Abstract
Detecting community structure is fundamental for uncovering the links between structure and function in complex networks and for practical applications in many disciplines such as biology and sociology. A popular method now widely used relies on the optimization of a quantity called modularity, which is a quality index for a partition of a network into communities. We find that modularity optimization may fail to identify modules smaller than a scale which depends on the total size of the network and on the degree of interconnectedness of the modules, even in cases where modules are unambiguously defined. This finding is confirmed through several examples, both in artificial and in real social, biological, and technological networks, where we show that modularity optimization indeed does not resolve a large number of modules. A check of the modules obtained through modularity optimization is thus necessary, and we provide here key elements for the assessment of the reliability of this community detection method.
Keywords: complex networks, modular structure, metabolic networks, social networks
Community detection in complex networks has attracted a lot of attention in recent years (for a review, see refs. 1 and 2). The main reason is that complex networks (3–7) are made of a large number of nodes and most previous quantitative investigations focused on statistical properties disregarding the roles played by specific subgraphs. Detecting communities (or modules) can be a way to identify substructures which could correspond to important functions. This is, for example, confirmed in the case of the World Wide Web, where communities are sets of Web pages dealing with the same topic (8). In biological networks, it is widely believed that the modular structure results from evolutionary constraints and plays a crucial role in biological functions (9–11), which makes community detection very relevant (12–14). Relevant community structures were also found in social networks (15–17), the Internet (18), food webs (19, 20), and in networks of sexual contacts (21, 22).
Loosely speaking, a community is a subgraph of a network whose nodes are more tightly connected with each other than with nodes outside the subgraph. A decisive advance in community detection was made by Newman and Girvan (23), who introduced a quantitative measure for the quality of a partition of a network into communities, the modularity. This measure essentially compares the number of links inside a given module with the expected value for a randomized graph of the same size and same degree sequence. If one chooses modularity as the relevant quality function, the problem of community detection becomes equivalent to modularity optimization. The latter is not trivial, as the number of possible partitions of a network into clusters increases at least exponentially with the size of the network, making exhaustive optimization computationally unfeasible even for relatively small graphs. Therefore, a number of algorithms have been devised to find a good optimization technique with the smallest computational cost possible. The fastest available procedures use greedy techniques (24, 25) and extremal optimization (26), which are, at the present time, the only algorithms capable of detecting communities in large networks. More accurate results are obtained through simulated annealing (27, 28), but this method is computationally very expensive.
Modularity optimization seems, therefore, to be a very effective method to detect communities, both in real and in artificially generated networks. However, modularity itself has not yet been thoroughly investigated, and only a few general properties are known. For example, it is known that the modularity value of a partition does not have a meaning by itself, but only when compared with the corresponding modularity expected for a random graph of the same size (29), as the latter may attain very high values due to fluctuations (27).
In this article, we present a critical analysis of modularity and of the applicability of modularity optimization to the problem of community detection. We show that modularity contains an intrinsic scale that depends on the total number of links in the network. Modules that are smaller than this scale may not be resolved, even in the extreme case where they are complete graphs connected by single bridges. The resolution limit of modularity actually depends on the degree of interconnectedness between pairs of communities and can reach values of the order of the size of the whole network. Tests performed on several artificial and real networks clearly show that this problem is likely to occur.
It is thus a priori impossible to tell whether a module (large or small), detected through modularity optimization, is indeed a single module or a cluster of smaller modules. This raises doubts about the effectiveness of modularity optimization in community detection, and more generally about the applicability of quality functions.
Modularity and the Notion of Community
The modularity of a partition of a network (23) can be written as
 |
where the sum is over the m modules of the partition, ls is the number of links inside module s, L is the total number of links in the network, and ds is the total degree of the nodes in module s. The first term of the summand in Eq. 1 is the fraction of links inside module s; the second term, in contrast, represents the expected fraction of links in that module, if links were located at random in the network (under the only constraint that the degree sequence coincides with the one of the original graph).
If, for a subgraph 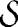 of a network, the first term is much larger than the second, it means that there are many more links inside
of a network, the first term is much larger than the second, it means that there are many more links inside 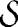 than one would expect by random chance. This means that
than one would expect by random chance. This means that 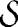 is, indeed, a module. The comparison with the null model (represented by the randomized network) leads to the quantitative definition of community embedded in Eq. 1. We conclude that, in a modularity-based framework, a subgraph
is, indeed, a module. The comparison with the null model (represented by the randomized network) leads to the quantitative definition of community embedded in Eq. 1. We conclude that, in a modularity-based framework, a subgraph 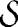 with ls internal links and total degree ds is a module if
with ls internal links and total degree ds is a module if
We can express the number of links lsout joining nodes of the module s to the rest of the network in terms of ls, i.e. lsout = als with a ≥ 0. Therefore, ds = 2ls + lsout = (a + 2)ls and the condition (Eq. 2) becomes
from which, rearranging terms, one obtains
If a = 0, the subgraph 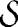 is a disconnected part of the network and is a module if ls < L, which is always true. If a is strictly positive, Eq. 4 sets an upper limit to the number of internal links that
is a disconnected part of the network and is a module if ls < L, which is always true. If a is strictly positive, Eq. 4 sets an upper limit to the number of internal links that 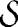 must have in order to be a module. This is counterintuitive, because it means that the definition of community implied by modularity depends on the size of the whole network, instead of involving a “local” comparison between the number of internal and external links of the module. For a < 2 one has 2ls > lsout, which means that the total internal degree of the subgraph is larger than its external degree: dsin > dsout. The attributes “internal” and “external” mean that the degree is calculated considering only internal or external links, respectively. In this case, the subgraph
must have in order to be a module. This is counterintuitive, because it means that the definition of community implied by modularity depends on the size of the whole network, instead of involving a “local” comparison between the number of internal and external links of the module. For a < 2 one has 2ls > lsout, which means that the total internal degree of the subgraph is larger than its external degree: dsin > dsout. The attributes “internal” and “external” mean that the degree is calculated considering only internal or external links, respectively. In this case, the subgraph 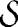 would be a community according to the “weak” definition given by Radicchi et al. (30).
would be a community according to the “weak” definition given by Radicchi et al. (30).
For a < 2, the right-hand-side of inequality (4) is in the interval [L/4, L]. A subgraph of size ls such that a < 2 and ls is less than a quantity in the interval [L/4, L] would then be a community both within the modularity framework and according to the weak definition of Radicchi et al. (30). Sufficient conditions for which these constraints are always met are then
In the following, we will only consider modules of this kind.
According to Eq. 2, a partition of a network into actual modules (i.e. subgraphs satisfying the condition Eq. 2) would have a positive modularity, as all summands in Eq. 1 are positive. On the other hand, it is possible to partition a network such that Q is negative. The network itself, considered as a partition with a single module, has modularity zero: in this case, in fact, l1 = L, d1 = 2L, and the only two terms of the unique module in Q cancel each other. Usually, a value of Q larger than 0.3–0.4 is a clear indication that the subgraphs of the corresponding partition are modules. However, the maximal modularity differs from one network to another and depends on the number of links of the network. Below, we will derive the expression of the maximal possible value QM(L) that Q can attain on a network with L links. We will prove that the upper limit for the value of modularity for any network is one and we will see why modularity is not scale independent.
The Most Modular Network
Here, we discuss the properties of the network with the highest possible modularity, which will then naturally lead to the problem of scales in modularity optimization. In ref. 2, the authors consider the interesting example of a network made of m identical complete graphs (cliques), disjoint from each other. In this case, the modularity is maximal for the partition of the network into the cliques and is given by the sum of m equal terms. In each clique there are l = L/m links, and the total degree is d = 2l, as there are no links connecting nodes of the clique to the other cliques. We thus obtain
which converges to one when the number of cliques goes to infinity. We note that this result is still valid even if the m connected components are not cliques. Also, the number of nodes of the network and within the modules does not affect modularity. If we have m modules, we just need to have L/m links inside the modules, as long as this is compatible with topological constraints such as connectedness.
A further interesting question is how to construct a connected network with N nodes and L links which maximizes modularity. To address this issue, we proceed in two steps: first, we consider the maximal value QM(m, L) for a partition with a fixed number of modules m; after that, we look for the number m* that maximizes QM(m, L).
Let us first consider a partition with m modules. Ideally, to maximize the contribution to modularity of each module, we should reduce the number of links connecting modules as much as possible. To keep the network connected, we must have at least m − 1 intercommunity links. For the sake of clarity and to simplify the mathematical expressions (without affecting the final result), we shall analyze the simple ring-like configuration illustrated in Fig. 1, which has m intercommunity links instead of m − 1.
Fig. 1.
Design of a connected network with maximal modularity. The modules (circles) must be connected to each other by the minimal number of links.
The modularity of such a network is
 |
where
 |
The expression of Eq. 7 reaches its maximum when all modules contain the same number of links, i.e. ls = l = L/m − 1, ∀s = 1, 2,…, m. The maximum is then given by
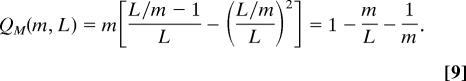 |
We have now to find the maximum of QM(m, L) when the number of modules m is variable. For this purpose, we treat m as a continuous variable and take the derivative of QM(m, L) with respect to m
which vanishes when . This point indeed corresponds to the absolute maximum QM(L) of the function QM(m, L). This result coincides with the one found by Guimerà et al. (27) for a one-dimensional lattice, but our proof is completely general and does not require preliminary assumptions on the type of network and modules.
Because m is actually integer, the maximum is reached when m equals one of the two integers closest to m*, but this is not important for our purpose and from now on we will stick to the real-valued expressions, their meaning being clear. The maximal modularity is then
and approaches one if the total number of links L goes to infinity. The corresponding number of links in each module is − 1. The fact that all modules have the same number of links does not imply that they have the same number of nodes. Again, modularity does not depend on the distribution of the nodes among the modules as long as the topological constraints are satisfied. For instance, if we assume that the modules are connected graphs, there must be at most nodes in each module. The crucial point here is that modularity has some intrinsic scale of order , which constrains the number and the size of the modules. For a given total number of nodes and links we could build many more than modules, but the corresponding network would be less “modular,” with a modularity lower than the maximum given by Eq. 11. This fact is the fundamental reason why small modules may not be resolved through modularity optimization, as it will be clear in the next section.
The Resolution Limit
We analyze a network with L links and with at least three modules (see Fig. 2), each of which satisfies the conditions given in Eq. 5. We focus on a pair of modules, 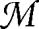 1 and
1 and 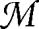 2, and distinguish three types of links: those internal to each of the two communities (l1 and l2, respectively), between
2, and distinguish three types of links: those internal to each of the two communities (l1 and l2, respectively), between 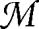 1 and
1 and 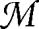 2 (lint) and between the two communities and the rest of the network
2 (lint) and between the two communities and the rest of the network 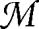 0 (l1out and l2out). To simplify the calculations, we express the numbers of external links in terms of l1 and l2, so lint = a1l1 = a2l2, l1out = b1l1 and l2out = b2l2, with a1, a2, b1, b2 ≥ 0. Because
0 (l1out and l2out). To simplify the calculations, we express the numbers of external links in terms of l1 and l2, so lint = a1l1 = a2l2, l1out = b1l1 and l2out = b2l2, with a1, a2, b1, b2 ≥ 0. Because 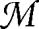 1 and
1 and 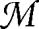 2 are modules by construction, we also have a1 + b1 ≤ 2, a2 + b2 ≤ 2 and l1, l2 < L/4 (see Eq. 5). We now consider two partitions A and B of the network. In partition A,
2 are modules by construction, we also have a1 + b1 ≤ 2, a2 + b2 ≤ 2 and l1, l2 < L/4 (see Eq. 5). We now consider two partitions A and B of the network. In partition A, 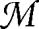 1 and
1 and 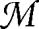 2 are taken as separate modules, and in partition B they are considered as a single community. The subdivision of the rest of the network,
2 are taken as separate modules, and in partition B they are considered as a single community. The subdivision of the rest of the network, 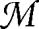 0, is arbitrary but identical in both partitions. We want to compare the modularity values QA and QB of the two partitions and, because modularity is a sum over the modules, the contribution of
0, is arbitrary but identical in both partitions. We want to compare the modularity values QA and QB of the two partitions and, because modularity is a sum over the modules, the contribution of 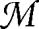 0 is the same in both partitions and is denoted by Q0. From Eq. 1, we obtain
0 is the same in both partitions and is denoted by Q0. From Eq. 1, we obtain
 |
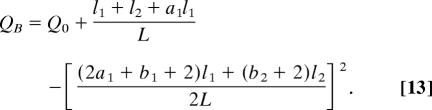 |
The difference ΔQ = QB − QA is
 |
As 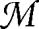 1 and
1 and 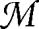 2 are both modules by construction, we expect a larger modularity for the partition where the two modules are separated, i.e. QA > QB, which in turn implies ΔQ < 0. From Eq. 14, we see that ΔQ is negative if
2 are both modules by construction, we expect a larger modularity for the partition where the two modules are separated, i.e. QA > QB, which in turn implies ΔQ < 0. From Eq. 14, we see that ΔQ is negative if
If a1 = a2 = 0, there are no links between 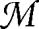 1 and
1 and 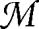 2 and the above condition is trivially satisfied. In contrast, if the two modules are connected to each other, something interesting happens. Each of the coefficients a1, a2, b1, and b2 must be less than two. The numbers of internal links l1 and l2 are both smaller than L/4 by construction and can be taken as small as we wish with respect to L. In this way, it is possible to choose l1 and l2 so that the inequality of Eq. 15 is not satisfied. In such a situation, we have ΔQ > 0 and the modularity of the configuration where the two modules are considered as a single community (B) is larger than the partition where the two modules are clearly identified (A). This implies that, by looking for the maximal modularity, there is the risk of missing important structures at smaller scales. To estimate the size of l1 and l2 at which modularity optimization could fail, we consider for simplicity the case in which
2 and the above condition is trivially satisfied. In contrast, if the two modules are connected to each other, something interesting happens. Each of the coefficients a1, a2, b1, and b2 must be less than two. The numbers of internal links l1 and l2 are both smaller than L/4 by construction and can be taken as small as we wish with respect to L. In this way, it is possible to choose l1 and l2 so that the inequality of Eq. 15 is not satisfied. In such a situation, we have ΔQ > 0 and the modularity of the configuration where the two modules are considered as a single community (B) is larger than the partition where the two modules are clearly identified (A). This implies that, by looking for the maximal modularity, there is the risk of missing important structures at smaller scales. To estimate the size of l1 and l2 at which modularity optimization could fail, we consider for simplicity the case in which 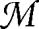 1 and
1 and 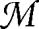 2 have the same number of links, l1 = l2 = l. The condition on l for the modularity to miss the two modules also depends on the “fuzziness” of the modules, as expressed by the values of the parameters a1, a2, b1, and b2. In order to find the range of potentially “dangerous” values of l, we consider the two following extreme cases
2 have the same number of links, l1 = l2 = l. The condition on l for the modularity to miss the two modules also depends on the “fuzziness” of the modules, as expressed by the values of the parameters a1, a2, b1, and b2. In order to find the range of potentially “dangerous” values of l, we consider the two following extreme cases
The two modules have a perfect balance between internal and external degree (a1 + b1 = 2, a2 + b2 = 2), so that they are on the edge of being communities in the weak sense defined in ref. 30.
The two modules have the smallest possible external degree, which means that there is a single link connecting them to the rest of the network and only one link connecting them to each other (a1 = a2 = b1 = b2 = 1/l).
In the first case, the maximum value of the coefficient of L in Eq. 15 is 1/4, obtained for a1 = a2 = 2 and b1 ≈ 0, b2 ≈ 0. Eq. 15 may thus not be satisfied for
which is a scale of the order of the size of the whole network. This result means that even a pair of large communities may not be resolved if they share enough links with the nodes outside them (in this case we speak of “fuzzy” communities). A more striking result emerges when we consider the other limit, when a1 = a2 = b1 = b2 = 1/l. In this case it is easy to check that Eq. 15 is not satisfied if the number of links inside the modules satisfies
 |
If we now assume that we have two (interconnected) modules with the same number of internal links l < lRmin < lRmax, the discussion above implies that the modules cannot be resolved through modularity optimization, even if they were complete graphs connected by a single link. As we have seen from Eq. 16, it is possible to miss modules of larger size, if they share more links with the rest of the network (and with each other). For l1 ≠ l2 the conclusion is similar but the scales lRmin,max are modified by simple factors.
Fig. 2.
Scheme of a network partition into three or more modules. The circles on the left represent two modules 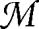 1 and
1 and 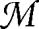 2, the oval on the right represents the rest of the network
2, the oval on the right represents the rest of the network 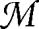 0, whose structure is arbitrary.
0, whose structure is arbitrary.
Consequences
To illustrate the consequences of our finding, we begin with two schematic examples. In Fig. 3A, we show a network consisting of a ring of cliques, connected through single links. Each clique is a complete graph Km with m nodes and has m(m − 1)/2 links. If we assume that there are n cliques (with n even), the network has a total of N = nm nodes and L = nm(m − 1)/2 + n links.
Fig. 3.
Schematic examples. (A) A network made out of identical cliques (which are here complete graphs with m nodes) connected by single links. If the number of cliques is larger than about , modularity optimization would lead to a partition where the cliques are combined into groups of two or more (represented by dotted lines). (B) A network with four pairwise identical cliques (complete graphs with m and p < m nodes, respectively); if m is large enough with respect to p (e.g., m = 20, p = 5), modularity optimization merges the two smallest modules into one (shown with a dotted line).
The network has a clear modular structure where the communities correspond to single cliques, and we expect that any detection algorithm should be able to detect these communities. The modularity Qsingle of this natural partition can be easily calculated and is equal to
On the other hand, the modularity Qpairs of the partition in which pairs of consecutive cliques are considered as single communities (as shown by the dotted lines in Fig. 3A) is
The condition Qsingle > Qpairs is satisfied only if
which can also be rewritten as . In this example, m and n are independent variables, and we can choose them such that the inequality of Eq. 20 is not satisfied. For instance, for m = 5 and n = 30, Qsingle = 0.876 and Qpairs = 0.888 > Qsingle. An efficient algorithm looking for the maximum modularity would find the configuration with pairs of cliques and not the actual modules. The difference Qpairs − Qsingle becomes even larger as n increases, for m fixed.
The example we considered was particularly simple and is not representative of situations found in real networks. However, the initial configuration that we considered above (Fig. 2) is absolutely general, and the results allow us to design arbitrarily many networks with obvious community structures for which modularity optimization will not recognize (some of) the real modules. Another example is shown in Fig. 3B, where the circles again represent cliques (i.e., complete graphs): the two on the left have m nodes each, the other two have p < m nodes. If we take m = 20 and p = 5, the maximal modularity of the network corresponds to the partition in which the two smaller cliques are merged (as shown by the dotted line in Fig. 3B). This trend of the optimal modularity to group small modules has already been empirically observed in ref. 31, but without a complete explanation.
In general, we cannot make any definitive statement about modules found through modularity optimization without a method which verifies whether the modules are indeed single communities or a combination of communities. It is then necessary to inspect the structure of each of the modules found. For example, if we take the network of Fig. 3A, with n = 30 and m = 5, we have seen that modularity optimization find modules which are pairs of connected cliques. By inspecting each of the modules of the “first generation” (by optimizing modularity, for example), we would ultimately find that each module is actually a set of two cliques.
We thus have seen that modules identified through modularity optimization may actually be combinations of smaller modules. During the process of modularity optimization, it is favorable to merge connected modules if they are sufficiently small. We showed in the previous section that any two interconnected modules, fuzzy or not, are merged if the number of links inside each of them does not exceed lRmin. This means that the largest structure one can form by merging a pair of modules of any type (including cliques) has at least 2lRmin internal links. By reversing the argument, we conclude that if modularity optimization finds a module 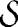 with lS internal links, it may be that the latter is a combination of two or more smaller communities if
with lS internal links, it may be that the latter is a combination of two or more smaller communities if
This example is an extreme case in which the internal partition of 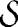 can be arbitrary, as long as the pieces are modules in the weak sense of (30). Under the condition in Eq. 21, the module could, in principle, be a cluster of loosely interconnected complete graphs.
can be arbitrary, as long as the pieces are modules in the weak sense of (30). Under the condition in Eq. 21, the module could, in principle, be a cluster of loosely interconnected complete graphs.
On the other hand, the upper limit of lS can be much larger than , if the substructures are, on average, more interconnected with each other, as we have seen with Eq. 16. In fact, fuzzy modules can be combined with each other even if they contain many more than lRmin links. The more interconnected the modules, the larger will be the resulting supermodule. In the extreme case in which all submodules are very fuzzy, the size lS of the supermodule could be in principle as large as that of the whole network, i.e., lS < L. This result comes from the extreme case where the network is split in two very fuzzy communities, with L/4 internal links each and L/2 between them. By virtue of Eq. 16, it is favorable (or just as good) to merge the two modules with the whole network as the resulting structure. This limit lS < L is always satisfied but suggests here that it is important to carefully analyze all modules found through modularity optimization, regardless of their size.
However, the probability that a very large module conceals substructures is small, because this only happens if all hidden submodules are very fuzzy communities, which is unlikely. Instead, modules with a size or smaller can result from an arbitrary merge of smaller structures, which may go from loosely interconnected cliques to very fuzzy communities. Modularity optimization is most likely to fail in these cases.
To illustrate this theoretical discussion, we analyze five examples of real networks: (i) the transcriptional regulation network of Saccharomyces cerevisiae (yeast), (ii) the transcriptional regulation network of Escherichia coli, (iii) a network of electronic circuits, (iv) a social network, and (v) the neural network of Caenorhabditis elegans. We obtained the lists of edges of the first four networks from www.weizman.ac.il/mcb/UriAlon, whereas the last one was found at http://cdg.columbia.edu.
In the transcriptional regulation networks, nodes represent operons, i.e., groups of genes that are transcribed on to the same mRNA. An edge is set between two nodes A and B if A activates B. These systems have been previously studied to identify motifs in complex networks (32). There are 688 nodes and 1,079 links for yeast and 423 nodes and 519 links for E. coli. Electronic circuits can be viewed as networks in which vertices are electronic components (capacitors, diodes, etc.) and connections are wires. This network maps one of the benchmark circuits of the so-called ISCAS'89 set; it has 512 nodes and 819 links. In the social network that we considered, the 67 nodes are people of a group and the 182 links represent positive sentiments (based on questionnaires) directed from one person to another. Finally, the neural network of C. elegans is made of 306 nodes (neurons), connected through 2,345 links (synapsis, gap junctions). Most of these networks are directed, but we will consider them as undirected.
We look for the maximum modularity by using simulated annealing and we adopt the same recipe introduced in ref. 13, which makes the optimization procedure very effective.
We found that the maximum modularity of all these networks is very high, with values Qmax ranging from 0.4081 (C. elegans) to 0.7519 (E. coli). The corresponding optimal partitions consist of 9 (yeast), 27 (E. coli), 11 (electronic), 10 (social), and 4 (C. elegans) modules (for E. coli, our results differ but are not inconsistent with those obtained in ref. 13 for a different database; these differences, however, do not affect our conclusions). In order to check if the communities have a substructure we used modularity optimization again, by constraining it to each of the modules found. In all cases, we found that most modules displayed a clear community structure with very high values of Q. The total number of submodules is 57 (yeast), 76 (E. coli), 70 (electronic), 21 (social), and 20 (C. elegans), and is far larger than the number of modules obtained at the maximum modularity. By restricting modularity optimization to a module, we neglect all links between the original communities and we have no guarantee that we accurately detect its substructure and that this is a safe way to proceed. Thus, we have to check whether all substructures we detected are real modules, i.e. if they satisfy the condition of Eq. 2; we find that it is indeed the case for all the networks considered here. Our results thus show that the search for the modularity optimum is not equivalent to the detection of communities defined through Eq. 2. The communities found through modularity optimization are in fact clusters of smaller modules. The modularity values corresponding to the partitions of the networks including the submodules are smaller than the peak modularities that we originally found through simulated annealing (see Table 1).
Table 1.
Results of the modularity analysis on real networks
| Network | No. of modules (Qmax) | Total no. of modules (Q) |
|---|---|---|
| Yeast | 9 (0.740) | 57 (0.677) |
| E. coli | 27 (0.752) | 76 (0.661) |
| Elect. circuit | 11 (0.670) | 70 (0.640) |
| Social | 10 (0.608) | 21 (0.532) |
| C. elegans | 4 (0.408) | 20 (0.319) |
In the second column, we report the number of modules detected in the partition obtained for the maximal modularity. However, these modules contain submodules; in the third column we report the total number of submodules we found and the corresponding value of the modularity of the partition, which is lower than the peak modularity initially found.
The networks that we have examined are fairly small but the problem we have discovered can only get worse if we increase the network size, especially when small communities coexist with large ones and the module size distribution is broad, which seems to happen in many cases (25, 33). As an example, we consider the recommendation network of the online seller Amazon.com. While buying a product, Amazon recommends items that have been purchased by people who bought the same product. In this way, it is possible to build a network in which the nodes are the items (books, music), and there is an edge between two items A and B if B was frequently purchased by buyers of A. Such a network was examined in ref. 25 and is very large, with 409,687 nodes and 2,464,630 edges. The authors analyzed the community structure by greedy modularity optimization, which is not necessarily accurate, but represents the only strategy currently available for large networks. They identified 1,684 communities whose size distribution is well approximated by a power law with exponent 2. From the size distribution, we estimated that >95% of the modules have sizes below the limit of Eq. 21, which implies that basically all modules deserve further investigation.
Conclusions
Here, we have analyzed in detail modularity and its applicability to community detection. We have found that the definition of community implied by modularity is actually not consistent with its optimization, which may favor network partitions with groups of modules combined into larger communities. We could say that, by enforcing modularity optimization, the possible partitions of the system are explored at a coarse level, so that modules smaller than some scale may not be resolved. The resolution limit of modularity does not depend on particular network structures, but results only from the comparison between the number of links of the interconnected communities and the total number of links of the network.
Our result implies that modularity optimization might miss important substructures of a network, as we have confirmed in real world examples. Our discussion suggests that it is not possible to rule out that modules of virtually any size may be clusters of modules, although the problem is most likely to occur for modules with a number of internal links of the order of or smaller. For this reason, it is crucial to check the structure of all detected modules, for instance by constraining modularity optimization on each single module, a procedure which is not safe but which might give useful indications.
The origin of the resolution scale lies in the fact that modularity is a sum of terms, where each term corresponds to a module. Finding the maximal modularity is then equivalent to looking for the ideal tradeoff between the number of terms in the sum, i.e., the number of modules, and the value of each term. An increase of the number of modules does not necessarily correspond to an increase in modularity because the modules would be smaller and so each term of the sum would be smaller. This is why, for some characteristic number of terms, modularity has a peak. The problem is that this “optimal” partition, imposed by mathematics, does not necessarily capture the actual community structure of the network, where communities may be very heterogeneous in size, especially if the network is large.
Quality functions other than modularity may have an intrinsic resolution scale that undermines their reliability. We believe that quality functions mathematically similar to modularity, i.e., such that the quality of a partition is given by the sum of the qualities of the individual modules, will have a resolution limit, because of the tradeoff described above. However, there are many possible ways to define the quality of a partition: for instance, one could take the average quality of the modules, instead of the sum, and obtain very different results. Besides, the null model one adopts to describe the absence of community structure could be global (this is the case for modularity, which uses a random graph with the same expected degree sequence) or local, i.e., determined by the properties of the module alone, regardless of the rest of the network. Because of the arbitrariness of the quality function, it is hard to address this issue in general. Nevertheless, for a given quality function, our results suggest that it is necessary to perform tests such as we did for the modularity in order to check for the existence of possible biases and resolution limits.
The fact that quality functions such as modularity can have an intrinsic resolution limit calls for a new theoretical framework that focuses on a local definition of community, rather than on definitions relying on a global null model. Quality functions are still helpful, but their role should probably be limited to the comparison of partitions with the same number of modules.
Acknowledgments
We thank A. Barrat, C. Castellano, V. Colizza, E. Flach, A. Flammini, J. Kertész, F. Menczer, and A. Vespignani for enlightening discussions and suggestions, and U. Alon for providing the network data.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Newman MEJ. Eur Phys J B. 2004;38:321–330. [Google Scholar]
- 2.Danon L, Díaz-Guilera A, Duch J, Arenas A. J Stat Mech. 2005 P09008. [Google Scholar]
- 3.Barabási A-L, Albert R. Rev Mod Phys. 2002;74:47–97. [Google Scholar]
- 4.Dorogovtsev SN, Mendes JFF. Evolution of Networks: From Biological Nets to the Internet and WWW. Oxford: Oxford Univ Press; 2003. [Google Scholar]
- 5.Newman MEJ. SIAM Rev. 2003;45:167–256. [Google Scholar]
- 6.Pastor-Satorras R, Vespignani A. Evolution and Structure of the Internet: A Statistical Physics Approach. Cambridge, UK: Cambridge Univ Press; 2004. [Google Scholar]
- 7.Boccaletti S, Latora V, Moreno Y, Chavez M, Hwang D-U. Phys Rep. 2006;424:175–308. [Google Scholar]
- 8.Flake GW, Lawrence S, Lee Giles C, Coetzee FM. IEEE Computer. 2002;35:66–71. [Google Scholar]
- 9.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. Nature. 1999;499:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 10.Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabási A-L. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 11.Papin JA, Reed JL, Palsson BO. Trends Biochem Sci. 2004;29:641–647. doi: 10.1016/j.tibs.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Holme P, Huss M, Jeong H. Bioinformatics. 2003;19:532–538. doi: 10.1093/bioinformatics/btg033. [DOI] [PubMed] [Google Scholar]
- 13.Guimerà R, Amaral LAN. Nature. 2005;433:895–900. doi: 10.1038/nature03288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palla G, Derényi I, Farkas I, Vicsek T. Nature. 2005;435:814–818. doi: 10.1038/nature03607. [DOI] [PubMed] [Google Scholar]
- 15.Girvan M, Newman MEJ. Proc Natl Acad Sci USA. 2002;99:7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lusseau D, Newman MEJ. Proc R Soc London B. 2004;271:S477–S481. doi: 10.1098/rsbl.2004.0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adamic L, Glance N. Proc 3rd Int Workshop on Link Discovery. Los Angeles: Information Sciences Inst, Univ of Southern California; 2005. pp. 36–43. [Google Scholar]
- 18.Eriksen K, Simonsen I, Maslov S, Sneppen K. Phys Rev Lett. 2003;90:148701. doi: 10.1103/PhysRevLett.90.148701. [DOI] [PubMed] [Google Scholar]
- 19.Pimm SL. Theor Popul Biol. 1979;16:144–158. doi: 10.1016/0040-5809(79)90010-8. [DOI] [PubMed] [Google Scholar]
- 20.Krause A-E, Frank KA, Mason DM, Ulanowicz RE, Taylor WW. Nature. 2003;426:282–285. doi: 10.1038/nature02115. [DOI] [PubMed] [Google Scholar]
- 21.Garnett GP, Hughes JP, Anderson RM, Stoner BP, Aral SO, Whittington WL, Handsfield HH, Holmes KK. Sexually Transmitted Diseases. 1996;23:248–257. doi: 10.1097/00007435-199605000-00015. [DOI] [PubMed] [Google Scholar]
- 22.Aral SO, Hughes JP, Stoner BP, Whittington WL, Handsfield HH, Anderson RM, Holmes KK. Am J Public Health. 1999;89:825–833. doi: 10.2105/ajph.89.6.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman MEJ, Girvan M. Phys Rev E. 2004;69 026113. [Google Scholar]
- 24.Newman MEJ. Phys Rev E. 2004;69 066133. [Google Scholar]
- 25.Clauset A, Newman MEJ, Moore C. Phys Rev E. 2004;70 066111. [Google Scholar]
- 26.Duch J, Arenas A. Phys Rev E. 2005;72 027104. [Google Scholar]
- 27.Guimerà R, Sales-Pardo M, Amaral LAN. Phys Rev E. 2004;70 doi: 10.1103/PhysRevE.70.025101. 025101(R) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reichardt J, Bornholdt S. Phys Rev E. 2006;74 016110. [Google Scholar]
- 29.Reichardt J, Bornholdt S. 2006. arXiv:cond-mat/0606220.
- 30.Radicchi F, Castellano C, Cecconi F, Loreto V, Parisi D. Proc Natl Acad Sci USA. 2004;101:2658–2663. doi: 10.1073/pnas.0400054101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muff S, Rao F, Caflisch A. Phys Rev E. 2005;72 doi: 10.1103/PhysRevE.72.056107. 056107. [DOI] [PubMed] [Google Scholar]
- 32.Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 33.Danon L, Díaz-Guilera A, Arenas A. 2006. arXiv:physics/0601144.





