transcription by directly phosphorylating nuclear substrates in tumor cells is extremely important to design specific inhibitors for cancer therapy (44). In this sense, we have been able to inhibit the release of the SMRT corepressor from the chromatin with the IKK inhibitor BAY11-7082 and to inhibit tumor cell growth. We have used this inhibitor to demostrate the therapeutic effects on implanted solid tumor fragments, in contrast with previous experiments with BAY11-7085 in HT29 cell-derived xenografts (30). Unfortunately this compound inhibits both IKKα and IKKβ activity and IκBα phosphorylation (28), blocking the classical NFκB pathway, and, although we demonstrate that NFκB-independent functions of IKK on Notch targets are inhibited by BAY11-7082, we cannot exclude that inhibition of basal NFκB activity may contribute to some of the anti-neoplastic effects observed with this compound. Based on our results, we propose that IKKα inhibitors designed to abrogate nuclear IKK functions, including SMRT phosphorylation, may be useful tools for colorectal cancer therapy.
Methods
Plasmids.
Expression vectors for Notch1-IC (45), hes1p-luc (46), flag-SMRT, flag-SMRT Ser2410Ala (20), and the kinase defective IKKα S176A/S180A mutant (47) have been described.
Antibodies.
α-IκBα (sc-1643), α-p65 (sc-109), α-p50 (sc-7178), α-p52 (sc-7386), α-IKKβ (sc-7330), α-HDAC1 (sc-7872), and α-Hes1 (sc-13844) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). α-IKKα(op-133) was purchased from Oncogen (La Jolla, CA). α-P-IKKα-Ser-180/IKKβ-Ser-181 (2681), α-SMRT (06-891), α-P-Ser-10-H3 (06-570) were purchased from Upstate (Charlottesville, VA) and α-αtubulin from Sigma (St. Louis, MO). Phosphospecific SMRT antibody α–P-S2410 has been described (20). Secondary antibodies conjugated with horseradish peroxidase (HRP) were from DAKO (Glostrup, Denmark, with AlexaFluor488 (A-11055) and 546 (A-11056) from Molecular Probes (Invitrogen, Carlsbad, CA) and Cy3-coupled tyramide was from PerkinElmer (Wellesley, MA).
Cell Lines and Culture Reagents.
HEK-293, HS27, HT-29, HT-29 M6, HCT-116, CaCo2, SW480, and MCF7 were cultured in DMEM 10% FBS. Human TNF-α is from Preprotech and was used at 40 ng/ml. BAY11-7082 was from Calbiochem (Darmstadt, Germany) (7082) and was used at 5–10 μM. Lipopolysaccharide (LPS) (Sigma) was used at 10 μg/ml.
Cell Fractionation.
Nuclei were isolated in 0.1% Nonidet P-40/PBS for 5 min on ice, followed by centrifugation at 720 × g and were lysed for 30 min in 50 mM Tris·HCl (pH 7.5), 150 mM NaCl, 1% Nonidet P-40, 5 mM EGTA, 5 mM EDTA, 20 mM NaF, and complete protease inhibitor mixture (Roche). The supernatant was the cytoplasmic fraction.
EMSA.
[γ-32P]ATP-labeled oligonucleotide containing NFκB-binding sites of the MMP9 promoter (5′-TGCCCCATGGAATTCCCCAA-3′) was incubated with 15 μg of nuclear cell extracts under standard conditions as described (48). Two micrograms of α-p65 (sc-109) was used for supershift experiments.
Protein Kinase Assays.
Nuclei from cells were isolated and lysed for 30 min at 4°C in 500 μl of PBS containing 0.5% Triton X-100, 1 mM EDTA, 100 mM Na-orthovanadate, 0.25 mM PMSF, and complete protease inhibitor mixture (Roche, Basel, Switerland). After centrifugation, supernatants were precleared for 2 h twice and incubated with 1 μg of the α-IKKα overnight at 4°C. After incubation with Protein A-Sepharose beads, precipitates were washed and assayed for kinase activity on GST-SMRTwt or GST-SMRTS2410A.
Immunofluorescence.
Slides were fixed with −20°C methanol and permeabilized in 10% FBS, 0.3% Triton X-100 (Pierce, Rockford, IL), and 5% nonfat milk in PBS. Samples were stained with α-hes1 (Santa Cruz Biotechnology) in 10% FBS, 5% nonfat milk in PBS and HRP-conjugated rabbit α-goat antibody (Dako) and developed with Cy3-coupled tyramide (PerkinElmer). Sections were mounted in Vectashield medium with DAPI (Vector, Brussels, Belgium).
Cell lines were fixed in 3% paraformaldehyde, permeabilized, incubated with primary and secondary antibodies, and mounted with Vectashield propidium iodide. Stainings were visualized in an Olympus BX-60 microscope or a Leica TCS-NT laser scanning confocal microscope with the ×63 Leitz Plan-Apo objective (NA 1.4). Images were edited on Adobe Photoshop.
IHC.
For histological analysis, samples were fixed in 4% paraformaldehyde, dehydrated, and parafin-embedded. Sections were hydrated and permeabilized, and antigen retrieval was achieved by boiling in 20 mM sodium citrate (pH 6.0) (2 min). HRP-conjugated secondary antibodies were detected with the diaminobenzidine peroxidase substrate kit (Dako Cytomation)
Human Samples and Tumor Xenografts.
Six human CRC were obtained from patients without previous cytotoxic therapy, cut into pieces of ≈2 mm3, and implanted s.c. in 5-week-old male nu/nu Swiss mice (Harlam, France). Animals were housed in a sterile environment, and all experiments were approved by the Institutional Animal Care Committee. All CRC-X were analyzed in <5 passage. Genotyping using a set of microsatellites of paired primary colorectal tumor and xenografts confirmed that the CRC-X maintain the original characteristics. Histologically, all tumors were classified as adenocarcinomas.
In Vivo Tumor-Growth Assay.
Equivalent 50- to 75-g CRC-X8 or CRC-X34 tumor pieces were s.c. transplanted into nu/nu swiss mice (n = 40). Ten days after transplantation, the control group received i.p. injections of DMSO whereas BAY11-7082-treated animals were injected with 5 μg/g dose, three times per week for 3 weeks as described (49). Tumors were measured weekly, and volume was calculated as V (mm3) = A(mm) × B2(mm2) × π/6, B being the smaller dimension.
Luciferase Assays.
Cells were transfected with calcium phosphate or lipofectamine (Invitrogen) with the indicated plasmids. Luciferase assays (Luciferase Assay System; Promega, Madison, WI) were performed 48 h after transfection as described (19).
ChIP Assay.
ChIP assay has been described (19). Briefly, chromatin from crosslinked cells was sonicated, incubated overnight with the indicated antibodies in RIPA buffer, and precipitated with protein G/A-Sepharose. All antibodies used have been tested for ChIP assay (refs. 18–20 and SI Fig. 6C). Cross-linkage of the coprecipitated DNA–protein complexes was reversed, and DNA was used as a template for semiquantitative PCR. All of the primers recognize human DNA sequence and were tested to avoid mouse DNA amplification. Primer sequences are in SI Fig. 12).
RT-PCR.
Total RNA from colorectal tumors was isolated by using TRIzol Reagent (Invitrogen), and RT-First Strand cDNA Synthesis kit (Amersham Pharmacia Biotech, GE Healthcare, Buckinghamshire, U.K.) was used. Densitometric analysis was performed with the Quantity One software from Bio-Rad (Hempstead, U.K.). Oligonucleotide sequences are in SI Fig. 12.
Flow Cytometry Analysis.
Cells were stained with AnnexinV-FITC and propidium iodide. Cells were analyzed on a FACScalibur (Becton Dickinson, San Jose, CA).
Supplementary Material
Acknowledgments
We thank M. Privalsky (University of California, Davis, CA) for kindly providing SMRT plasmids, M. Karin (University of California at San Diego, La Jolla, CA) for providing IKKα and IKKα S176A/S180A plasmids, Serveis Cientifico-Tècnics, UB-Bellvitge, for confocal microscopy and flow cytometry technical support, M. Gumà for statistical analysis, and J. Inglés-Esteve and L. Miele for helpful discussions. L.E. is an Investigator from the Carlos III program (ISCIII/02/3027) and A.V. from the Ramón y Cajal program. C.A. is a recipient of MEyC Predoctoral Fellowship BES-2002-0028, V.F.-M. and A.R.-M. are recipients of Department of Universities, Research and the Information Society (DURSI) Predoctoral Fellowships 2005-FI00458 and 2002-SI00791, and F.V. and A.A. are recipients of a Beca de Formación en Investigación (BEFI) fellowship. This work was supported by Instituto de Salud Carlos III Grant PI041890, Fundació Marató TV3 Grant 051730, and National Institutes of Health Grant R01 CA104397.
Abbreviations
- IKK
IκB kinase
- IHC
immunohistochemistry
- CRC-X
colorectal cancer xenograft.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0606476104/DC1.
References
- 1.Vogelstein B, Kinzler KW. Trends Genet. 1993;9:138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 2.Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, Gould KA, Dove WF. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 3.van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, Clevers H. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 4.Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 5.Leong KG, Karsan A. Blood. 2006;107:2223–2233. doi: 10.1182/blood-2005-08-3329. [DOI] [PubMed] [Google Scholar]
- 6.Visvader J, Begley CG. Trends Biochem Sci. 1991;16:330–333. doi: 10.1016/0968-0004(91)90137-k. [DOI] [PubMed] [Google Scholar]
- 7.Cuevas IC, Slocum AL, Jun P, Costello JF, Bollen AW, Riggins GJ, McDermott MW, Lal A. Cancer Res. 2005;65:5070–5075. doi: 10.1158/0008-5472.CAN-05-0240. [DOI] [PubMed] [Google Scholar]
- 8.Fan X, Mikolaenko I, Elhassan I, Ni X, Wang Y, Ball D, Brat DJ, Perry A, Eberhart CG. Cancer Res. 2004;64:7787–7793. doi: 10.1158/0008-5472.CAN-04-1446. [DOI] [PubMed] [Google Scholar]
- 9.Kawamata S, Du C, Li K, Lavau C. Oncogene. 2002;21:3855–3863. doi: 10.1038/sj.onc.1205487. [DOI] [PubMed] [Google Scholar]
- 10.Iso T, Sartorelli V, Chung G, Shichinohe T, Kedes L, Hamamori Y. Mol Cell Biol. 2001;21:6071–6079. doi: 10.1128/MCB.21.17.6071-6079.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuroda K, Tani S, Tamura K, Minoguchi S, Kurooka H, Honjo T. J Biol Chem. 1999;274:7238–7244. doi: 10.1074/jbc.274.11.7238. [DOI] [PubMed] [Google Scholar]
- 12.Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. EMBO J. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomita K, Hattori M, Nakamura E, Nakanishi S, Minato N, Kageyama R. Genes Dev. 1999;13:1203–1210. doi: 10.1101/gad.13.9.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunisato A, Chiba S, Nakagami-Yamaguchi E, Kumano K, Saito T, Masuda S, Yamaguchi T, Osawa M, Kageyama R, Nakauchi H, et al. Blood. 2003;101:1777–1783. doi: 10.1182/blood-2002-07-2051. [DOI] [PubMed] [Google Scholar]
- 15.Suh J, Payvandi F, Edelstein LC, Amenta PS, Zong WX, Gelinas C, Rabson AB. Prostate. 2002;52:183–200. doi: 10.1002/pros.10082. [DOI] [PubMed] [Google Scholar]
- 16.Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, Kagnoff MF, Karin M. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 17.Karin M. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 18.Anest V, Hanson JL, Cogswell PC, Steinbrecher KA, Strahl BD, Baldwin AS. Nature. 2003;423:659–663. doi: 10.1038/nature01648. [DOI] [PubMed] [Google Scholar]
- 19.Aguilera C, Hoya-Arias R, Haegeman G, Espinosa L, Bigas A. Proc Natl Acad Sci USA. 2004;101:16537–16542. doi: 10.1073/pnas.0404429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoberg JE, Yeung F, Mayo MW. Mol Cell. 2004;16:245–255. doi: 10.1016/j.molcel.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto Y, Verma UN, Prajapati S, Kwak YT, Gaynor RB. Nature. 2003;423:655–659. doi: 10.1038/nature01576. [DOI] [PubMed] [Google Scholar]
- 22.Anest V, Cogswell PC, Baldwin AS., Jr J Biol Chem. 2004;279:31183–31189. doi: 10.1074/jbc.M404380200. [DOI] [PubMed] [Google Scholar]
- 23.Park KJ, Krishnan V, O'Malley BW, Yamamoto Y, Gaynor RB. Mol Cell. 2005;18:71–82. doi: 10.1016/j.molcel.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong SH, Privalsky ML. Mol Cell Biol. 2000;20:6612–6625. doi: 10.1128/mcb.20.17.6612-6625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jang MK, Goo YH, Sohn YC, Kim YS, Lee SK, Kang H, Cheong J, Lee JW. J Biol Chem. 2001;276:20005–20010. doi: 10.1074/jbc.M010211200. [DOI] [PubMed] [Google Scholar]
- 27.Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- 28.Pierce JW, Schoenleber R, Jesmok G, Best J, Moore SA, Collins T, Gerritsen ME. J Biol Chem. 1997;272:21096–21103. doi: 10.1074/jbc.272.34.21096. [DOI] [PubMed] [Google Scholar]
- 29.Hoberg JE, Popko AE, Ramsey CS, Mayo MW. Mol Cell Biol. 2006;26:457–471. doi: 10.1128/MCB.26.2.457-471.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scaife CL, Kuang J, Wills JC, Trowbridge DB, Gray P, Manning BM, Eichwald EJ, Daynes RA, Kuwada SK. Cancer Res. 2002;62:6870–6878. [PubMed] [Google Scholar]
- 31.Gilmore T, Gapuzan ME, Kalaitzidis D, Starczynowski D. Cancer Lett. 2002;181:1–9. doi: 10.1016/s0304-3835(01)00795-9. [DOI] [PubMed] [Google Scholar]
- 32.Rayet B, Gelinas C. Oncogene. 1999;18:6938–6947. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- 33.Yang J, Richmond A. Cancer Res. 2001;61:4901–4909. [PubMed] [Google Scholar]
- 34.Biswas DK, Shi Q, Baily S, Strickland I, Ghosh S, Pardee AB, Iglehart JD. Proc Natl Acad Sci USA. 2004;101:10137–10142. doi: 10.1073/pnas.0403621101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang W, Abbruzzese JL, Evans DB, Larry L, Cleary KR, Chiao PJ. Clin Cancer Res. 1999;5:119–127. [PubMed] [Google Scholar]
- 36.Gasparian AV, Yao YJ, Kowalczyk D, Lyakh LA, Karseladze A, Slaga TJ, Budunova IV. J Cell Sci. 2002;115:141–151. doi: 10.1242/jcs.115.1.141. [DOI] [PubMed] [Google Scholar]
- 37.Bonizzi G, Karin M. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 38.Krappmann D, Scheidereit C. EMBO Rep. 2005;6:321–326. doi: 10.1038/sj.embor.7400380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jepsen K, Rosenfeld MG. J Cell Sci. 2002;115:689–698. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- 40.Hermanson O, Jepsen K, Rosenfeld MG. Nature. 2002;419:934–939. doi: 10.1038/nature01156. [DOI] [PubMed] [Google Scholar]
- 41.Hong SH, Yang Z, Privalsky ML. Mol Cell Biol. 2001;21:7172–7182. doi: 10.1128/MCB.21.21.7172-7182.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song L, Zlobin A, Ghoshal P, Zhang Q, Houde C, Weijzen S, Jiang Q, Nacheva E, Yagan D, Davis E, et al. Cancer Res. 2005;65:4554–4561. doi: 10.1158/0008-5472.CAN-04-4108. [DOI] [PubMed] [Google Scholar]
- 43.Sassone-Corsi P, Mizzen CA, Cheung P, Crosio C, Monaco L, Jacquot S, Hanauer A, Allis CD. Science. 1999;285:886–891. doi: 10.1126/science.285.5429.886. [DOI] [PubMed] [Google Scholar]
- 44.Greten FR, Karin M. Cancer Lett. 2004;206:193–199. doi: 10.1016/j.canlet.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 45.Espinosa L, Ingles-Esteve J, Aguilera C, Bigas A. J Biol Chem. 2003;278:32227–32235. doi: 10.1074/jbc.M304001200. [DOI] [PubMed] [Google Scholar]
- 46.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 47.Delhase M, Hayakawa M, Chen Y, Karin M. Science. 1999;284:309–313. doi: 10.1126/science.284.5412.309. [DOI] [PubMed] [Google Scholar]
- 48.Jorda M, Olmeda D, Vinyals A, Valero E, Cubillo E, Llorens A, Cano A, Fabra A. J Cell Sci. 2005;118:3371–3385. doi: 10.1242/jcs.02465. [DOI] [PubMed] [Google Scholar]
- 49.Mabuchi S, Ohmichi M, Nishio Y, Hayasaka T, Kimura A, Ohta T, Saito M, Kawagoe J, Takahashi K, Yada-Hashimoto N, et al. J Biol Chem. 2004;279:23477–23485. doi: 10.1074/jbc.M313709200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.