Abstract
We previously proposed that a balance between nucleotide excision and template RNA degradation plays an important role in nucleoside reverse transcriptase inhibitor (NRTI) resistance. To explore the predictions of this concept, we analyzed the role of patient-derived C-terminal domains of HIV-1 reverse transcriptase (RT) in NRTI resistance. We found that when the polymerase domain contained previously described thymidine analog resistance mutations, mutations in the connection domain increased resistance to 3′-azido-3′-deoxythymidine (AZT) from 11-fold to as much as 536-fold over wild-type RT. Mutational analysis showed that amino acid substitutions E312Q, G335C/D, N348I, A360I/V, V365I, and A376S were associated strongly with the observed increase in AZT resistance; several of these mutations also decreased RT template switching, suggesting that they alter the predicted balance between nucleotide excision and template RNA degradation. These results indicate that mutations in the C-terminal domain of RT significantly enhance clinical NRTI resistance and should be considered in genotypic and phenotypic drug resistance studies.
Keywords: drug resistance, excision, recombination, RNase H, thymidine analog mutations
Nucleoside reverse transcriptase inhibitors (NRTIs) constitute a major class of clinically effective antiretroviral drugs (1). HIV-1 populations possess high genetic diversity, which allows them to acquire rapidly resistance to NRTIs and other inhibitors, limiting the effectiveness of antiviral drugs in controlling viral replication and combating AIDS (2). Resistance to the NRTIs 3′-azido-3′-deoxythymidine (AZT), 2,3-didehydro-2,3-dideoxythymidine (d4T), dideoxyinosine 2′,3′-dideoxyinosine, 2′,3′-dideoxycytidine, abacavir, and tenofovir is associated with thymidine analog resistance mutations (TAMs) that are located in the polymerase (pol) domain of HIV-1 reverse transcriptase (RT) (1).
We recently observed that AZT treatment increases the frequency of RT template switching in single-cycle assays and that mutations in the RNase H (rh) domain of HIV-1 RT confer high-level resistance to AZT and d4T (3). RT template switching occurs through a proposed mechanism called dynamic copy choice (4), which postulates that a balance between the rates of DNA synthesis and RNA degradation is an important determinant of RT template switching: slowing DNA synthesis increases RT template switching, whereas reducing RNA degradation decreases RT template switching (4, 5). Based on these observations and predictions of the dynamic copy choice model, we proposed a previously undescribed mechanism for NRTI resistance, which states that a balance between degradation of HIV-1 RNA by rh and nucleotide excision from a terminated primer is an important determinant of NRTI resistance. Thus, a reduced rate of RNA degradation is proposed to increase the time period available for excision of incorporated NRTIs, leading to an increase in NRTI resistance.
To investigate whether this proposed mechanism contributes to NRTI resistance arising during antiviral therapy, we performed extensive genotypic and phenotypic analyses of RTs derived from seven NRTI-experienced and seven treatment-naïve patients. We found that the C-terminal domains of RT from treatment-experienced patients, but not treatment-naïve patients, substantially increased resistance to AZT as much as 536-fold over wild-type RT when the pol domain contained TAMs. We identified several amino acid substitutions in the connection (cn) domain that were strongly associated with the observed increase in AZT resistance. Additionally, the changes in AZT resistance correlated with a reduction in the RT template-switching frequency, reflecting a shift in the balance between polymerization and RNA degradation.
Results
C-Terminal Domains from NRTI Treatment-Experienced Patients Increase AZT Resistance.
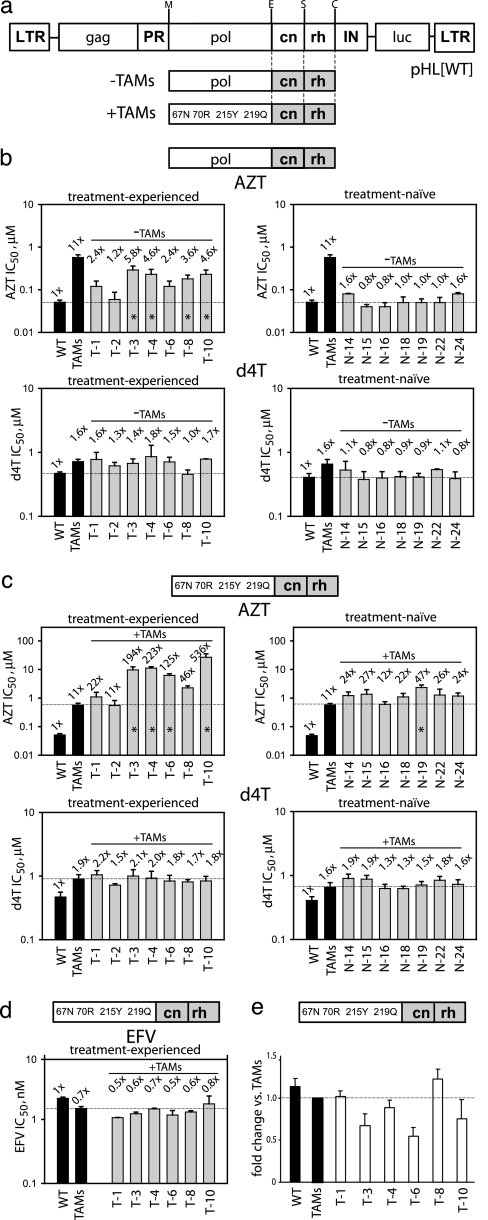
Susceptibility to antiretroviral drugs was determined in a single-replication-cycle drug susceptibility assay (6) by using an HIV-1 vector containing either the wild-type pol domain (pHL[WT]; WT control) or a pol domain with a cluster of four TAMs (D67N, K70R, T215Y, and K219Q) (pHL[TAMs]; TAMs control) (Fig. 1a). The AZT concentration needed to reduce luciferase activity to 50% (IC50) was determined to be 0.05 ± 0.006 μM for WT (1×) and 0.57 ± 0.09 μM for the TAMs control (11× over WT).
Fig. 1.
Drug resistance and replicative capacity associated with the C-terminal RT domains derived from treatment-experienced and treatment-naïve patients. (a) Schematic representation of the HIV-1 vector pHL[WT] used for antiretroviral drug resistance testing. Patient RT subdomains (gray boxes) were subcloned either in the context of a wild-type pol domain (−TAMs) or a TAMs (D67N, K70R, T215Y, and K219Q)-containing pol domain (+TAMs). LTR, long-terminal repeat; gag, gag gene; PR, protease gene; IN, integrase gene; luc, luciferase gene. The letters above the vector designate restriction sites: M, MscI; E, Eco47III; S, SpeI; C, ClaI. (b) AZT and d4T IC50 values for treatment-experienced and treatment-naïve patient C-terminal RT domains in the context of a wild-type pol domain. (c) AZT and d4T IC50 values for treatment-experienced and treatment-naïve patient C-terminal RT domains in the context of a TAMs-containing pol domain. (d) EFV IC50 values for treatment-experienced patient C-terminal RT domains in the context of a TAMs-containing pol domain. In b–d, the fold changes in IC50 values vs. wild-type virus (WT) are shown above each bar. (e) Replicative capacity of virus containing treatment-experienced patient C-terminal RT domains in the context of a TAMs-containing pol domain. Statistically significant differences (∗) in IC50 values or replicative capacities were measured vs. WT (b, dashed reference line) or vs. TAMs control (c–e, dashed reference line). Error bars represent SEM from 3 to 10 replicates per experiment.
The C-terminal RT domain (amino acids 289–560) from NRTI treatment-experienced and treatment-naïve patients was subcloned in the context of a wild-type pol domain (Fig. 1a, −TAMs,) and the effects on AZT sensitivity were determined [Fig. 1b Upper and supporting information (SI) Table 1]. Several of the C-terminal domains from treatment-experienced patients (T-3, T-4, T-8, and T-10) contributed small (3.6- to 5.8-fold) but statistically significant increases in AZT resistance above WT (P < 0.05). As expected, C-terminal domains of treatment-naïve patients (N-14, N-15, N-16, N-18, N-19, N-22, and N-24) did not exhibit significantly increased AZT resistance above WT (P > 0.3).
We then subcloned C-terminal domains from treatment-experienced and treatment-naïve patients in the context of a pol domain containing TAMs (Fig. 1a, +TAMs) and determined the effects on AZT resistance (Fig. 1c Upper and SI Table 2). The C-terminal domains derived from treatment-experienced patients T-3, T-4, T-6, and T-10 exhibited 125- to 536-fold increases in AZT resistance relative to WT. These values were significantly higher than the 11-fold increase in AZT resistance observed with the TAMs control (P < 0.0001). The C-terminal domains from patients T-1, T-2, and T-8 did not exhibit significantly increased AZT resistance above the TAMs control (P > 0.6). Most of the C-terminal domains from treatment-naïve patients did not exhibit significant increases in AZT resistance (22- to 27-fold) above the TAMs control, except from patient N-19, which exhibited a significant 47-fold increase in AZT resistance (P < 0.0001). Examination of the original pol domain from this patient showed the presence of mutation T215C, suggesting that the patient's virus likely was exposed at one time to NRTIs (7).
The C-terminal domains from both treatment-experienced and treatment-naive patients did not show an increase in d4T resistance in the context of a wild-type (Fig. 1b Lower and SI Table 1) or TAMs-containing (Fig. 1c Lower and SI Table 2) pol domain (P > 0.4). Similarly, no significant changes in resistance to the nonnucleoside RT inhibitor efavirenz (EFV) were observed for patient-derived C-terminal domains in the context of a TAMs-containing pol domain relative to the TAMs control (P > 0.1) (Fig. 1d and SI Table 2).
AZT Resistance Associated with the C-Terminal Domain Is Not Correlated with Replicative Capacity.
To determine whether increases in AZT resistance associated with the C-terminal domains from treatment-experienced patients were correlated with their effects on replicative capacity, we infected 293T cells with equivalent amounts of virus derived from pHL[TAMs] constructs containing the patient's C-terminal RT domains in the context of a TAMs-containing pol domain, as determined by p24 capsid quantities in virion preparations. Replicative capacity was measured by relative luciferase light units. No significant differences in replicative capacity were observed (P > 0.1), and the replicative capacity was not correlated (by linear regression analysis, r2 = 0.2807; data not shown) with the observed increases in AZT resistance (Fig. 1e).
The Increase in AZT Resistance Is Primarily Associated with the cn Domain.
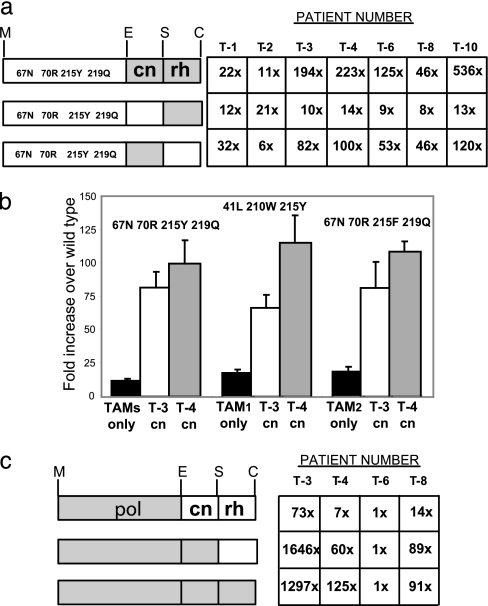
The C-terminal domains of treatment-experienced and treatment-naïve patients contained on average 16 and 12 aa substitutions, respectively, relative to reference strain pNL4–3 (8). To determine which amino acid substitutions were responsible for the increase in AZT resistance, we subcloned patient-derived C-terminal fragments containing either the rh domain (amino acids 424–560) or the cn domain (amino acids 289–423) into pHL[TAMs] and determined the effects on AZT sensitivity (Fig. 2a). We found that the rh domain fragments did not significantly increase AZT resistance compared with the 11-fold increase observed with the TAMs control. In contrast, the cn domain fragments from treatment-experienced patients T-3, T-4, T-6, T-8, and T-10 increased AZT resistance 46- to 120-fold relative to WT (P < 0.05). The increases in AZT resistance observed with the entire C-terminal domains were generally 2- to 4-fold higher than the increases observed with only the cn domains. Thus, the increases in AZT resistance associated with the C-terminal domains could be largely attributed to the fragments containing the cn domains.
Fig. 2.
The effects of the pol, cn, and rh domains on AZT resistance levels. (a) The resistance levels to AZT for viruses containing the patient-derived C-terminal domains, rh domains, or cn domains in conjunction with TAMs. The left side schematically shows RT divided into pol, cn, and rh domains. Gray boxes indicate patient-derived amino acid sequences; white boxes indicate wild-type amino acid sequences. The amino acids representing the cluster of TAMs are shown in the white box corresponding to pol. The numbers in the table represent the mean of fold differences in resistance to AZT over WT for the corresponding combinations of domains from three or more experiments. (b) AZT resistance level for the combination of patients' connection domains with different clusters of AZT resistance mutations in pol. AZT resistance is shown as the fold difference in AZT IC50 over WT (mean of three or more experiments ± SEM). Black bars correspond to the combination of TAMs with WT cn domain; white and gray bars correspond to the combinations of TAMs with T-3 and T-4 cn domains, respectively. The first group of TAMs is D67N, K70R, T215Y, and K219Q; the second group (TAM1) is M41L, L210W, and T215Y; the third group (TAM2) is D67N, K70R, T215F, and K219Q. (c) AZT resistance levels for the patient's entire RT, patient's RT with the rh domain replaced by WT, and the combination of the patient's pol domain with WT C-terminal RT domain. All designations are the same as for a. The following mutations were present in the pol domain from each patient: T-3 = V60I, K64H, D67N, T69N, K70R, V106I, K122E, I135T, Y188L, T215F, D218E, K219Q, K233Q, and L228H; T-4 = M41L, K43E, E44A, D67N, L100I, K102R, K103N, V118I, K122E, D123N, T139K, D177N, M184V, G196E, L210W, R211T/A, T215Y, and K219N; T-6 = V35L, M41L, E44D, D67N, L74V, R83K, L100I, K103N, K122P, A158S, Q174K, D177E, M184V, E194D, and T215Y; and T-8 = M41L, L74I, K103N, V108I, V118I, L210W, and T215Y.
Patient-Derived cn Domains Increase AZT Resistance in the Presence of Different TAMs Combinations.
To determine whether increases in AZT resistance observed with the C-terminal domains obtained from treatment-experienced patients depended on the TAMs present in the pol domain, we tested AZT resistance in the presence of two additional TAMs combinations associated with the TAMs1 and TAMs2 pathways described in refs. 9–11 (Fig. 2b). Cn domains from WT HIV-1, T-3, or T-4 in combination with a pol domain containing D67N, K70R, T215Y, and K219Q resulted in 11-, 82-, and 100-fold increases in AZT resistance over WT (1-fold; IC50 = 0.045 ± 0.003 μM), respectively. Similarly, the combination of WT HIV-1, T-3, and T-4 cn domains with TAMs1 (M41L, L210W, and T215Y) or TAMs2 (D67N, K70R, T215F, and K219Q) pathway cluster mutations resulted in 17-, 66-, and 115-fold or 18-, 81-, and 108-fold increases in AZT resistance over WT, respectively. Therefore, the addition of patient-derived cn domains to all analyzed TAMs combinations resulted in statistically significant increases in AZT IC50 values (P < 0.005).
The cn Domains from Treatment-Experienced Patients Increase AZT Resistance in the Context of Patient-Derived pol Domains.
We determined whether the cn domains from treatment-experienced patients increased AZT resistance in the context of pol domains obtained from the same patients. We subcloned the pol, pol plus cn, or pol plus cn plus rh domains from patients T-3, T-4, T-6, and T-8 into pHL[WT] and determined AZT sensitivity (Fig. 2c). The pol domain of patient T-3 increased AZT resistance 73-fold vs. WT (1-fold); addition of the cn and the cn plus rh domains increased AZT resistance 1,646- and 1,297-fold, respectively. Similarly, the pol domain from patient T-4 or T-8 increased AZT resistance 7- or 14-fold vs. WT, respectively; the addition of the cn and the cn plus rh domains from patient T-4 or T-8 increased AZT resistance 60- and 125-fold or 89- and 91-fold, respectively. These results indicated that the cn domains from patients T-3, T-4, and T-8 increased AZT resistance in the context of the pol domain derived from the same patient.
Surprisingly, the pol domain obtained from patient T-6 did not increase AZT resistance, even with the inclusion of the cn or the cn plus rh domains. In an effort to understand why the cn domain from T-6 increased AZT resistance when the pol domain contained TAMs (D67N, K70R, T215Y, and K219Q), but not when the pol domain was obtained from the same patient, we analyzed the amino acid sequence of the pol domain from patient T-6 (Fig. 2). The pol domain of patient T-6 contained the M184V and L74V substitutions in addition to the TAMs, both of which have been shown to increase AZT sensitivity by decreasing the efficiency of nucleotide excision (12–15). This result suggested that the cn domain mutations were likely to have been selected during the treatment of patient T-6 and the AZT-sensitizing mutations M184V and L74V were selected later, perhaps in response to treatment with 2′,3′-dideoxy-3′thiacytidine (3TC), abacavir and/or dideoxyinosine 2′,3′-dideoxyinosine (ddI) (SI Table 3). The observation that the cn domain from T-6 did not increase AZT resistance indicated that these cn domain mutations could not overcome the AZT-sensitizing effects of M184V plus L74V. Patient-derived T-3, T-4, and T-8 pol domains also showed the presence of TAMs (Fig. 2), with T-4 also containing M184V and T-8 also containing L74I. These mutations likely decreased the enhancing effects of the cn domain on AZT resistance (compared with patient T-3 lacking these mutations), but they were not as severe as the M184V plus L74V double mutation in T-6.
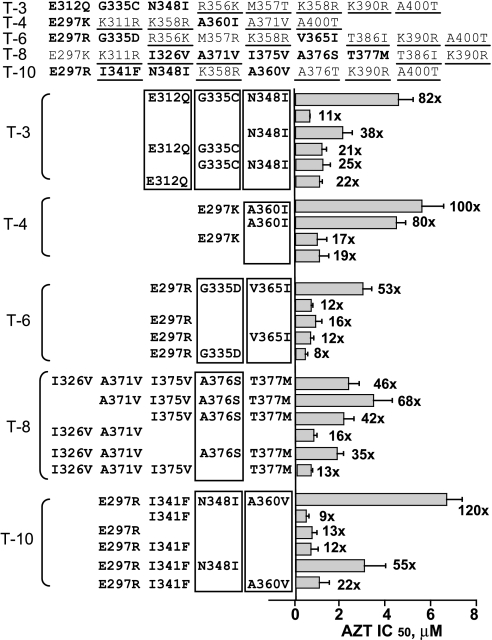
Identification of Mutations in the cn Domain That Are Associated with AZT Resistance.
The cn domains from treatment-experienced patients T-3, T-4, T-6, T-8, and T-10 contained 6 to 10 aa substitutions relative to the reference NL4-3 sequence. To identify the mutations responsible for the increase in AZT resistance, we performed mutational analysis to revert one or more of the substituted amino acids in the patient cn domains to the reference NL4-3 and determined AZT resistance (Fig. 3). For example, the cn domain obtained from patient T-3 increased AZT resistance 82-fold relative to WT. Reversion of amino acid substitutions E312Q, G335C, and N348I resulted in a reduction in AZT resistance to only 11-fold over WT, the same as the TAMs control (11-fold), indicating that the 82-fold increase in AZT resistance was associated with one or more of these three substitutions. Reversion of the E312Q and G335C substitutions reduced AZT resistance to 38-fold increase relative to WT, indicating that these substitutions were not sufficient to account for the 82-fold increase in AZT resistance. Reversion of the N348I substitution reduced AZT resistance to a 21-fold increase, indicating that the N348I substitution largely, but not completely, accounted for the increase in AZT resistance. Similar to the N348I reversion, the E312Q and the G335C plus N348I reversions showed 25- and 22-fold increases in AZT resistance, respectively. Taken together, these results indicated that all three substitutions E312Q, G335C, and N348I contributed to the increase in AZT resistance associated with the cn domain of patient T-3. Similar mutational analysis indicated that the A360I substitution was responsible for the 100-fold increase in AZT resistance observed with patient T-4 cn domain, the G335D and V365I substitutions were responsible for the 53-fold increase associated with patient T-6 cn domain, the A376S substitution was responsible for the 46-fold increase noted with patient T-8 cn domain, and the combination of substitutions N348I plus A360V was responsible for the 120-fold increase associated with patient T-10 cn domain. These results indicated that at least eight different amino acid substitutions at six positions in the cn domain could increase AZT resistance in the context of a pol domain containing TAMs.
Fig. 3.
Identification of mutations that enhance AZT resistance in the C-terminal domain of RT from treatment-experienced patients. Each line named T-3, T-4, T-6, T-8, and T-10 in the top portion indicates the patient's cn domain amino acids (297–423) that are different from wild-type pNL4–3. The underlined amino acids were present in one or more cn domains from treatment-naïve patients; amino acids indicated in bold were reverted to the wild type in the mutational analysis. The results of mutational analysis are shown below, and each group is labeled with the patient number (T-3 to T-10). Only the amino acids targeted for mutational analysis are shown. The top line in each group represents the control that contains all mutations in this patients' cn domain. The absence of amino acids in subsequent lines indicates reversion to the WT amino acid. The bars next to each line correspond to the AZT resistance level from three or more experiments ± SEM; the number is the fold difference in AZT IC50 over WT (1-fold).
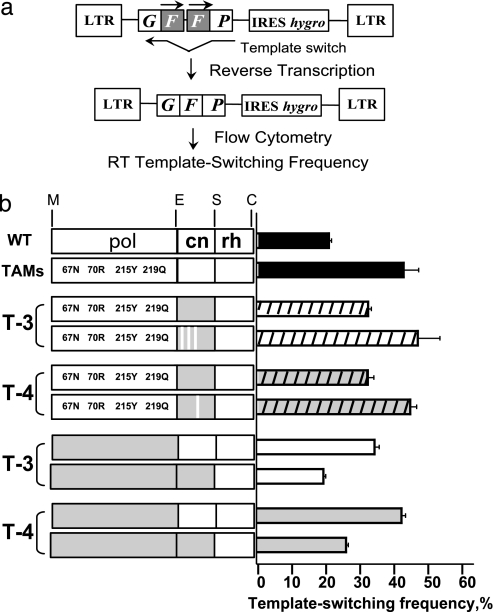
The cn Domains of Treatment-Experienced Patients Reduce RT Template Switching.
We previously proposed that reductions in rh activity would increase resistance to AZT by increasing the time period available for nucleotide excision (3). We and others have observed that reducing rh activity also results in a decrease in RT template switching (4, 5, 16, 17). To determine whether mutations in the cn domains obtained from treatment-experienced patients reduce rh activity, we determined their effects on RT template switching by using a previously described direct-repeat deletion assay (5). Briefly, an HIV-1 GFFP vector that contains two overlapping fragments of the green fluorescent protein (GFP) gene was mobilized with a helper construct and a vesicular stomatitis virus envelope protein expression construct, and allowed to undergo one cycle of replication (Fig. 4a). During reverse transcription, RT undergoes template-switching events within homologous repeats composed of the GFP overlapping portions to functionally reconstitute the gfp gene. Quantitation of the GFP-positive infected cells provides a measure of the frequency of RT template switching in a single replication cycle.
Fig. 4.
Effect of RT amino acid sequence on template-switching frequency. (a) Simplified structure of HIV-1-GFFP provirus with two overlapping fragments of the gfp gene (gray box). HIV-1-GFFP provirus mobilized with a helper construct undergoes one cycle of replication; during reverse transcription, template-switching events within homologous repeats functionally reconstitute the GFP gene. Quantitation of the GFP-positive infected cells by flow cytometry provides a measure of the frequency of RT template switching in a single replication cycle. (b) The left side schematically shows RT divided into pol, cn, and rh domains. Gray boxes represent patient-derived amino acid sequences; white boxes represent WT amino acid sequences. White stripes in the gray cn domain boxes represent a reversion of the patient-derived mutant amino acid back to WT. The bars on the right represent the template-switching frequency (% GFP reconstitution) as measured in the direct-repeat-deletion assay (mean of two or more experiments ± SEM.).
A helper construct containing a patient-derived cn domain was used to mobilize the HIV-1 GFFP vector from a stable cell line. The virus produced was used to infect 293T cells, which then were selected for hygromycin resistance and analyzed for GFP expression by using flow cytometry. Mobilization of the HIV-1 GFFP vector by using the TAMs control vector resulted in ≈43% GFP-positive infected cells vs. 21% for WT (P < 0.006) (Fig. 4b). This result is consistent with our previous observation that RT mutations that affect the balance of reverse transcription by slowing pol activity cause an increase in template switching (5). The TAMs presumably slow the rate of reverse transcription and, consequently, increase template switching.
When the vector pHL[TAMs] containing the cn domain from patient T-3 was used to mobilize the GFFP vector, the frequency of GFP-positive infected cells was 32% (P < 0.05, vs. TAMs control), indicating that inclusion of the cn domain from T-3 reduced the RT template-switching frequency. When we used the vector in which mutations E312Q, G335C, and N348I were reverted to WT, the frequency of GFP-positive infected cells increased to 47%, which was not different from TAMs control (P > 0.8), indicating that the cn domain mutations associated with AZT resistance also were associated with a reduction in RT template switching. Similarly, the cn domain obtained from patient T-4 also was associated with a reduction in RT template-switching frequency to 32% (P < 0.05, vs. TAMs control). The reversion of the A360I substitution, which was associated with an increase in AZT resistance, increased the RT template-switching frequency to 45%, which is not different from TAMs control (P > 0.9).
We also determined the effects of the cn domains from patients T-3 and T-4 on RT template-switching frequency in the context of the pol domains obtained from the same respective patients. The results indicated that the T-3 and T-4 patient-derived cn domains were associated with reductions in RT template-switching frequencies from 35% to 20% and from 43% to 26% in the context of the patient-derived pol domains, respectively (P < 0.001, within each pair).
Discussion
The results of these studies show that several mutations in the cn domain of HIV-1 RT are strongly associated with an enhancement of AZT resistance when the pol domain contains TAMs. Because five of seven randomly chosen treatment-experienced patients possessed AZT resistance-associated mutations in the cn domain, which were not found in the cn domains of seven treatment-naïve patients, it is likely that cn domain mutations contribute to NRTI resistance in a large percentage of treatment-experienced patients. Larger studies are needed to determine the prevalence of cn domain mutations in HIV-1-infected patients undergoing antiviral therapy.
The absence of AZTR-associated mutations in the rh domain could reflect the small number of patients analyzed in this study, and analysis of additional patients could reveal such mutations. It is also possible that mutations within the cn domain are less deleterious for viral replication than mutations in the rh domain and, as a result, are preferentially selected in response to antiviral therapy.
Our observation that the cn domain mutations reduced the frequency of RT template switching is consistent with the hypothesis that these mutations reduce rh activity and, thereby, increase the time period available for nucleotide excision. One attractive hypothesis is that these mutations adversely affect the rh primer grip, which helps position the template primer at the rh active site and, therefore, reduce the rate of RNA template degradation. This hypothesis is supported by the fact that residue A360, which is associated with enhanced AZT resistance, is part of the rh primer grip, and substitutions at this position were shown to reduce rh activity in biochemical studies (18, 19). The other amino acids that are associated with AZT resistance are not close to the rh primer grip, and the mechanism by which they increase AZT resistance is not apparent from their position in the crystal structure. Interestingly, a G333E substitution was reported to be associated with AZT resistance (20); this substitution and the G335C/D substitutions identified in this study might increase AZT resistance through a similar mechanism.
Although these data are consistent with the cn domain mutations reducing rh activity and, thereby, enhancing AZTR, other mechanisms also could explain the decrease in RT template switching and increase in AZTR. Specifically, the cn domain mutations could alter the affinity of the RT for the template primer, enhance nucleotide excision, and reduce template switching. The cn domain mutations also could alter the structure of the RT and/or the template-primer at the pol active site as observed for murine leukemia virus RT (21) and directly enhance nucleotide excision.
Very few studies have focused on the potential role of the C-terminal domains of RT in antiviral drug resistance. Commercial genotypic and phenotypic assays traditionally have characterized and reported on the first 300 aa of RT; consequently, very little genotypic data are widely available for analysis of the cn domain mutations and their prevalence in treatment-experienced patients. Based on the results reported here, we propose that the C-terminal domains of RT should be considered in future genotypic and phenotypic analyses of clinical antiviral drug resistance.
Methods
Plasmids, Cloning, and Mutagenesis.
pHCMV-G expresses the vesicular stomatitis virus envelope protein (22). Vector pHL[WT] was constructed by introducing two restriction enzyme sites, Eco47III (flanking RT amino acid 288/289) and SpeI (flanking RT amino acid 423/424), into pNLuc, an HIV-1-based vector expressing the firefly luciferase reporter gene and all of the HIV-1 proteins except Nef and Env (23). In addition, pHL[WT] contains a natural MscI site at the beginning of RT (flanking RT amino acid 25/26) and a natural ClaI site at the end of rh (flanking integrase amino acid 4/5). To ensure these sites were unique, we eliminated the MscI, ClaI, and SpeI sites in the integrase, luciferase, and gag genes, respectively. pHL[TAMs] was constructed by introducing mutations D67N, K70R, T215Y, and K219Q into the pHL[WT] RT domain. All mutagenesis was carried out by site-directed mutagenesis (QuikChange XL Site-Directed Mutagenesis Kit, Stratagene, La Jolla, CA), and the presence or absence of each mutation was verified by sequencing. Viral RNA and cDNA preparation was described in ref. 24. Patient RT pol (MscI to Eco47III), cn (Eco47III to SpeI) and/or rh (SpeI to ClaI) domain combinations were subcloned into pHL[WT] or pHL[TAMs] via PCR amplification of patient cDNAs with specific primer sets.
Clinical Patient Samples and Antiviral Drugs.
Clinical samples were collected from patients at the Critical Care Medicine Department of the National Institute of Allergy and Infectious Diseases (NIAID) at the Clinical Center of the National Institutes of Health (NIH), Bethesda, MD, and was approved by the Institutional Review Board of NIAID. Blood samples were collected, with written consent, from seven NRTI treatment-experienced and seven treatment-naïve patients. Whole-blood samples were processed and centrifuged within 4 h of collection to separate plasma, and aliquots were stored at –80°C. Treatment-experienced patients were labeled with a number preceded by “T.” Treatment-naïve patients were labeled with a number preceded by “N.” The patient treatment histories are summarized in SI Table 3. NRTI inhibitors AZT and d4T were obtained from Sigma–Aldrich (St. Louis, MO). The nonnucleoside RT inhibitor EFV was obtained from the NIH AIDS Research & Reference Reagent Program.
Cells, Transfection, Virus Production, and Single-Replication Cycle Drug Susceptibility Assay.
Human 293T cells (American Type Culture Collection, Manassas, VA) and the 293T-based cell line GN-HIV-GFFP (5) were maintained at 37°C and 5% CO2 in Dulbecco's Modified Eagle's Medium (CellGro, Herdon, VA) supplemented with 10% FCS (HyClone, Logan, UT), penicillin (50 units/ml; GIBCO, Carlsbad, CA) and streptomycin (50 μg/ml; GIBCO). Hygromycin (Calbiochem, San Diego, CA) selection was performed at a final concentration of 270 μg/ml. Virus production and drug susceptibility testing was carried out as described in refs. 3 and 6.
Replicative Capacity Assay.
Virus containing the entire C-terminal RT domain from patient clones was harvested from 293T cells, and normalized p24 capsid (HIV-1 p24 ELISA Kit; PerkinElmer, Shelton, CT) values were used to infect target 293T cells in a single-replication-cycle assay. Luciferase light units were measured 48 h after infection to determine replicative capacity.
Determination of RT Template Switching Frequency.
RT template-switching frequency was determined by using the direct-repeat-deletion assay as described in ref. 5. For the mobilization of provirus from the GN-HIV-GFFP cell line, cotransfection with vesicular stomatitis virus envelope protein and the corresponding pHL[WT]-based helper construct were used.
Statistical Analysis.
Group comparisons were quantified by using a one-way ANOVA (SAS, JMP software). Dunnett's test was used to compare a control group with several other groups. Additional multiple comparisons were completed by using the Matlab software system (Mathworks, Natick, MA) with the Kruskal–Wallis nonparametric ANOVA. Bonferroni's adjustment was used where appropriate to control for type I errors. In some cases, Student's t test was used to describe differences (SIGMAPLOT 8.0 software). P values of <0.05 were regarded as significant.
Supplementary Material
Acknowledgments
We especially thank W.-S. Hu for intellectual input throughout the project and J. Mellors for valuable discussions. We also thank Eric Freed for critical comments during manuscript preparation and A. Arthur for expert editorial assistance. This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Abbreviations
- AZT
3′-azido-3′-deoxythymidine
- cn
connection domain
- d4T
2,3-didehydro-2,3-dideoxythymidine
- NRTI
nucleoside reverse transcriptase inhibitor
- pol
polymerase
- rh
RNase H
- RT
reverse transcriptase
- TAM
thymidine analog resistance mutation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609642104/DC1.
References
- 1.Vivet-Boudou V, Didierjean J, Isel C, Marquet R. Cell Mol Life Sci. 2006;63:163–186. doi: 10.1007/s00018-005-5367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Havlir DV. Proc Natl Acad Sci USA. 2002;99:4–6. doi: 10.1073/pnas.022629399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikolenko GN, Palmer S, Maldarelli F, Mellors JW, Coffin JM, Pathak VK. Proc Natl Acad Sci USA. 2005;102:2093–2098. doi: 10.1073/pnas.0409823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Svarovskaia ES, Delviks KA, Hwang CK, Pathak VK. J Virol. 2000;74:7171–7178. doi: 10.1128/jvi.74.15.7171-7178.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikolenko GN, Svarovskaia ES, Delviks KA, Pathak VK. J Virol. 2004;78:8761–8770. doi: 10.1128/JVI.78.16.8761-8770.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petropoulos CJ, Parkin NT, Limoli KL, Lie YS, Wrin T, Huang W, Tian H, Smith D, Winslow GA, Capon DJ, Whitcomb JM. Antimicrob Agents Chemother. 2000;44:920–928. doi: 10.1128/aac.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Lerma JG, Nidtha S, Blumoff K, Weinstock H, Heneine W. Proc Natl Acad Sci USA. 2001;98:13907–13912. doi: 10.1073/pnas.241300698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanna GJ, Johnson VA, Kuritzkes DR, Richman DD, Brown AJL, Savara AV, Hazelwood JD, D'Aquila RT. J Infect Dis. 2000;181:904–911. doi: 10.1086/315329. [DOI] [PubMed] [Google Scholar]
- 10.Marcelin AG, Delaugerre C, Wirden M, Viegas P, Simon A, Katlama C, Calvez V. J Med Virol. 2004;72:162–165. doi: 10.1002/jmv.10550. [DOI] [PubMed] [Google Scholar]
- 11.Yahi N, Tamalet C, Tourres C, Tivoli N, Ariasi F, Volot F, Gastaut JA, Gallais H, Moreau J, Fantini J. J Clin Microbiol. 1999;37:4099–4106. doi: 10.1128/jcm.37.12.4099-4106.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gotte M, Arion D, Parniak MA, Wainberg MA. J Virol. 2000;74:3579–3585. doi: 10.1128/jvi.74.8.3579-3585.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. J Virol. 2002;76:3248–3256. doi: 10.1128/JVI.76.7.3248-3256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miranda LR, Gotte M, Liang F, Kuritzkes DR. Antimicrob Agents Chemother. 2005;49:2648–2656. doi: 10.1128/AAC.49.7.2648-2656.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frankel FA, Marchand B, Turner D, Gotte M, Wainberg MA. Antimicrob Agents Chemother. 2005;49:2657–2664. doi: 10.1128/AAC.49.7.2657-2664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang CK, Svarovskaia ES, Pathak VK. Proc Natl Acad Sci USA. 2001;98:12209–12214. doi: 10.1073/pnas.221289898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brincat JL, Pfeiffer JK, Telesnitsky A. J Virol. 2002;76:88–95. doi: 10.1128/JVI.76.1.88-95.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Julias JG, McWilliams MJ, Sarafianos SG, Alvord WG, Arnold E, Hughes SH. J Virol. 2003;77:8548–8554. doi: 10.1128/JVI.77.15.8548-8554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rausch JW, Lener D, Miller JT, Julias JG, Hughes SH, Le Grice SFJ. Biochemistry. 2002;41:4856–4865. doi: 10.1021/bi015970t. [DOI] [PubMed] [Google Scholar]
- 20.Kemp SD, Shi CF, Bloor S, Harrigan PR, Mellors JW, Larder BA. J Virol. 1998;72:5093–5098. doi: 10.1128/jvi.72.6.5093-5098.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mbisa JL, Nikolenko GN, Pathak VK. J Virol. 2005;79:419–427. doi: 10.1128/JVI.79.1.419-427.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yee JK, Miyanohara A, Laporte P, Bouic K, Burns JC, Friedmann T. Proc Natl Acad Sci USA. 1994;91:9564–9568. doi: 10.1073/pnas.91.20.9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiernan RE, Ono A, Englund G, Freed EO. J Virol. 1998;72:4116–4126. doi: 10.1128/jvi.72.5.4116-4126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmer S, Kearney M, Maldarelli F, Halvas EK, Bixby CJ, Bazmi H, Rock D, Falloon J, Davey RT, Dewar RL, et al. J Clin Microbiol. 2005;43:406–413. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.