Abstract
To determine which of seven library design algorithms best introduces new protein function without destroying it altogether, seven combinatorial libraries of green fluorescent protein variants were designed and synthesized. Each was evaluated by distributions of emission intensity and color compiled from measurements made in vivo. Additional comparisons were made with a library constructed by error-prone PCR. Among the designed libraries, fluorescent function was preserved for the greatest fraction of samples in a library designed by using a structure-based computational method developed and described here. A trend was observed toward greater diversity of color in designed libraries that better preserved fluorescence. Contrary to trends observed among libraries constructed by error-prone PCR, preservation of function was observed to increase with a library's average mutation level among the four libraries designed with structure-based computational methods.
Keywords: GFP, library design, protein design, protein engineering, high-throughput screening
Protein sequence space is so vast that one can easily imagine the optimal sequence for a particular application will never be sampled by random mutation and recombination. Structure-based computational protein design tools seek to screen that sequence space more thoroughly than can be screened in the laboratory, but are currently based on approximate representations of candidate sequences and an incomplete understanding of the relationships between structure and function. Although many algorithms used to screen sequences in silico aim to identify a single optimal sequence (1–5), others aim instead to optimize the composition of a library of sequences (6–13). Provided that resources exist to synthesize and screen such libraries, library design algorithms compensate for the approximations built into them by increasing the number of attempts at designing the desired function. Viewed from a complementary perspective, such algorithms aim to sample sequence space more effectively than methods that randomly generate sequence diversity.
Designed libraries can be synthesized for roughly the same cost as a designed sequence by recognizing the opportunities in gene synthesis for the combinatorial shuffling of sequence diversity (14–17). Although many algorithms have now been proposed to design such combinatorial libraries (7–9, 11, 12), few computationally designed libraries have been characterized experimentally (9, 18, 19), and, to our knowledge, there have been no controlled experiments comparing these methods with each other or with libraries of randomly generated sequence diversity. The results of such a comparison would be hard to predict, especially because none of these methods models protein function explicitly. Instead, these algorithms attempt to model protein stability as a surrogate for protein function on the assumption that libraries with a greater fraction of well folded proteins are more likely to contain variants with the desired function.
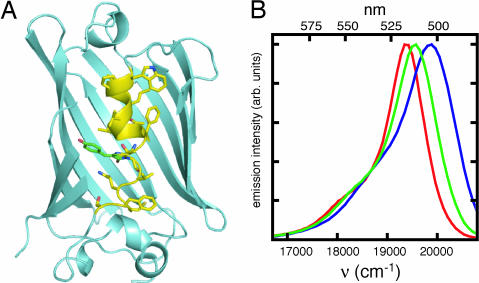
Here, we evaluate seven designed combinatorial libraries of GFPs, including one with mutations picked at random. Preservation and diversity of function were judged by using distributions of brightness and color, respectively, compiled from measurements made in vivo with a monochromator-based plate reader. GFP from Aequorea victoria modified by S65T (20) (GFP-S65T) was chosen as a reference sequence for each design algorithm because this variant is less extensively engineered than other variants whose structures have been solved to similarly high resolution. Positions 57–72 were targeted for this test because they form the longest contiguous stretch of core positions in the GFP-S65T structure (21). The structure of GFP-S65T is illustrated in Fig. 1A, with the targeted positions shown in yellow. Because random core mutations are generally more disruptive than random surface mutations (22, 23), it was assumed that targeting core positions would provide better differentiation of designed libraries according to preservation and diversity of function criteria. Contiguity was imposed to allow an economical and high-fidelity cassette-based library synthesis [see supporting information (SI) Text]. Where possible, libraries were controlled for both theoretical size and the precise distribution of mutation levels within each library, because one would expect these factors to affect library quality when controlled for the same method of design.
Fig. 1.
Structure of GFP-S65T and spectra of variants. (A) The front side of this cylindrical protein has been clipped to spotlight residues 57–72 in its core. Side chain atoms for targeted positions 57–65 and 67–72 are illustrated in CPK colors, with carbon in yellow. The chromophore of GFP is shown in CPK colors, with carbon in green. This figure was composed from a 1.45 Å-resolution structure of GFP containing the S65T and Q80R mutations (PDB ID code 1q4a) (22). (B) Extremes of function. Of the 11,575 spectra measured, 701 were at least one-half as bright as spectra of cultures known to express GFP-S65T. Of these, the redmost spectrum was sampled from the epPCR library (red), and the bluemost spectrum was sampled from the CORBIT library (blue). The spectrum of a culture expressing GFP-S65T is shown in green. The three spectra have been normalized to the same peak intensity. arb, arbitrary
We show that the corresponding design algorithms perform quite differently in this test. Four of the seven libraries were designed with structure-based computational methods: two with an algorithm introduced here (see Methods) and two with algorithms described in refs. 7 and 9. Among these four libraries, we observe that preservation of function increases with a library's average mutation level, contrary to the trends observed for libraries constructed by error-prone PCR (epPCR) (24, 25). Across all seven libraries, we observe a trend toward greater diversity of function in designed libraries with greater preservation of function. An additional library generated by epPCR amplification of the entire GFP-S65T gene exhibited much less dispersion of function than designed libraries with similar preservation of function.
Results
Library Composition.
The seven combinatorial libraries with compositions listed in Table 1 were designed, synthesized, and characterized as described in Methods and in SI Text. Briefly, the labels DBISORBIT, DBISORBIT 44, CORBIT, and SCMFORBIT 322 represent the four libraries designed by using structure-based computational methods that draw on the ORBIT suite of protein design tools (1–3). The DBISORBIT and DBISORBIT 44 libraries were designed by using an algorithm whose principal innovations can be summarized as a diversity benefit applied to interacting sets of amino acids (DBIS). The CORBIT library was designed with a consensus (C) method based on the work of Hayes et al. (9). The SCMFORBIT 322 library was designed by using a self-consistent mean field (SCMF) calculation to direct combinatorial saturation mutagenesis as suggested by Voigt et al. (7). The CMSA and SE/CMSA libraries were each designed with the same multiple sequence alignment (MSA) of naturally occurring fluorescent proteins (26). Both were designed by using a consensus method derived from the work of Hayes et al. (9), but the latter is distinguished by directing mutations to positions that have the largest site entropies (SEs). Mutations in the Random library were picked with a random number generator. To approach 95% confidence that the true extremes of function in each library would be sampled, we aimed to sample most designed libraries by three times their theoretical size (27). Considering also that one-half hour was needed to acquire each set of 96 high-resolution emission spectra, these constraints dictated that theoretical library sizes should be close to 500. Although this size is orders-of-magnitude smaller than most libraries screened for binding (28) or low-resolution fluorescence properties (29, 30), it is especially relevant to difficult-to-screen functions such as improved enzymatic activity with nonfluorogenic substrates. It was assumed that the best differentiation between design algorithms would be achieved by applying them in ways that maximized the average number of mutations per sequence, yet each combinatorial library was constrained to include the sequence of GFP-S65T so that none would be rendered completely nonfunctional because of a uniquely disruptive mutation. Thus, most designed libraries tested here (DBISORBIT, CORBIT, CMSA, SE/CMSA, and Random) have a theoretical size of 29 and an average of 4.5 mutations per sequence. The DBISORBIT 44 and SCMFORBIT 322 libraries have unique sizes and average mutation levels that are conveyed by the labels we have given them. For example, the SCMFORBIT 322 label indicates that this library was made by combinatorial saturation mutagenesis at two positions by using 32-fold degenerate codons.
Table 1.
Library designs
| Pos | DBISORBIT | DBISORBIT44 | CORBIT | SCMFORBIT322 | CMSA | SE/CMSA | Random |
|---|---|---|---|---|---|---|---|
| 57 | W | W | W | W | W | W | W |
| 58 | PA | PAST | PT | all | P | PH | PQ |
| 59 | TS | T | TS | T | TI | T | TN |
| 60 | L | L | L | L | L | L | L |
| 61 | VL | VALS | VL | V | V | VI | VD |
| 62 | TA | TAGS | TA | T | TA | TA | TN |
| 63 | T | T | TA | T | TA | TA | T |
| 64 | F | F | F | F | FL | FL | F |
| 65 | TA | T | TA | T | TS | TS | TK |
| 67 | G | G | G | G | G | G | G |
| 68 | VA | V | V | V | VF | VF | VM |
| 69 | QL | QELV | QL | Q | QR | QR | QE |
| 70 | C | C | C | all | C | C | C |
| 71 | FL | F | FL | F | FY | F | FY |
| 72 | SA | S | SA | S | SA | SA | SI |
The first amino acid listed at each position is that of GFP-S65T. Underlined amino acids are mutations designed as described in Methods.
It is interesting to note the extent to which the compositions of the designed libraries reflect the fact that evolution disfavors ionizable side chains in protein cores. The MSA used to design the CMSA and SE/CMSA libraries illustrates this trend, with a notable exception being the unusually high degree of conservation at position 69 for a buried basic side chain (26). The scoring function used for structure-based design was parameterized specifically to prevent the desolvation of hydrophilic side chains in protein cores under most circumstances (31). Thus, the DBISORBIT 44 library introduces only one acidic side chain among its 12 mutations distributed over four positions, and the DBISORBIT and CORBIT libraries do not introduce any ionizable side chains anywhere. Although the SCMFORBIT 322 library was designed by using the same scoring function as these three other libraries, imposing saturation mutagenesis for this one library makes it introduce many mutations that are strongly disfavored by this scoring function. Thus, the SCMFORBIT 322 library introduces ionizable side chains at core positions with greater frequency than each library tested except the Random library.
Preservation of Function.
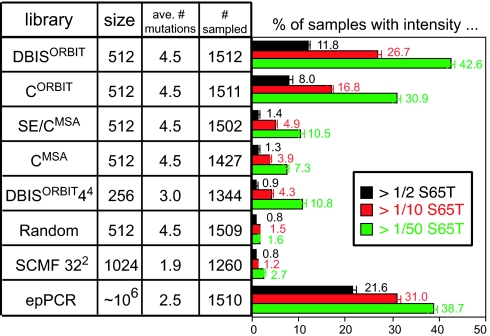
For each of the designed libraries, and for the epPCR library, emission spectra were recorded for ≈1,500 bacterial cultures expressing GFP variants. We define the brightness and color of each spectrum sampled as its integrated emission intensity and average position, respectively. Because it is not clear how best to define a functional sample, we have quantified each library's preservation of function in three ways. For each library, the percentage of samples that have at least one-half, one-tenth, and one-fiftieth the brightness of cultures expressing GFP-S65T are presented as bar graphs in Fig. 2. By all three of these measures, most of the designed libraries performed considerably better than the Random library. Only 1.6% of samples from the Random library had at least one-fiftieth the brightness of cultures expressing GFP-S65T. Although the SCMFORBIT 322 library had a larger fraction of functional samples than the Random library by this most inclusive definition of function, the SCMFORBIT 322 library had a similar fraction by the most exclusive definition. The relatively poor performance of these two libraries is probably due in part to the relatively large frequencies with which these libraries introduce ionizable side chains to the protein core.
Fig. 2.
Preservation of function. A sample is variously defined as being functional if its emission intensity is at least one-half (black), one-tenth (red) or one-fiftieth (green) the intensity of cultures expressing GFP-S65T. Designed libraries are listed from top to bottom according to preservation of function calculated by the most exclusive definition. The theoretical library size, the average (ave.) number of mutations, and the number of clones sampled are listed for each library. A complementary illustration of preservation of function is provided as SI Fig. 5.
By all three of these measures, the DBISORBIT library performed best of all. More than 10% and 40% of its samples were at least one-half and one-fiftieth as bright as cultures expressing GFP-S65T, respectively. The CORBIT library performed nearly as well. The SE/CMSA, CMSA, and DBISORBIT 44 libraries performed similarly to each other, with ≈1% and 10% of samples being at least one-half and one-fiftieth as bright, respectively, as cultures expressing GFP-S65T. The Q69R mutation, because it introduces an ionizable side chain to the protein core, would seem to be responsible for much of the weaker performance of the MSA-based libraries, compared with the DBISORBIT and CORBIT libraries, which instead introduce the Q69L mutation. However, even if it is assumed that the Q69R mutation always disrupts function and that the Q69L mutation never disrupts function, less striking differences among these libraries must account for at least half the observed differences in performance.
Multiple epPCR libraries were synthesized by using different mutation rates. Only the library that appeared to have a fraction of functional samples similar to that of the DBISORBIT library was characterized in detail to compare average mutation levels and diversity of function under this constraint. Despite the fact that random mutations are generally tolerated at surface positions better than at core positions (22, 23), the average number of nonsynonymous mutations for genes in this epPCR library was determined by sequencing to be 2.5, roughly half the average of 4.5 mutations per gene for the core-directed DBISORBIT library.
Diversity of Function.
Because the dimmest samples have colors biased by emission from molecules other than GFP, here, we consider only those samples with at least one-half the brightness of cultures expressing GFP-S65T. Of the 11,575 spectra sampled, 701 met this criterion. The redmost and bluemost of these spectra are illustrated in Fig. 1B.
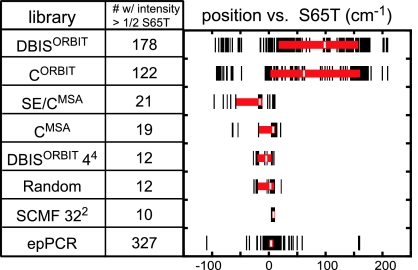
The diversity of function for a library of fluorescent proteins may be associated with either its extremes of color or its dispersion of color. The former we define as the difference between the positions of the redmost and bluemost spectra in a library. Fig. 3 illustrates the set of colors sampled for each library with black marks, such that the separation between leftmost and rightmost marks illustrates a library's performance according to this extremes-of-function metric. Dispersion of function we define as the difference between the positions of the spectra that lie one quartile above and below the median for a library. In Fig. 3, this median is illustrated with a white bar on top of a red box that illustrates the positions of the first and third quartiles.
Fig. 3.
Diversity of function. Considering only those spectra with (w/) at least one-half the intensity of cultures expressing GFP-S65T, this plot illustrates the set of colors sampled from each library (black marks), the median of each set (white bar), and the first and third quartiles (red box). Positions are calculated relative to GFP-S65T standards as described in SI Text. Designed libraries are listed from top to bottom according to preservation of function calculated by the most exclusive definition of function. A complementary illustration of diversity of function is provided as SI Fig. 5.
The seven designed libraries are thus seen to cluster into four performance categories based on these complementary metrics. The DBISORBIT and CORBIT libraries outperform all of the other designed libraries by having both the largest separation between extremes and the greatest dispersion. The SE/CMSA and CMSA libraries constitute the next category by having greater separation between extremes than the DBISORBIT 44 and Random libraries, although they have similar dispersion. The SCMFORBIT 322 library then constitutes the last category by having both the smallest separation between extremes and the least dispersion. By the extremes-of-function metric, the epPCR library performs better than each of the designed libraries except the DBISORBIT and CORBIT libraries; however, by the dispersion-of-function metric, the epPCR library performs worse than each of the designed libraries except the SCMFORBIT 322 library.
A complementary illustration of the preservation and diversity of function sampled from each library is provided as SI Fig. 5. For each library, the width of each spectrum sampled is plotted against its color with a circle of area proportional to its brightness. Although SI Fig. 5 does not characterize the libraries with the statistical rigor of Figs. 2 and 3, it does provide additional support for the clustering and ranking of the designed libraries described above. It also reveals a striking correlation between emission line shape and emission color among the brightest samples in each library. We have investigated the physical mechanisms that may be responsible for this trend with additional measurements (T.P.T., C.L.V., M. A. Mena, D.N., B. D. Olafson, P. S. Daugherty, and S.L.M., unpublished work).
Discussion
Fig. 2 illustrates that preservation of function increases with average mutation level among the four libraries designed by using structure-based computational methods. The opposite trend has been observed for protein libraries synthesized by epPCR (24, 25) and would suggest that, constrained to a particular library size, the designed library with the lowest mutation rate should yield the largest fraction of functional samples. It is thus notable that the poor performance of the SCMFORBIT 322 library in this respect may have more to do with the overarching strategy that enforced its low mutation rate, combinatorial saturation mutagenesis, than the computational method used to select positions for mutation. A library defined by combinatorial saturation mutagenesis would have to tolerate ≈12 different amino acids per position to preserve function as well as the DBISORBIT and CORBIT libraries. Finding any two core positions in GFP-S65T that could accept such great diversity, let alone two between positions 57 and 72, would seem to be an especially difficult problem.
Fig. 3 illustrates that diversity of function tends to increase with preservation of function among the seven designed libraries. This result justifies an approach to library design in which protein stability is modeled as a surrogate for protein function (7–9, 11, 12), as long as mutations are directed toward positions likely to perturb function. Moreover, this result suggests that improvements in modeling protein stability should yield designed libraries that sample a wider array of protein functions.
A frequently desired trait among GFP variants has been red-shifted emission (29, 32, 33). Although the vast majority of the bright variants sampled from the epPCR library have emission spectra nearly identical to cultures expressing GFP-S65T, the one sample from this library with a substantial red-shift did have the redmost spectrum sampled in our test. The corresponding GFP gene was sequenced and was determined to have the V224I and M233K mutations. Only the V224I mutation is in the core of the protein and close to the chromophore, suggesting that it is primarily responsible for the observed red shift. The fact that neither of these mutations involves the positions targeted in the test underscores the way the performance of a designed library is intrinsically limited by the quality of the information in the design, such as the choice of positions targeted for mutation. Nevertheless, the far greater number of almost identically red-shifted samples from the DBISORBIT and CORBIT libraries indicates that our best information at present is a valuable tool with which to complement epPCR for sampling diverse functions.
Even though red-shifted emission is frequently desired for GFPs, other measures described here may be more relevant to the extrapolation of these results to other protein engineering projects. Such projects typically aim to increase the stability of an enzyme, its rate of catalysis, or the affinity of a protein for a ligand (28, 34). Because denatured GFP does not fluoresce (35), one interpretation of Fig. 2 is that the algorithms that preserved function best did so by disrupting the global structure of GFP the least. According to this interpretation, we would predict that the algorithms used to design the DBISORBIT and CORBIT libraries would also perform best when attempting to stabilize an enzyme with core-directed mutations. However, the relative performance of the MSA-based methods might be expected to increase in this case if the covariances among amino acid frequencies important for protein stability can be extracted from evolutionary noise (13, 36, 37).
The emission spectrum of GFP is a reporter on the local structure of its chromophore. In other words, a more varied sampling of spectral properties is equivalent to a more varied sampling of structures at the “active site” of GFP. Thus, based on Figs. 2 and 3, we can predict that the algorithms used to design the DBISORBIT and CORBIT libraries will provide the most diverse sampling of active-site structures in functional enzymes. Structure-based computational methods should thus prove especially useful for relatively low-throughput screening projects in which libraries made by epPCR, even those with low mutation rates, cannot be screened thoroughly.
Binding between a protein and its ligand might also be improved most efficiently by sampling with the greatest frequency those perturbations to the structure of the binding interface that do not completely disrupt the global structure of the complex. Thus, if a structure of the bound complex is available, in this case, too, we would recommend using structure-based computational methods of library design to suggest a small number of mutations at each of many buried positions in or near the binding interface. However, if binding to a novel ligand is desired, it may be necessary to disrupt the structure of the protein more significantly than when improvements in binding to a known ligand are desired. In this case, the kinds of mutations suggested by these algorithms may be overly conservative, especially if the new ligand has a different charge. Because selections for protein binding frequently have much greater throughput than the plate reader-based screen we have implemented here, it is worth noting that the DBIS algorithm can be used to design libraries of practically any size.
In summary, we have shown that small combinatorial libraries can exhibit considerable diversity of function if designed well. Based on the design and results of this test, we recommend complementing more widely used strategies for generating functional diversity, such as epPCR and combinatorial saturation mutagenesis, with a strategy that defines a combinatorial library by a single conservative mutation at each of many positions close to a protein's active site. We have found structural information as used by the DBIS algorithm or the method of Hayes et al. (9) to be more successful than limited evolutionary information in identifying compatible conservative mutations. Although currently limited by the need for an accurate structure, the utility of the structure-based design algorithms should improve as methods improve for docking ligands onto proteins and for determining protein structures from protein sequences. Indeed the great promise of these methods for library design is that they might be used to implement a knowledge-based approach to engineering totally novel functions for which no natural protein exhibits even the slightest glimmer of the desired function. In the meantime, this approach to protein engineering should prove especially useful for investigations of protein structure–function relationships (T.P.T., C.L.V., M. A. Mena, D.N., B. D. Olafson, P. S. Daugherty, and S.L.M., unpublished work), where, ideally, large numbers of differently functional variants would be related by the same small set of mutations.
Methods
The DBIS Algorithm.
One of the fundamental innovations of the DBIS algorithm is that it aims to explicitly model the interactions among sets of amino acids at the positions targeted for design. Set singles and pairs energies are constructed analogous to rotamer singles and pairs energies in structure-based computational protein design (1). Thus, the exact optimization algorithms used to determine the global minimum energy conformation (GMEC) from a rotameric representation of the sequence design problem (38, 39) can be used instead to determine the global minimum energy combinatorial library (GMEL) from a set-based representation of the combinatorial library design problem.
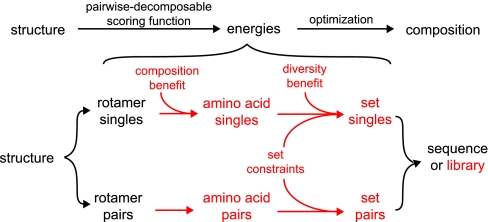
Fig. 4 illustrates the main components of the generalized DBIS algorithm. A symmetric matrix of rotamer singles and pairs energies is first calculated by using a template structure and rotamer library (1–3). This rotameric representation of the sequence design problem is then projected onto a smaller matrix with one row and one column for each combination of amino acid and targeted position (see below). These amino acid singles and pairs energies are then combined to build the set-based representation of the combinatorial library design problem by filling a matrix with one row and one column for each set of amino acids considered at each position in the library design. The number of these sets can be reduced from the 220 − 1 unique sets of 1 to 20 amino acids any number of ways: here, we have imposed both a set size constraint to limit sets to specific numbers of amino acids and a genetic code constraint to limit even these sets to those combinations of amino acids that can be introduced with degenerate codons during primer synthesis. To impose a composition constraint, such that the composition of the library is biased toward the inclusion or exclusion of a specific sequence (e.g., the wild-type sequence), we have applied benefits to some amino acid singles energies. Lastly, a diversity benefit that increases with set size is introduced to the set singles energies to favor larger sets over smaller sets during optimization.
Fig. 4.
The DBIS algorithm. The flow chart at the top illustrates the core procedure shared by many algorithms used for the structure-based computational design of either proteins or combinatorial libraries. The flow chart at the bottom illustrates the main components of the generalized DBIS algorithm. If the components shown in red were eliminated, the remaining components would be sufficient to design a single sequence instead of a library.
Here, we have implemented the generalized DBIS algorithm such that a library's energy is equal to an arithmetic average of conformational energies calculated for each sequence in the library, adjusted for composition and diversity benefits. Optimizing library composition thus corresponds to minimizing this energy. For rotamer r at each position i, the energy of point mutation, Epm(ir), is evaluated as
where Erot(ir) and Erot(ir, jcurrent) are rotamer singles and pairs energies, respectively, and jcurrent is the rotamer defined by the amino acid at position j in the template structure. Within the set of rotamers r at position i corresponding to amino acid a, ir ∈ ia, the rotamer that minimizes Epm(ir) is represented as imin,a. If there exists some ir ∈ ia that has survived a previous rotamer pruning step (see SI Text), the amino acid singles energy for amino acid a at position i, Eaa(ia), is then set equal to
where the composition benefit Ecomp(ia) has a user-defined value that biases optimization toward or away from libraries that include amino acid a at position i. Otherwise Eaa(ia) is set equal to the cutoff value used to prune rotamers, 20 kcal/mol, such that these amino acids are effectively eliminated from the calculation; a value similar to some of the better rotamer singles energies could conceivably improve library design for some applications by complementing the conservative nature of our structure-based method with a desired degree of randomness. Assignment of the amino acid energies in this manner effectively prunes the rotamers in the calculation to no more than one rotamer per amino acid per position. In SI Text, we show that high-scoring sequences in core design tend to use a very small subset of rotamers and that minimizing Epm(ir) is an effective way to identify this subset.
If there exists some ir ∈ ia and some js ∈ε jb that have survived the rotamer pruning step, the amino acid pairs energy, Eaa(ia, jb), is then set equal to
Otherwise Eaa(ia, jb) is set equal to the cutoff value used to prune rotamers, 20 kcal/mol, such that these amino acids are effectively eliminated from the calculation; a value similar to some of the better rotamer pairs energies could conceivably improve library design for some applications by complementing the conservative nature of our structure-based method with a desired degree of randomness.
For the set of amino acids a represented by x, a set singles energy, Eset(ix), is calculated at each position i as
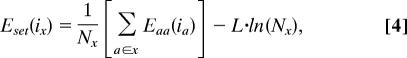 |
where Nx is the number of amino acids in set x, and L is a factor used to control the size of the optimal library. We refer to the second term in this equation as a diversity benefit and to L as a diversity benefit scale factor. Faced with two libraries of the same size, the logarithmic form of the diversity benefit will tend to favor the one with sequence diversity distributed over a greater number of positions. A quadratic form would have the opposite effect and may be more desirable, depending on one's application. Of course, the functional form for the diversity benefit is inconsequential when only two set sizes are considered in a design, as was the case in designing the DBISORBIT and DBISORBIT 44 libraries (see below). For sets x and y at positions i and j, the set pairs energy is then calculated as
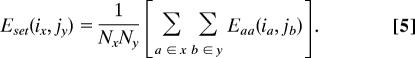 |
The composition of the optimal combinatorial library was thus defined by the optimal combination of these set singles and pairs energies. In designing the DBISORBIT and DBISORBIT 44 libraries, we first imposed Ecomp(ia) = 0 at all positions; if the GMEL for the value of L that gives the desired library size did not include the GFP-S65T sequence, we iteratively altered Ecomp(ia) in −5 kcal/mol increments for the missing GFP-S65T residues until this sequence was recovered in the designed library.
Library Design Methods.
Composition, set-size, and genetic-code constraints were enforced for all tested design algorithms to facilitate comparisons among them. The genetic-code constraint allowed each library to be constructed at minimal cost and effectively applied some of the physicochemical information that may exist in the genetic code to the process of design (it is notable that there were large differences in performance among libraries, although each shared this constraint). Relaxing the genetic-code constraint would change the composition of each designed library substantially and could alter the observed performance ranking.
One set of rotamer singles and pairs energies (calculated as described in SI Text) was used in four different ways to design the DBISORBIT, DBISORBIT 44, CORBIT, and SCMFORBIT 322 libraries. In order for the DBIS algorithm to yield a library of 29 sequences that included GFP-S65T, all values of Ecomp(ia) were set equal to 0, except Ecomp(63T) = −10 kcal/mol, and Ecomp(69Q) = −5 kcal/mol; the only sets considered at each position were the 95 unique sets of either one or two amino acids that can be defined by the use of mixed bases during primer synthesis; L was set equal to 6.5. In order for the DBIS algorithm to yield a library of 44 sequences that included GFP-S65T, all values of Ecomp(ia) were set equal to 0 except Ecomp(63T) = −10 kcal/mol, and Ecomp(69Q) = −10 kcal/mol; the only sets considered at each position were the 113 unique sets of either one or four amino acids that can be defined by the use of mixed bases during primer synthesis; L was set equal to 4.6.
The SCMFORBIT 322 library was designed by applying the method of Voigt et al. (7) in the following way. Each rotamer was first assigned a probability equal to the inverse of the number of rotamers at its position. The self-consistent mean-field solution was then calculated for an initial temperature of 50,000 K. As the temperature was lowered in 100 K increments, the solution from each previous temperature was used as the initial configuration for the next temperature. Saturation mutagenesis was directed to the two positions with site entropies >1.0 at a final temperature of 1,000 K.
The CORBIT library was designed by applying the consensus method of Hayes et al. (9) in the following way. The GMEC for this design problem was used as the initial configuration for a Monte Carlo trajectory through conformation space. One million steps were used for each of 100 cycles during which temperature oscillated between 4,000 K and 150 K. Only the 1,010 unique amino acid sequences with the best energies sampled were retained for further analysis. At 9 of 15 positions, there appeared at least one mutation that could be introduced to GFP-S65T by a single nucleotide substitution. The CORBIT library was thus defined by the one such mutation that appeared with the greatest frequency at each of these nine positions. (At 1,000 sequences a unique library could not be defined by this method because both alanine and threonine appeared with equal frequency at position 58.) Three apparent deficiencies of this consensus method were addressed by developing the DBIS algorithm: first, Monte Carlo-based sampling of the energy landscape is by its nature both inexhaustive and random; second, disruptive combinations of amino acids might arise when a library is designed without accounting for correlations in an alignment; and third, even if correlations were accounted for, any alignment with enough sequences to truly reflect global trends in these correlations would likely be too large to be practical.
The CMSA and SE/CMSA libraries were each designed with the same alignment of naturally occurring fluorescent proteins according to similar consensus methods. Of the 48 GFP homologs aligned by Shagin et al. (26), we used only the 36 homologs labeled as either GFPs, YFPs, cyan fluorescent proteins or red fluorescent proteins. To design the CMSA library, a consensus method derived from the one used by Hayes et al. (9) was used. At 12 of the positions between 57 and 72, there appeared at least one mutation that could be introduced to GFP-S65T by a single nucleotide substitution. The nine positions that had at least one such mutation represented at least four times were mutated to whichever of these mutations occurred with the greatest frequency at each position. Because two such mutations occurred with greatest frequency at positions 62 and 72, we elected, in each case, to introduce the mutation that happened to be shared with the DBISORBIT library. The approach used to design the CMSA library thus directs mutations away from the positions that exhibit the least conservation. To explore the possibility that these least-conserved positions might tolerate mutation best, the SE/CMSA library was designed by directing mutations to the 9 positions (of 12) that had the greatest site entropies,
where p(ia) is the frequency of amino acid a at position i, and the sum is taken over all amino acids for which p(ia) ≠ 0. The mutations introduced at these positions were chosen by the same considerations used to design the CMSA library. We did not use any design algorithms that used pair-wise correlations among the mutations in the MSA, because this alignment was rather small and there may be considerable evolutionary noise in such correlations (36, 37).
The Random library was designed by using a Python script to pick one mutation at random at each of the nine positions mutated in the DBISORBIT library.
Procedures used to synthesize and characterize libraries, including data analysis and error estimation, are provided in SI Text.
Supplementary Material
Acknowledgments
We thank Patrick Daugherty (University of California, Santa Barbara, CA) for providing many of the primers used to assemble GFP-S65T and a pBAD-derived vector engineered with SfiI recognition sequences; Christina Smolke for the use of her plate reader; Marco Mena, Michelle Meyer, and Frances Arnold for advice in designing this project; and Marie Ary for useful comments on the manuscript. This research was supported by the Howard Hughes Medical Institute and the Army Research Office. T.P.T. was supported by National Institutes of Health Grant F32-GM07438. C.L.V. was supported by a National Science Foundation Graduate Research Fellowship.
Abbreviations
- DBIS
diversity benefit applied to interacting sets
- epPCR
error-prone PCR
- MSA
multiple sequence alignment
- SCMF
self-consistent mean-field
- SE
site entropy.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609647103/DC1.
References
- 1.Dahiyat BI, Mayo SL. Science. 1997;278:82–87. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- 2.Gordon DB, Marshall SA, Mayo SL. Curr Opin Struct Biol. 1999;9:509–513. doi: 10.1016/s0959-440x(99)80072-4. [DOI] [PubMed] [Google Scholar]
- 3.Street AG, Mayo SL. Structure Fold Des. 1999;7:R105–R109. doi: 10.1016/s0969-2126(99)80062-8. [DOI] [PubMed] [Google Scholar]
- 4.Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 5.Dwyer MA, Looger LL, Hellinga HW. Science. 2004;304:1967–1971. doi: 10.1126/science.1098432. [DOI] [PubMed] [Google Scholar]
- 6.Saven JG, Wolynes PG. J Phys Chem B. 1997;101:8375–8389. [Google Scholar]
- 7.Voigt CA, Mayo SL, Arnold FH, Wang ZG. Proc Natl Acad Sci USA. 2001;98:3778–3783. doi: 10.1073/pnas.051614498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voigt CA, Martinez C, Wang ZG, Mayo SL, Arnold FH. Nat Struct Biol. 2002;9:553–558. doi: 10.1038/nsb805. [DOI] [PubMed] [Google Scholar]
- 9.Hayes RJ, Bentzien J, Ary ML, Hwang MY, Jacinto JM, Vielmetter J, Kundu A, Dahiyat BI. Proc Natl Acad Sci USA. 2002;99:15926–15931. doi: 10.1073/pnas.212627499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larson SM, England JL, Desjarlais JR, Pande VS. Protein Sci. 2002;11:2804–2813. doi: 10.1110/ps.0203902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore GL, Maranas CD. Proc Natl Acad Sci USA. 2003;100:5091–5096. doi: 10.1073/pnas.0831190100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Endelman JB, Silberg JJ, Wang ZG, Arnold FH. Protein Eng Des Sel. 2004;17:589–594. doi: 10.1093/protein/gzh067. [DOI] [PubMed] [Google Scholar]
- 13.Socolich M, Lockless SW, Russ WP, Lee H, Gardner KH, Ranganathan R. Nature. 2005;437:512–518. doi: 10.1038/nature03991. [DOI] [PubMed] [Google Scholar]
- 14.Ness JE, Kim S, Gottman A, Pak R, Krebber A, Borchert TV, Govindarajan S, Mundorff EC, Minshull J. Nat Biotechnol. 2002;20:1251–1255. doi: 10.1038/nbt754. [DOI] [PubMed] [Google Scholar]
- 15.Coco WM, Encell LP, Levinson WE, Crist MJ, Loomis AK, Licato LL, Arensdorf JJ, Sica N, Pienkos PT, Monticello DJ. Nat Biotechnol. 2002;20:1246–1250. doi: 10.1038/nbt757. [DOI] [PubMed] [Google Scholar]
- 16.Hogrefe HH, Cline J, Youngblood GL, Allen RM. BioTechniques. 2002;33:1158–1165. doi: 10.2144/02335pf01. [DOI] [PubMed] [Google Scholar]
- 17.Hiraga K, Arnold FH. J Mol Biol. 2003;330:287–296. doi: 10.1016/s0022-2836(03)00590-4. [DOI] [PubMed] [Google Scholar]
- 18.Meyer MM, Silberg JJ, Voigt CA, Endelman JB, Mayo SL, Wang ZG, Arnold FH. Protein Sci. 2003;12:1686–1693. doi: 10.1110/ps.0306603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otey CR, Silberg JJ, Voigt CA, Endelman JB, Bandara G, Arnold FH. Chem Biol. 2004;11:309–318. doi: 10.1016/j.chembiol.2004.02.018. [DOI] [PubMed] [Google Scholar]
- 20.Heim R, Cubitt AB, Tsien RY. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 21.Jain RK, Ranganathan R. Proc Natl Acad Sci USA. 2004;101:111–116. doi: 10.1073/pnas.2534352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rennell D, Bouvier SE, Hardy LW, Poteete AR. J Mol Biol. 1991;222:67–88. doi: 10.1016/0022-2836(91)90738-r. [DOI] [PubMed] [Google Scholar]
- 23.Markiewicz P, Kleina LG, Cruz C, Ehret S, Miller JH. J Mol Biol. 1994;240:421–433. doi: 10.1006/jmbi.1994.1458. [DOI] [PubMed] [Google Scholar]
- 24.Shafikhani S, Siegel RA, Ferrari E, Schellenberger V. BioTechniques. 1997;23:304–310. doi: 10.2144/97232rr01. [DOI] [PubMed] [Google Scholar]
- 25.Bloom JD, Silberg JJ, Wilke CO, Drummond DA, Adami C, Arnold FH. Proc Natl Acad Sci USA. 2005;102:606–611. doi: 10.1073/pnas.0406744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shagin DA, Barsova EV, Yanushevich YG, Fradkov AF, Lukyanov KA, Labas YA, Semenova TN, Ugalde JA, Meyers A, Nunez JM, et al. Mol Biol Evol. 2004;21:841–850. doi: 10.1093/molbev/msh079. [DOI] [PubMed] [Google Scholar]
- 27.Patrick WM, Firth AE, Blackburn JM. Protein Eng. 2003;16:451–457. doi: 10.1093/protein/gzg057. [DOI] [PubMed] [Google Scholar]
- 28.Hoogenboom HR. Nat Biotechnol. 2005;23:1105–1116. doi: 10.1038/nbt1126. [DOI] [PubMed] [Google Scholar]
- 29.Wang L, Jackson WC, Steinbach PA, Tsien RY. Proc Natl Acad Sci USA. 2004;101:16745–16749. doi: 10.1073/pnas.0407752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cormack BP, Valdivia RH, Falkow S. Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 31.Street AG, Mayo SL. Fold Des. 1998;3:253–258. doi: 10.1016/S1359-0278(98)00036-4. [DOI] [PubMed] [Google Scholar]
- 32.Heim R, Tsien RY. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 33.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 34.Cherry JR, Fidantsef AL. Curr Opin Biotechnol. 2003;14:438–443. doi: 10.1016/s0958-1669(03)00099-5. [DOI] [PubMed] [Google Scholar]
- 35.Bokman SH, Ward WW. Biochem Biophys Res Commun. 1981;101:1372–1380. doi: 10.1016/0006-291x(81)91599-0. [DOI] [PubMed] [Google Scholar]
- 36.Noivirt O, Eisenstein M, Horovitz A. Protein Eng Des Sel. 2005;18:247–253. doi: 10.1093/protein/gzi029. [DOI] [PubMed] [Google Scholar]
- 37.Fodor AA, Aldrich RW. Proteins. 2004;56:211–221. doi: 10.1002/prot.20098. [DOI] [PubMed] [Google Scholar]
- 38.Desmet J, Demaeyer M, Hazes B, Lasters I. Nature. 1992;356:539–542. doi: 10.1038/356539a0. [DOI] [PubMed] [Google Scholar]
- 39.Gordon DB, Hom GK, Mayo SL, Pierce NA. J Comput Chem. 2003;24:232–243. doi: 10.1002/jcc.10121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.