Abstract
Nonribosomal peptide synthetases (NRPS), polyketide synthases (PKS), and hybrid NRPS/PKS are of particular interest, because they produce numerous therapeutic agents, have great potential for engineering novel compounds, and are the largest enzymes known. The predicted masses of known enzymatic assembly lines can reach almost 5 megadaltons, dwarfing even the ribosome (≈2.6 megadaltons). Despite their uniqueness and importance, little is known about the organization of these enzymes within the native producer cells. Here we report that an 80-kb gene cluster, which occupies ≈2% of the Bacillus subtilis genome, encodes the subunits of ≈2.5 megadalton active hybrid NRPS/PKS. Many copies of the NRPS/PKS assemble into a single organelle-like membrane-associated complex of tens to hundreds of megadaltons. Such an enzymatic megacomplex is unprecedented in bacterial subcellular organization and has important implications for engineering novel NRPS/PKSs.
Keywords: bacillaene, nonribosomal peptide, polyketide, Streptomyces
Nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) enzymes have received much attention in recent years, in large part because of their potential to be engineered to direct the synthesis of novel compounds with therapeutic value. This engineering potential stems from the fact that the individual modules of these enzymes are organized into assembly lines in which the order of modules determines the identity of the polyketide or polypeptide product (1, 2). Each domain in the assembly line specifies the incorporation of an individual substrate, because the nascent peptide or polyketide is passed sequentially between domains. The structures of many individual NRPS and PKS domains have been solved (3), and the structures of mammalian and fungal isoforms of the PKS-like fatty acid synthases were recently reported (4, 5), bringing into focus a picture of individual enzymatic assembly line architecture. However, much of our current knowledge of NRPSs and PKSs comes from studies in which these proteins are expressed individually in heterologous hosts, leaving much to learn about the quaternary organization and cell biology of these massive enzymes in the context of their native producer organisms.
The genome of Bacillus subtilis contains the pks gene cluster, which encodes a hybrid NRPS/PKS (Figs. 1A and 4) that was considered nonfunctional until recently. A recent report has assigned the pks gene cluster to production of bacillaene based on the analysis of an orthologous gene cluster from Bacillus amyloliquefaciens FZB42 called bae (6). In concurrent work, we have solved the structure of bacillaene from B. subtilis and proposed a model for its biosynthesis (28). Bioinformatic analyses of the bacillaene synthase gene cluster reveal features in common with unconventional PKSs from streptomycetes, myxobacteria, and cyanobacteria (see Fig. 4). Examples of these features include three trans-acting acyltransferase (AT) domains that introduce substrates to the assembly line (7) and a six-protein subcluster that converts a carbon–oxygen double bond to a β-methyl group using a variation of isoprene biosynthetic logic (8–10). The bacillaene synthase is a rare example of an unusual active hybrid NRPS/PKS in an extensively studied model organism.
Fig. 1.
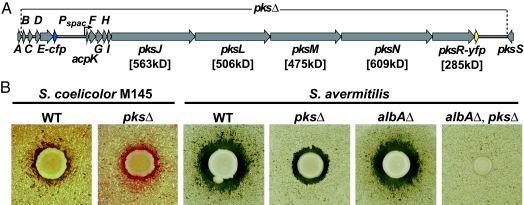
Genetic organization and activity of the B. subtilis bacillaene synthase. (A) A simplified schematic of the bacillaene (pks) gene cluster indicating the points of fusion to the fluorescent protein genes. The extent of pks gene deletion in the pksΔ strain and the location of pksE-cfp (blue arrow) and pksR-yfp (yellow arrow) fusions are indicated. In only the cells containing a fusion of cfp to the pksE ORF, genes downstream of pksE-cfp are expressed under isopropyl β-d-thiogalactoside control of the Pspac promoter. Predicted molecular weights of large multimodular proteins are listed below the gene names (genolist.pasteur.fr/SubtiList). (B) Mutations in the individual pks genes or a deletion of the entire pks gene cluster reveal growth and developmental phenotypes in coculture with Streptomycetes (SI Text). (Left) B. subtilis WT and pksΔ cocultured with S. coelicolor M145 on YEME pH7 agar. A red ring (undecylprodigiosin produced by S. coelicolor) is visible around the pksΔ but not the WT strain. (Right) NRPS/PKS activity produced by B. subtilis inhibits S. avermitilis growth in coculture. The activity is lost in a pksΔ strain. The loss of PK activity in a pksΔ strain is obscured by the effect of subtilosin (product of the sbo-alb gene cluster). Total loss of activity is seen by comparison of pks+, albAΔ and pksΔ, albAΔ double mutant strains.
Fig. 4.
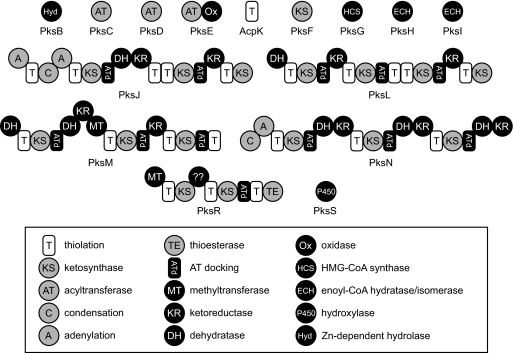
The predicted function for the enzymes and enzymatic modules encoded by the bacillaene gene cluster. The proteins PksB-I, AcpK, and PksS are free-standing enzymes thought to function in trans to the multimodular proteins. PksJ, -L, -M, -N, and -R are multimodular NRPS/PKS (PksJ and -N) or PKS (PksL, -M, and -N) proteins forming the core assembly line synthase. The predicted function of each individual module or freestanding enzyme is listed in the legend. The unusual C-terminal KS modules in PksJ and -L and the N-terminal DH modules in PksL and -M are likely to mediate interprotein transfer of nascent bacillaene.
The constituent subunits of the bacillaene synthase are encoded in 16 pks genes, which occupy an ≈80-kb region of the chromosome, encompassing nearly 2% of the genome. As a multimodular assembly line, the bacillaene synthase proteins in single copy would comprise a core synthase of nearly 2.5 megadaltons. The conservation of this massive gene cluster and its encoded synthase in multiple Bacillus species argues that bacillaene has an important ecological function, and the production of bacillaene by B. subtilis suggests that this genetically manipulable host has an untapped metabolic potential for producing complex natural products.
We have a long-standing interest in understanding the molecular mechanisms that underlie interspecies interactions in the microbial world, which has led us to study the production and function of small-molecule metabolites that mediate these interactions (11–15). We are particularly interested in the interactions between members of the genera Bacillus and Streptomyces because species of both of these genera coexist in soils and are prolific producers of antimicrobial compounds (16, 17). Here we report that a phenotype associated with mutations of the B. subtilis pks gene cluster is visible as a perturbation in the development of cocultured Streptomyces on an agar plate. Using this bacterial interspecies interaction as a simple phenotypic assay, we have pursued in vivo studies in B. subtilis to gain insight into the cell biology of complex secondary metabolite production. We show that the PKS proteins in B. subtilis assemble into a megacomplex of bacillaene synthases. We show that multiple proteins of the PKS complex colocalize to a single mass in the cytoplasm and contrast this observation with the diffuse localization pattern of the related surfactin NRPS proteins. The PKS megacomplex is visible by electron microscopy and suggests that the PKS and possibly other proteins form an organelle-like structure unprecedented in subcellular organization of the bacterial cell.
Results and Discussion
A B. subtilis pks Phenotype Visible in an Interspecies Interaction.
When cocultivated on agar plates, B. subtilis and Streptomyces coelicolor undergo characteristic developmental pathways that lead to secondary metabolite production and sporulation. At early time points during coincubation, a colony of B. subtilis growing on top of a lawn of S. coelicolor produces a rather unremarkable mixed culture (Fig. 1B). When we screened random transposon insertion mutants of B. subtilis under conditions of cocultivation with S. coelicolor, several B. subtilis mutants displayed a striking phenotype: they induced the premature synthesis in the adjacent S. coelicolor of the red antibiotic undecylprodigiosin [ref. 18; Fig. 1B and supporting information (SI) Fig. 5]. Nucleotide sequence analyses revealed that each of the B. subtilis mutants harbored a transposon insertion in one of the pks genes; in total, we recovered six independent mutants with transposon insertions in 5 of the 16 pks genes (SI Table 1). We subsequently generated a deletion of the entire pks gene cluster, pksΔ (Fig. 1A) and found that the complete deletion caused a phenotype indistinguishable from that observed with the individual transposon mutants.
Armed with the pks deletion strain, we investigated its effects on the interactions between B. subtilis and other Streptomyces species. We found that the pks gene cluster is responsible for the production of a diffusible metabolite that inhibits the growth of Streptomyces avermitilis. The pks phenotypic effect was initially obscured by the fact that B. subtilis produces another antibiotic active against Streptomyces avermitilis. Using a second screen, we identified the other activity by a mutant in the albA locus, which disrupted the production of the lantibiotic subtilosin encoded by the sbo-alb genes (SI Text; ref. 19). The pks phenotype in interaction with S. avermitilis is made clear when comparing an albAΔ single mutant to a pksΔ, albAΔ double mutant strain (Fig. 1B). These results provided a clear indication that the B. subtilis pks genes produce an active bacillaene synthase (20), prompting us to pursue questions regarding the organization of synthase enzymes in their natural producer.
Subcellular Localization of the PKS Proteins.
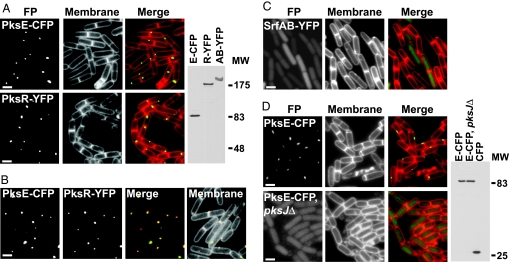
B. subtilis has emerged as an excellent model organism for bacterial cytology, which gave us a unique opportunity to investigate the cell biology of the enormous NRPS/PKS biosynthetic system. We began by constructing C-terminal fusions of cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) to individual Pks proteins in the synthase (Fig. 1A). Cells producing these fusion proteins retained inhibitory activity against S. avermitilis, indicating that the fusion proteins were functional (SI Fig. 5). Cells harboring fusions of YFP with PksR, a large multimodular component of the synthase, and CFP with PksE, a trans-acting AT (7), were grown to late-exponential phase and examined by fluorescence microscopy. Quite surprisingly, we found that PksR-YFP and PksE-CFP each localize to a single focus within the B. subtilis cells (Fig. 2A). The same was observed for a PksJ-CFP fusion (SI Fig. 6). The focus of fluorescence was not fixed at the same location in each cell; medial as well as polar positioning was observed in different cells. The majority of cells had a single focus of fluorescence (69%, n = 876, PksR-YFP), although a minor fraction of cells showed two foci (6%, n = 876, PksR-YFP), and some cells had no detectable fluorescent signal (25%, n = 876, PksR-YFP). In these cells, the pks gene fusions are expressed under their native promoters, and proteins of predicted size for each Pks fusion are detectable by Western blot (Fig. 2A). No fluorescence was observed in cells from cultures in early exponential phase, suggesting that NRPS/PKS synthesis or assembly is subject to growth-phase regulation. We examined the PksR-YFP protein by immunoblot analysis during a time course and found that protein abundance of PksR-YFP increases with cell density, consistent with growth-phase regulation of the bacillaene synthase (SI Fig. 7).
Fig. 2.
Localization of Pks proteins in B. subtilis 3610. (A) The PksE-CFP (Upper) and PksR-YFP fusions (Lower) localize to a discrete spot. (Left) Fluorescence of Pks protein fusion (FP); (Center) fluorescence from membrane stained with the dye TMA-DPH (Molecular Probes, Eugene, OR); (Right) overlay of fluorescence images. (Images are the same in C and D). On the far right, an immunoblot of PksE-CFP, PksR-YFP and SrfAB-YFP (see C, below) probed with anti-GFP antibody shows synthesis of the full-length fusion proteins. (B) Colocalization in a strain simultaneously producing PksE-CFP and PksR-YFP. (Left) Fluorescence from fusion proteins as noted above. PksE-CFP and PksR-YFP images are merged to show colocalization at points of overlap in yellow (Merge). Membranes are stained with TMA-DPH as in A (Membranes). (C) The SrfAB-YFP protein is diffuse in the cytoplasm. (D) PksE-CFP localization occurs with an intact pks gene cluster (Upper) but not with a disruption of pksJ (Lower). Production of PksE-CFP is identical in both strains by immunoblot (far right). Purified CFP (right lane) is included to demonstrate that the PksE-CFP fusion is stable in the pksJ mutant background. (Scale bars, 1 μm.)
We next compared the localization of PksE-CFP and PksR-YFP in the same strain. As predicted, we observed a pattern of overlapping localization, consistent with concerted action of the proteins as part of the holoenzyme synthase. Indeed, fluorescence microscopy of a strain harboring both PksR-YFP and PksE-CFP shows that the proteins colocalize (Fig. 2B). Similar colocalization was also apparent using the PksJ-CFP with PksR-YFP fusions (SI Fig. 6). These observations support a model in which the Pks proteins, possibly assembled as functional enzymatic complexes, are organized as a single megacomplex per cell.
To compare localization of the Pks proteins with proteins of a similar size, we fused CFP and YFP to the four proteins of the surfactin NRPS [SrfAA (≈402 kDa), SrfAB (≈401 kDa), SrfAC (≈144 kDa), and SrfAD (≈27 kDa)] (genolist.pasteur.fr/SubtiList). In contrast to the Pks proteins, the surfactin synthase proteins do not localize to a point but are diffuse throughout the cytoplasm (Fig. 2C and SI Fig. 6). The level of fluorescence intensity varies between cells in a field expressing SrfA protein fusions to CFP and YFP, consistent with a previous report that described heterogeneity within a population of B. subtilis cells (21). Differential patterns of localization for Pks fusion proteins and those of the surfactin synthetase suggest that the localization patterns of the NRPS/PKS and NRPS proteins are physiologically relevant for production of small molecules.
The fluorescent fusion proteins also allowed us to test some of the requirements for the assembly of the megacomplex. We reasoned that the PksE trans-AT, a relatively small protein (≈86 kDa), might require the presence of the large core synthase proteins PksJ, L, M, N, and R to localize properly. We therefore investigated the localization of a fluorescent PksE protein fusion in a strain lacking the giant subunits (Fig. 1A). We engineered a B. subtilis strain carrying the PksE-CFP fusion and a transposon insertion in the pksJ gene (SI Text). Disrupting the pksJ gene caused PksE-CFP to mislocalize and become diffuse in the cytoplasm (Fig. 2D). However, normal localization of PksE-CFP was observed in a strain carrying a transposon disruption in pksR, the last gene in the cluster (SI Fig. 6). The requirement of the multimodular enzyme assembly line proteins for localization of PksE-CFP indicates that localization of the large Pks proteins determines the pattern for localization of the trans-acting enzymes.
A PKS Megacomplex Visible by Cryoelectron Microscopy.
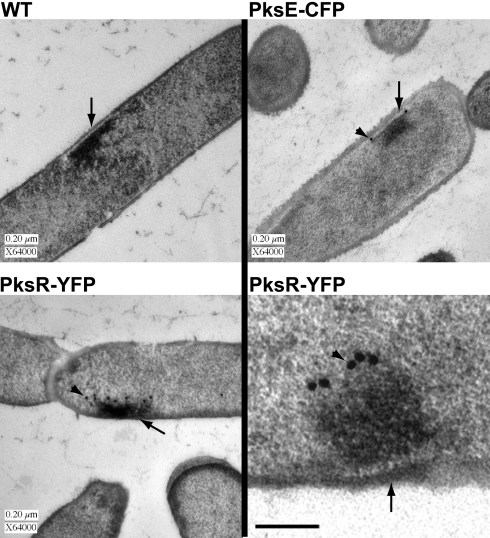
Given that our data suggest that all of the Pks proteins reside at a single site, we reasoned that this megacomplex might be large and distinct enough to be observed using EM. WT (unmodified strain NCIB3610) cells or cells containing either PksR-YFP or PksE-CFP were cryopreserved using high-pressure freezing and freeze-substitution fixation to provide accurate preservation of subcellular structures, and thin sections were examined by EM (22). The WT, PksR-YFP, and PksE-CFP samples all showed a large electron-dense mass in the cytoplasm (Fig. 3 and SI Fig. 8). In all cases, the mass is juxtaposed to the cell membrane, and the position within the cell varies from the midcell to the pole. The mass, which differs in size and shape from cell to cell, does not appear to have a discrete border with the cytoplasm, suggesting that it is not membrane enclosed (see SI Fig. 8 for additional images). This mass has not been reported previously in EM analyses of B. subtilis. We hypothesized that previous EM analyses using laboratory strains of B. subtilis would not have detected this structure due to the presence of a mutated copy of the sfp gene (sfp°), which encodes the phosphopantetheinyl transferase required for assembly line enzyme function. However, a large electron-dense mass was observed by EM in PY79 (sfp°) cells using our cryopreservation techniques, indicating that a functional Sfp enzyme is not required for megacomplex formation (SI Fig. 9). In addition, we have noted that localization of Pks protein fusions is visible by fluorescence in the PY79 strain. An alternative explanation for lack of detection by EM previously is that our method of cryopreservation maintains the integrity of these structures, which might be destroyed using classical fixation protocols.
Fig. 3.
Assembly of a megasynthase complex in B. subtilis. Transmission electron microscopy (TEM) of thin sections from cryopreserved B. subtilis cells reveals an electron-dense mass near the plasma membrane in WT (unmodified strain NCIB3610), PksE-CFP, and PksR-YFP cells. ImmunoEM localization of the PksR-YFP and PksE-CFP proteins to the mass is visible as circular electron-dense dots of 20-nm gold particles conjugated to the secondary antibody (arrowheads). Arrows indicate the NRPS/PKS megacomplex mass. (Scale bar, 0.2 μm.) (Bottom Right) A higher-magnification image of PksR-YFP localized to the megacomplex is displayed. The arrow points to the cell wall and plasma membrane. Arrowhead points to a gold particle. (Scale bar, 0.1 μm.)
To determine whether the mass we see by EM in a WT B. subtilis cell corresponds to the fluorescent focus of NRPS/PKS observed in our fluorescence microscopy analyses, we used immunoEM detection of PksR-YFP and PksE-CFP. Using an anti-GFP primary antibody and a gold-labeled secondary antibody, we observed that PksR-YFP and PksE-CFP localize to the electron-dense mass in the B. subtilis cytoplasm (Fig. 3 and SI Fig. 8). The presence of the large mass in WT B. subtilis and the localization of Pks proteins to this mass suggest that we have identified a proteinaceous subcellular structure whose function, at least in part, is the biosynthesis of secreted bacillaene.
To investigate whether assembly of the megacomplex depends upon the Pks proteins, we compared WT B. subtilis to a pksΔ strain by EM. Limiting our analysis to cells sectioned longitudinally in our EM thin sections, we compared 43 cells of the WT strain to 43 cells of the pksΔ strain. In this analysis, 21 of 43 WT thin sections had a visible mass with similar characteristics to masses that were positive in our immunoEM localization of PksR-YFP and PksE-CFP (SI Fig. 9). By contrast, the pksΔ thin sections lacked these structures, having only 3 of the 43 pksΔ cells with a mass that we could not unambiguously rule out. The presence of these masses in WT but not pksΔ cells indicates that assembly of this structure requires the Pks proteins. We hypothesize that the mass is an organelle-like assemblage of primarily NRPS/PKS synthases and possibly other proteins.
Given the massive size of this macromolecular assemblage, we sought to estimate the amount of NRPS/PKS protein in a single cell using quantitative Western blots (SI Text). We calculated that, on average, a single cell contains 50–150 copies of the PksR protein and ≈5,000–10,000 copies of the PksE protein. It is likely that not all of the PksE proteins are present in the complex, because we can detect some fluorescent signal in the cytoplasm. The accuracy of the estimated mass of the megacomplex is limited by variation in size of the structure (as seen by fluorescence intensity and EM) from cell to cell and by the differences in immunoblot transfer between small purified GFP (27-kDa) and the much larger PksR-YFP (312-kDa) and PksE-CFP (104-kDa) protein fusions. However, the ratio of 10–100 × the amount of the trans-AT, PksE, over the multimodular protein, PksR, is consistent with the predicted function of the proteins. Based on the relative stoichiometry of core synthase proteins to trans-acting enzymes, the mass of a single complete synthase may be well over 2.5 megadaltons. An assemblage of many synthases would range from 10 to 100 megadaltons.
Assembly Line Association of PKS Multimodular Proteins PksM, PksN, and PksR.
In accord with the assembly line organization, we hypothesized that individual B. subtilis NRPS/PKSs consist of a core synthase composed of the PksJ-R proteins and multiple copies of trans-acting enzymes. To probe the association of individual synthase components in the cell, we used an anti-GFP antibody conjugated to Sepharose beads to immunoprecipitate PksR-YFP and associated NRPS/PKS proteins from cell lysates. Coimmunoprecipitated proteins were either trichloroacetic acid-precipitated or excised from silver-stained acrylamide gels and identified by tandem MS analysis. A WT strain without a fusion served as a negative control. Peptide fragments of the PksN and PksM proteins were identified in the MS analyses, consistent with these proteins forming individual assembly lines with PksR (SI Table 2). As predicted by a colinear order of assembly (Fig. 1A), peptides of PksN were more abundant in MS analysis than peptides of PksM. We did not identify PksJ and PksL in the coimmunoprecipitation experiments. The absence of canonical linker domains at the C termini of PksJ and PksL suggests that the split modules at the PksJ-L and PksL-M junctions are organized by nonlinker protein–protein interactions (6, 23, 28). Thus, the interactions between PksJ-PksL and PksL-PksM proteins may not form a complex of sufficient stability for immunoprecipitation. We have proposed a biosynthetic scheme for bacillaene that is supported by the collinear assembly of PksJ-R (28). Based on our knowledge of the bacillaene structure, the C-terminal KS domains on PksJ and PksL would serve only for transfer of an acyl-S-T intermediate, as opposed to incorporation of an additional substrate. In contrast, the interaction between PksN and PksR would require action of the PksN C-terminal DH and KR modules on an acyl-S-T intermediate attached to the N-terminal PksR-T domain (Fig. 4). Given this scheme, we speculate that PksN, PksM, and PksR form a complex that we can identify by immunoprecipitation, but PksJ-PksL or PksL-PksM do not. Thus, in the absence of canonical C- and N- terminal linkers, the requirements for substrate processing and transfer between multimodular proteins in a synthase may be important determinants of protein complex formation and synthase assembly.
It remains to be determined how the collection of individual NRPS/PKS assembly lines in a cell organizes into a single megacomplex. One possibility is that a specific portion of every assembly line is associated with a membrane subdomain, either directly or by interaction with a membrane protein. This kind of membrane association has been observed in proteins from other assembly line enzymes (24, 25). Another possibility is that the individual assembly line complexes associate with each other to form a megacomplex. In this scenario, membrane association of the megacomplex could occur through a subset of the intact assembly lines, but megacomplex assembly would be driven by interactions between domains within individual synthases. Intriguingly, there are predicted coiled-coil motifs within several of the ATd domains in the Pks synthase (Fig. 4). We speculate that the coiled-coil domains may play a role in ordering multimodular proteins within a single assembly line or in mediating the potential association of two complete assembly lines.
In sum, we are reporting several previously uncharacterized features of the organization and capacity of B. subtilis to make antibiotics. The proteins of the bacillaene synthase are incorporated into an organelle-like cluster juxtaposed to the membrane. Formation and localization of this megacomplex are consistent with assembly of a biosynthetic factory, potentially coupled to dedicated secretion machinery to avoid toxic buildup of bacillaene in the cytoplasm. It will be important to determine the identity of the exporter protein(s) as well as the biochemical determinants for localization of the Pks proteins and assembly of the megacomplex. Although lantibiotic synthase factories anchored at the cytoplasmic face of bacterial membranes have been reported (26, 27), our findings provide insights into quaternary organization of natural product biosynthetic factories in antibiotic producers. Finally, we highlight that the combination of the bacillaene structure and the observed synthase organization in the model organism B. subtilis holds great potential for understanding enzymatic synthesis of hybrid PK-NRP metabolites.
Materials and Methods
General Methods.
Media used are as follows: LB was used for routine culture: 1% Bacto Tryptone/0.5% yeast extract (Difco, Sparks, MD)/0.5% NaCl (Mallinckrodt, Hazelwood, MO). YEME-pH7 was used for coculture plating assays: 1% Bacto Malt Extract/0.4% yeast extract/0.4% d-glucose, pH 7.0/buffered with 100 mM morpholinepropane sulfonic acid/5 mM potassium phosphate. Solid media contained 1.5% agar. CH medium was used for microscopy: 1% casein hydrolysate (Oxoid, Hampshire, U.K.)/0.47% l-glutamate/0.16% l-asparagine/0.12% l-alanine/1 mM KH2PO4/25 mM NH4Cl/0.22 mg/ml Na2SO4/0.2 mg/ml NH4NO3/1 μg/ml FeCl3·6H2O/25 mg/liter CaCl2·2H2O/50 mg/liter MgSO4/15 mg/liter MnSO4·H2O/20 μg/ml l-tryptophan, pH 7.0. Antibiotics were used at the following concentrations: spectinomycin, 100 μg/ml; erythromycin plus lincomycin (mls), 1 and 25 μg/ml; kanamycin, 25 μg/ml. Isopropyl β-d-thiogalactoside was added to media to a final concentration of 1 mM.
Bacterial cultures.
B. subtilis coculture with S. coelicolor and S. avermitilis was carried out by spreading ≈1 × 106 Streptomyces spores per plate, allowing the plates to dry, and subsequently spotting 2.5 μl of B. subtilis from an overnight culture. Plates were incubated at 30°C for 2–4 days for phenotypic analysis. To culture B. subtilis for microscopy, 5 ml of CH medium was inoculated with a single colony. After overnight incubation at 30°C, the cells were diluted into 25 ml of fresh CH medium to an optical density of OD600 = 0.1 and cultured at 30°C to an OD600 ≥ 1.2.
Protein analysis.
Western blotting was performed by using cell lysates prepared as follows. Ten-milliliter cell pellets from an OD600 ≥ 1.2 culture were lysed by incubation in 1 ml of lysis buffer at 4°C for 20 min. Lysis buffer: 50 mM Tris, pH 7.5/200 mM NaCl/0.5 mM EDTA/5 mM MgCl2/1 mg/ml lysozyme/1 mM PMSF/1 mM DTT. After lysozyme treatment, cells were disrupted by sonication for 4 × 20-sec pulses. After sonication, lysates were treated with 10 μg of DNase I and 100 μg of RNase A for 20 min on ice. Thirty microliters of crude lysate was added to 30 μl of 2× sample buffer. Samples were loaded on 7.5% acrylamide gels for electrophoresis.
Strain and Plasmid Construction.
Strains used are listed in SI Table 3, plasmid construction is described in SI Text, and the primers used are listed in SI Table 4.
Fluorescence Microscopy.
Fluorescence microscopy was done on an Olympus (Melville, NY) BX61 phase-contrast microscope equipped with an UplanF1 ×100 objective and a CoolSnapHQ digital camera (Photometrics, Tucson, AZ). Image acquisition and analysis were performed by using Metamorph software Version 6.1 (Universal Imaging, Glendale, WI). Exposure times were 2,000 ms for CFP and YFP and 200 ms for TMA-DPH. TMA-DPH (Molecular Probes) was used at a final concentration of 10 μM.
Electron Microscopy.
Cells were cultured in 50 ml of CH medium at 30°C to OD600 = 1.2. One-milliliter samples for EM were pelleted by centrifugation, and aliquots of cell pellets were transferred to the 0.2-mm deep well of aluminum planchettes (Technotrade International, Manchester, NH) and high-pressure-frozen in a BAL-TEC (Oerlikon, Balzers, Liechtenstein) HPM-010 High Pressure Freezer. The frozen samples were freeze-substituted in 0.25% glutaraldehyde/0.1% uranyl acetate in acetone at −80°C for 3 days, then gradually warmed to −20°C, infiltrated with Lowicryl HM20 embedding resin (EMS, Hatfield, PA), and finally polymerized in BEEM capsules under UV illumination at −45°C. Sections with a nominal thickness of 60 nm were collected on formvar-coated copper or nickel (for immunolabeling) EM grids. For immunolocalization of the GFP-fusion proteins, sections on nickel grids were floated on blocking buffer consisting of 1% nonfat dry milk in PBS with 0.1% Tween 20 for 30 min, incubated for 2 h on anti-GFP primary antibody (J. Kahana and P. Silver, Harvard University, Cambridge, MA) diluted 1:150 in blocking buffer, rinsed in PBST, then exposed to goat-anti-rabbit 20-nm gold secondary antibody (Ted Pella, Inc., Redding, CA) for 1 h and rinsed. All grids were stained with uranyl acetate and lead citrate, then viewed in a Philips CM10 transmission electron microscope (FEI Company, Hillsboro, OR) operating at 80 KV. Images were recorded with a Gatan BioScan (Pleasanton, CA) digital camera or on Kodak (Rochester, NY) 4489 electron microscope film.
Supplementary Material
Acknowledgments
We thank Thomas Giddings, Jr., at the University of Colorado, Boulder, CO, for EM Miling Yan for genetic screening; Nathalie Campo for sharing experimental expertise; Ross Tomaino for MS analyses; and members of the D.Z.R. and R.K. laboratories for advice. This work was supported by National Institutes of Health Grants GM20011 (to C.T.W.) and GM58213 (to R.K.). M.A.F. was supported by a fellowship from the Hertz Foundation, and P.D.S. was supported by National Science Foundation Postdoctoral Fellowship in Microbial Biology DBI-0200307.
Abbreviations
- NRPS
nonribosomal peptide synthetase
- PKS
polyketide synthase
- AT
acyltransferase
- CFP
cyan fluorescent protein
- YFP
yellow fluorescent protein.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609073103/DC1.
References
- 1.Du L, Sanchez C, Shen B. Metab Eng. 2001;3:78–95. doi: 10.1006/mben.2000.0171. [DOI] [PubMed] [Google Scholar]
- 2.Walsh CT. Science. 2004;303:1805–1810. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- 3.Fischbach MA, Walsh CT. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 4.Jenni S, Leibundgut M, Maier T, Ban N. Science. 2006;311:1263–1267. doi: 10.1126/science.1123251. [DOI] [PubMed] [Google Scholar]
- 5.Maier T, Jenni S, Ban N. Science. 2006;311:1258–1262. doi: 10.1126/science.1123248. [DOI] [PubMed] [Google Scholar]
- 6.Chen XH, Vater J, Piel J, Franke P, Scholz R, Schneider K, Koumoutsi A, Hitzeroth G, Grammel N, Strittmatter AW, et al. J Bacteriol. 2006;188:4024–4036. doi: 10.1128/JB.00052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng YQ, Tang GL, Shen B. Proc Natl Acad Sci USA. 2003;100:3149–3154. doi: 10.1073/pnas.0537286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards DJ, Marquez BL, Nogle LM, McPhail K, Goeger DE, Roberts MA, Gerwick WH. Chem Biol. 2004;11:817–833. doi: 10.1016/j.chembiol.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 9.Chang Z, Sitachitta N, Rossi JV, Roberts MA, Flatt PM, Jia J, Sherman DH, Gerwick WH. J Nat Prod. 2004;67:1356–1367. doi: 10.1021/np0499261. [DOI] [PubMed] [Google Scholar]
- 10.Calderone CT, Kowtoniuk WE, Kelleher NL, Walsh CT, Dorrestein PC. Proc Natl Acad Sci USA. 2006;103:8977–8982. doi: 10.1073/pnas.0603148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogan DA, Kolter R. Science. 2002;296:2229–2232. doi: 10.1126/science.1070784. [DOI] [PubMed] [Google Scholar]
- 12.Hogan DA, Vik A, Kolter R. Mol Microbiol. 2004;54:1212–1223. doi: 10.1111/j.1365-2958.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- 13.Romano JD, Kolter R. J Bacteriol. 2005;187:940–948. doi: 10.1128/JB.187.3.940-948.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Straight PD, Willey JM, Kolter R. J Bacteriol. 2006;188:4918–4925. doi: 10.1128/JB.00162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weaver VB, Kolter R. J Bacteriol. 2004;186:2376–2384. doi: 10.1128/JB.186.8.2376-2384.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baltz RH. SIM News. 2005;55:186–196. [Google Scholar]
- 17.Stein T. Mol Microbiol. 2005;56:845–857. doi: 10.1111/j.1365-2958.2005.04587.x. [DOI] [PubMed] [Google Scholar]
- 18.Tsao SW, Rudd BA, He XG, Chang CJ, Floss HG. J Antibiot (Tokyo) 1985;38:128–131. doi: 10.7164/antibiotics.38.128. [DOI] [PubMed] [Google Scholar]
- 19.Zheng G, Yan LZ, Vederas JC, Zuber P. J Bacteriol. 1999;181:7346–7355. doi: 10.1128/jb.181.23.7346-7355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel PS, Huang S, Fisher S, Pirnik D, Aklonis C, Dean L, Meyers E, Fernandes P, Mayerl F. J Antibiot (Tokyo) 1995;48:997–1003. doi: 10.7164/antibiotics.48.997. [DOI] [PubMed] [Google Scholar]
- 21.Kearns DB, Losick R. Genes Dev. 2005;19:3083–3094. doi: 10.1101/gad.1373905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giddings TH. J Microsc. 2003;212:53–61. doi: 10.1046/j.1365-2818.2003.01228.x. [DOI] [PubMed] [Google Scholar]
- 23.Tang GL, Cheng YQ, Shen B. Chem Biol. 2004;11:33–45. doi: 10.1016/j.chembiol.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 24.Simunovic V, Gherardini FC, Shimkets LJ. J Bacteriol. 2003;185:5066–5075. doi: 10.1128/JB.185.17.5066-5075.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain M, Cox JS. PLoS Pathogenet. 2005;1:e2. doi: 10.1371/journal.ppat.0010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engelke G, Gutowski-Eckel Z, Hammelmann M, Entian KD. Appl Environ Microbiol. 1992;58:3730–3743. doi: 10.1128/aem.58.11.3730-3743.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiesau P, Eikmanns U, Gutowski-Eckel Z, Weber S, Hammelmann M, Entian KD. J Bacteriol. 1997;179:1475–1481. doi: 10.1128/jb.179.5.1475-1481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butcher RA, Schroeder F, Fischbach MA, Straight PD, Kolter R, Walsh CT, Clardy J. Proc Natl Acad Sci USA. 2007 doi: 10.1073/pnas.0610503104. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.