Abstract
CCA-adding enzymes build and repair the 3′-terminal CCA sequence of tRNA. These unusual RNA polymerases use either a ribonucleoprotein template (class I) or pure protein template (class II) to form mock base pairs with the Watson–Crick edges of incoming CTP and ATP. Guided by the class II Bacillus stearothermophilus CCA-adding enzyme structure, we introduced mutations designed to reverse the polarity of hydrogen bonds between the nucleobases and protein template. We were able to transform the CCA-adding enzyme into a (U,G)-adding enzyme that incorporates UTP and GTP instead of CTP and ATP; we transformed the related Aquifex aeolicus CC- and A-adding enzymes into UU- and G-adding enzymes and Escherichia coli poly(A) polymerase into a poly(G) polymerase; and we transformed the B. stearothermophilus CCA-adding enzyme into a poly(C,A) polymerase by mutations in helix J that appear, based on the apoenzyme structure, to sterically limit addition to CCA. We also transformed the B. stearothermophilus CCA-adding enzyme into a dCdCdA-adding enzyme by mutating an arginine that interacts with the incoming ribose 2′ hydroxyl. Most importantly, we found that mutations in helix J can affect the specificity of the nucleotide binding site some 20 Å away, suggesting that the specificity of both class I and II enzymes may be dictated by an intricate network of hydrogen bonds involving the protein, incoming nucleotide, and 3′ end of the tRNA. Collaboration between RNA and protein in the form of a ribonucleoprotein template may help to explain the evolutionary diversity of the nucleotidyltransferase family.
Keywords: nucleotidyltransferase, tRNA
The CCA-adding enzyme [ATP(CTP):tRNA nucleotidyltransferase (NTR)] builds and repairs tRNA by adding the nucleotide sequence CCA to the 3′ terminus of immature or damaged tRNA (1). Although this unusual RNA polymerase has no nucleic acid template, it constructs the CCA sequence one nucleotide at a time using CTP and ATP as substrates (1). All CCA-adding enzymes and poly(A) polymerases belong to the NTR superfamily, which can be divided into two distinct classes according to sequence motifs in the catalytic domain (2–6). The class I and class II CCA-adding enzymes exhibit little if any homology outside the NTR domain (6). Class I NTRs include archaeal CCA-adding enzymes, eukaryotic poly(A) polymerases, and probably eukaryotic terminal uridylyltransferases (7, 8); class II NTRs include eukaryotic and eubacterial CCA-adding enzymes as well as eubacterial poly(A) polymerases (6).
We found previously that a single NTR motif adds all three nucleotides (9, 10), that tRNA does not rotate or translocate along the enzyme during addition of C75 and A76 (11), and that a single active subunit in these dimeric or tetrameric enzymes can carry out all three steps of CCA addition (9). We therefore proposed that the growing 3′ end of tRNA refolds progressively to reposition the new 3′ hydroxyl identically relative to the single active site (10, 12). This prediction was confirmed by cocrystal structures of the class I archaeal Archaeoglobus fulgidus CCA-adding enzyme with oligonucleotide mimics of two different substrates (tRNA-NC + CTP and tRNA-NCC + ATP), as well as with mature tRNA-NCCA (where N is the discriminator base), which demonstrated that class I enzymes use a ribonucleoprotein template where the growing 3′ terminus of tRNA works together with the protein to “template” addition of the next nucleotide (13). In contrast, cocrystal structures of the class II eubacterial Bacillus stearothermophilus enzyme in complex with CTP and ATP revealed that class II enzymes use a pure protein template that forms mock base pairs with the Watson–Crick edges of CTP and ATP (14). The hydrogen bonds between the incoming nucleotide and the protein template are almost identical to those seen in standard Watson–Crick G:C and A:U base pairs, enabling the enzyme to distinguish C and A from U and G (14, 15).
Guided by the structure of the B. stearothermophilus CCA-adding enzyme (BstCCA), we designed mutations in the protein template that changed the nucleotide specificity of the enzyme. We also made mutations in helix J that transformed the enzyme into a polynucleotide polymerase by altering residues predicted to limit addition to three nucleotides (14). Most importantly, we found that mutations in helix J often affect the specificity of the distant nucleotide binding site, suggesting that a flexible network of hydrogen bonds (orchestrated by the NTR domain between the protein, the RNA substrate, and the incoming nucleotide) may explain interconversion of CCA-adding enzymes and poly(A) polymerases over evolutionary time (6) as well as the unusual diversity of the NTR superfamily.
Results and Discussion
Structure-Based Design of a (U,G)-Adding Enzyme.
Unlike the class I archaeal CCA-adding enzyme from Ar. fulgidus in which the 3′ end of the tRNA substrate constitutes part of the ribonucleoprotein template for CTP and ATP addition (13), the class II eubacterial BstCCA enzyme uses a pure protein template (Fig. 1) and can bind CTP or ATP in the absence of tRNA substrate (14). As shown in Fig. 2, the protein template forms mock base pairs with the Watson–Crick edges of the incoming CTP and ATP nucleotides, and the polarity of the resulting hydrogen bonds excludes UTP and GTP (15). Moreover, because a single protein template in the apoenzyme binds both CTP and ATP, the nucleotide specificity switch from CTP to ATP most likely reflects a subtle change in the geometry of the binding site (14). We therefore asked whether, if the Watson–Crick edge of protein template were redesigned to recognize UTP and GTP, the enzyme would add UUG to tRNA-N.
Fig. 1.
Nucleotide binding site of the class II eubacterial BstCCA enzyme. The site is composed of a catalytic head (helices A–E) and neck (helices F–L) domain (14). Stick models of protein side chains are shown with nitrogen blue and oxygen red; carbon is yellow in the polypeptide and light blue in the nucleotide; and hydrogen bonds are shown as black dashed lines. Some connecting loops have been omitted for clarity. This and all other images were drawn in PyMOL (16).
Fig. 2.
Transforming the CCA-adding enzyme into a (U,G)-adding enzyme. (Upper) Watson–Crick interactions between ATP or CTP and the protein template of the BstCCA enzyme (14). (Lower) Protein template redesigned to recognize UTP and GTP. Representation is as in Fig. 1, except that carbon is violet in the nucleotide and hydrogen bonds are shown as white dashed lines. Mutant side chains were positioned by the PyMOL Mutagenesis Wizard, using lowest energy rotamer conformations (16).
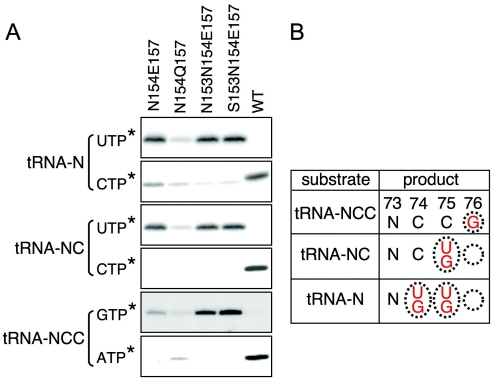
Guided by the structure of the BstCCA enzyme (14), we designed two double and two triple mutants intended to reverse the polarity of hydrogen bonds between the phylogenetically conserved protein template [supporting information (SI) Fig. 5] and the bases (Fig. 2). To reverse the polarity of the hydrogen bonds themselves, we changed donors into acceptors and vice versa: D154 was changed to N, and the highly conserved R157 was changed to E or Q, generating double mutants N154E157 and N154Q157. To maintain the proper orientation of the mutant side chains in the N154E157 double mutant, we also changed E153, the partner of R157, to N or S, generating triple mutants N153N154E157 and S153N154E157 (NNE and SNE in Fig. 2).
The double and triple mutants were tested for altered or compromised specificity by standard gel assay using natural tRNA substrates (Fig. 3A). The two triple mutants functioned as predicted, incorporating UTP and GTP but excluding CTP and ATP (also see SI Fig. 6); however, the double mutants were severely damaged. N154E157 failed to discriminate completely against CTP at position 74, whereas mutant N154Q157 was nearly inactive (Fig. 3A and SI Table 1). Compared with the double mutant N154E157, the triple mutants exhibited improved selectivity for UTP over CTP by 2- to 4-fold (Fig. 3A Upper) and improved efficiency of GTP incorporation (Fig. 3A Lower). Although it is difficult to say why N154Q157 is severely compromised, these data suggest that supporting residues N153 or S153 do indeed stabilize the conformation of E157 in the mutant, just as E153 stabilizes the conformation of R157 in the cocrystal structures of wild type (Fig. 2) (14).
Fig. 3.
Nucleotide specificity of (U,G)-adding enzymes. (A) BstCCA enzymes were assayed under standard conditions in the linear range with tRNA substrates, all four unlabeled nucleotides, and the labeled NTP indicated by an asterisk. (B) Nucleotide specificity for triple mutants N153N154E157 and S153N154E157, double mutant N154E157, and single mutant A157 based on assays under standard conditions (SI Figs. 6D and 7). Addition of U74U75 and G74G75 suggests that the enzyme can also add U74G75 or G74U75. For clarity, residual activities of <10% are neglected. Black letters, substrate; red letters, added nucleotides; empty circle, no addition.
The two triple mutants add two nucleotides to tRNA-N and one nucleotide to tRNA-NC but do not discriminate between UTP and GTP; however, both triple mutants discriminate against UTP at position 76 and add only 3′-terminal GTP to tRNA-NCC (Fig. 3B and SI Fig. 6D). Thus, the scrunching pocket can accommodate any two nucleotides at positions 74 and 75, but only C74C75 can pack appropriately for A76 addition, consistent with observations that the 3′-terminal sequence of the growing tRNA substrate can affect the efficiency (15) or specificity of subsequent nucleotide addition (H.D.C., unpublished observations). We conclude that mutations in the nucleotide binding site do not affect the counting mechanism that limits addition to three nucleotides and only partially compromise the position-dependent specificity switch from pyrimidine to purine nucleotides.
We also made the single mutant R157A in an absolutely conserved arginine (SI Fig. 5), which should in principle eliminate two of three hydrogen bonds to the incoming CTP nucleobase and one of two hydrogen bonds to the incoming ATP nucleobase (14); however, R157A readily added a single C, A, U, or G to tRNA-N or tRNA-NC (SI Fig. 6), suggesting that the primary function of the protein template is not to improve either Km or kcat for nucleotide, but to ensure nucleotide specificity.
To better understand the phenotypes of these mutant enzymes, we determined an apparent Km and kcat for all four nucleotides using the natural substrates tRNA-NC and tRNA-NCC (SI Table 1 and SI Fig. 6C). Km and kcat for N154Q157 are both severely compromised, but N154E157 (despite a potential charge repulsion between glutamates E153 and E157) incorporates UTP as well as wild type incorporates CTP. Most surprisingly, N154E157 incorporates GTP poorly but incorporates UTP well (Fig. 3A and SI Table 1); moreover, introduction of the S153 substitution to generate S153N154E157 has almost no effect on the Km or kcat for UTP yet increases the kcat for GTP by 9-fold while decreasing Km by only 2-fold. The simplest explanation for a much greater improvement in kcat for GTP than for UTP is that GTP requires an additional conformation change, after binding, before catalysis can take place. Alternatively, because the ratio of kcat/Km for addition of N75 compared with N76 is 4.3 for wild type but 149 for the S153N154E157 mutant, introduction of S153 may compromise an interaction made by helix F during N76 but not N74 or N75 addition.
Structure-Based Redesign of CC- and A-Adding Enzymes.
In most eubacteria, CCA addition is carried out by a single CCA-adding enzyme, but in the deeply branching eubacteria Aquifex aeolicus and Deinococcus radiodurans (17, 18), and possibly in the Gram-positive eubacterium Bacillus halodurans (19), related CC- and A-adding enzymes work sequentially to add CCA. All of these eubacterial CCA-, CC-, and A-adding enzymes, as well as eubacterial poly(A) polymerase and eukaryotic CCA-adding enzymes, belong to the same class II NTR superfamily (6, 17, 19, 20); moreover, a sequence alignment of the nucleotide binding region (helices F and G) in class II enzymes (SI Fig. 5) reveals that the protein template residues in the BstCCA enzyme (E153, D154, and R157) are conserved in the A. aeolicus CC-adding enzyme (E164, D165, and R168) and A-adding enzyme (E583, D584, and R587). We therefore attempted to transform the A. aeolicus CC-adding enzyme into a UU-adding enzyme and the A-adding enzyme into a G-adding enzyme by reversing the polarity of the hydrogen bonds between the incoming nucleotides and the protein template as in structure-based redesign of the (U,G)-adding enzyme (Fig. 2).
We assayed the redesigned UU- and G-adding enzymes with three different tRNA substrates and all four nucleotides (for tRNA-NC and tRNA-NCC, see SI Fig. 7A; data not shown for tRNA-N). As designed, the UU-adding enzyme efficiently adds U74 and U75, prefers UTP over CTP by >4-fold, and excludes ATP and GTP; the G-adding enzyme adds only 3′-terminal G excluding ATP, CTP, and UTP. The UU- and G-adding enzymes exhibit even greater nucleotide specificity than the redesigned (U,G)-adding enzyme (compare Fig. 2 and SI Fig. 7A); the UU-adding enzyme adds only two U's; the G-adding enzyme adds only a single G; and both redesigned enzymes retain the position-dependent pyrimidine and purine nucleotide incorporation of the wild-type enzymes, consistent with the view that A. aeolicus CC- and A-adding enzymes are frozen steps along the BstCCA reaction pathway. The scrunching pocket of the A-adding enzyme would be frozen in the conformation that binds tRNA-NCC and dictates A addition, whereas the scrunching pocket of the CC-adding enzyme would be sufficiently flexible to bind both tRNA-N and tRNA-NC but could not adopt the conformation required for A addition.
Surprisingly, the catalytic efficiencies of the A. aeolicus CC-adding enzyme for CTP and A-adding enzyme for ATP were 16- and 26-fold lower than the equivalent catalytic efficiencies of the BstCCA enzyme for CTP and ATP (compare SI Tables 1 and 2). One might have thought that the price of binding both CTP and ATP and undergoing a position-dependent nucleotide specificity switch would be to decrease catalytic efficiency compared with dedicated CC- and A-adding enzymes; however, the opposite appears to be the case, and the effect is almost entirely on apparent Km (which varies by 5- to 29-fold), not kcat (which varies by <2-fold). Similarly unexpected is the observation that kcat/Km for GTP addition by the G-adding enzyme is actually 5.3-fold improved relative to kcat/Km for ATP addition by the wild-type A-adding enzyme (SI Table 2). Taken together, these observations are more consistent with duplication of a CCA-adding gene to give CC- and A-adding activities than with evolution of a CC- or A-adding enzyme into a true CCA-adding activity as originally proposed (18). The decreased catalytic efficiency of the A. aeolicus CC- and A-adding enzymes might then reflect the compromises required for the new nucleotide binding sites to exclude one NTP when the enzyme originally bound both, and for the new tRNA binding sites to exclude tRNAs of nearly comparable length when the site originally bound three different substrates (tRNA-N, tRNA-NC, and tRNA-NCC). Alternatively, if the concentration of tRNA precursors is significantly higher in A. aeolicus than in B. stearothermophilus, the increased Km of the A. aeolicus CC- and A-adding enzymes for CTP and ATP compared with the BstCCA may simply reflect lack of selection for a lower Km.
Transforming the CCA-Adding Enzyme into a Poly(C,A) Polymerase.
Unlike poly(A) polymerases, CCA-adding enzymes not only add two different nucleotides in a defined order but terminate addition after three nucleotides (1, 12, 14). The cocrystal structures of two class II enzymes [the BstCCA enzyme in complex with CTP or ATP (14) and the A. aeolicus class II A-adding enzyme in complex with tRNA-NCC and a nonreactive ATP analog (21)] suggest that three residues in helix J (R194, M197, and E198) may constrain the growing 3′ end of tRNA and/or help to thread it into the active site (Fig. 4A and B). Moreover, sequence alignment of class II NTR helix J regions reveals two residues (R194 and E198) that are invariant in all CCA-adding enzymes and poly(A) polymerases and four residues that are invariant in eubacterial poly(A) polymerases (A193, L195, E/D197, and K201) (SI Fig. 8A). Although M197 is not conserved among CCA-adding enzymes, it approaches the tRNA backbone (Fig. 4A), and we therefore asked whether mutation of three residues (R194, M197, and E198) to alanine (Fig. 4B) could transform the BstCCA enzyme into a poly(C,A) polymerase.
Fig. 4.
Transformation of the CCA-adding enzyme into a poly(C,A) polymerase. (A) Helix J constrains the growing 3′ end of tRNA. Coordinates are from the A. aeolicus A-adding cocrystal structure (21), but homologous residues are labeled according to the BstCCA sequence (14), including M197, which is N197 in A. aeolicus. The protein and nucleotides are represented as in Fig. 1. The 3′ end of tRNA-NCC (green) may interact with nearby β-turn (blue). tRNA backbone phosphates are shown in orange. Incoming ATP is shown although cocrystals were obtained by soaks with the nonhydrolyzable analogue AMPcPP (α,β-methyleneadenosine 5′-triphosphate). Modeling of nucleotides C74 and C75 is somewhat uncertain because 3′-terminal tRNA electron density was weak (21). (B) Mutant A194A197A198 removes constraints on the growing 3′ end of tRNA. The tRNA acceptor stem is shown in tan using a sticks representation. 3′-terminal nucleotides A73, C74, and C75 are in magenta, cyan, and orange, respectively, and the incoming ATP is in light blue. Side chains that constrain the 3′ end of tRNA are shown in a space-filling representation: blue, nitrogen; yellow, carbon; red, oxygen; orange, sulfur. Mutant side chains are positioned as in Fig. 2. (C) Runaway polymerization by the A194A197A198 enzyme. Addition of CTP to tRNA-NC or tRNA-N and ATP to tRNA-NCC assayed as in Fig. 3 but using uniformly labeled tRNA substrates and 200 μM CTP or ATP.
We found that the triple mutant A194A197A198 will add either poly(C) or poly(A) to tRNA-N, tRNA-NC, or tRNA-NCC with 3′ tails averaging 20 nucleotides but as large as 100 (Fig. 4C and SI Fig. 9). The double mutants A194A197 and A194A198 do not cause runaway polymerization, nor do mutants A198A201 and A194A198A201. These data suggest that M197 must be mutated to relieve the scrunching constraint and that residue 201 is too far from the tRNA backbone to influence scrunching as the structure implies (21). Most intriguingly, four of the mutants (A194A197, A194A198, A198A201, and A194A198A201) as well as R194E and E194E197E201 (data not shown) leave the scrunching pocket largely intact, limiting nucleotide addition to two or at most three nucleotides, but none of these mutants (including A194A197A198) can distinguish between CTP and ATP (compare Fig. 4C and SI Fig. 9).
These data suggest two key conclusions. First, interactions between the growing 3′ end of tRNA and helix G may be communicated through a network of hydrogen bonds involving the 3′ end of tRNA and residues belonging to both helices (primarily R194 and E198 of helix J with D154, R160, and R163 of helix G) that are disrupted in the mutants (Fig. 4A). Communication between the 3′ end of tRNA and the nucleotide binding site could also explain why combining either of the nucleotide binding site mutations N153N154E157 or S153N154E157 (which allow UTP and GTP incorporation) with the helix J mutation A194A197A198 (which allows runaway polymerization) did not generate poly(U) or poly(G) polymerases as expected, but proteins that were unable to add any NTP to tRNA substrates (data not shown). Second, the class II scrunching mechanism may not involve a scrunching pocket as in class I enzymes (13, 22), but rather a channel across the face of helix J (Fig. 4A). When the 3′ end of tRNA is threaded through R194, M197, and E198, scrunching would be largely intact because the distance between the end of tRNA acceptor stem and the growing 3′ primer terminus is highly constrained; when the threading constraint is removed by mutation, the enzyme would become an RNA polymerase because the growing 3′ end could repeatedly loop out to allow attack on a new incoming nucleotide bound in the active site.
The equivalent triple mutation A194A197A198, which confers runaway polymerization on the BstCCA enzyme, was also introduced into the CC- and A-adding enzymes of A. aeolicus and D. radiodurans (data not shown). In each case, the CC-adding enzymes were transformed into poly(C) polymerases and the A-adding enzymes were transformed into poly(A) polymerases as expected. However, although three of these four mutant polymerases retained the original nucleotide specificity for CTP or ATP, the A. aeolicus CC-adding mutant was a poly(C,A) polymerase that used both CTP and ATP. These data are also consistent with an interaction between the scrunching apparatus centering on helix J and the nucleotide selection apparatus centering on helix G.
Using swap experiments between CCA-adding enzymes and the N and C termini of the class II Escherichia coli poly(A) polymerase (23, 24), Betat et al. (25) identified a region between residues 219 and 245 in the CCA-adding enzyme that determined whether the chimeric enzymes behaved as a CCA-adding enzyme or a poly(A) polymerase. The ability of this region to confer CCA-adding activity on the N terminus of poly(A) polymerase is most surprising and is consistent with the notion that scrunching, counting, and the nucleotide specificity switch in class II CCA-adding enzymes may be localized, intrinsic characteristics of the NTR active site; however, we hesitate to interpret the results of Betat et al. (25) structurally because no structure has been determined for E. coli poly(A) polymerase; the helix M regions of the E. coli poly(A) polymerase and CCA-adding enzymes exhibit little if any homology; and helix M appears to be remote from the tRNA acceptor stem in the cocrystal structure of the A. aeolicus A-adding enzyme (21).
Transforming a Poly(A) Polymerase into a Poly(G) Polymerase.
The eubacterial CCA-adding enzymes and poly(A) polymerases both belong to the class II NTR superfamily (6, 20), and sequence alignment suggests, quite remarkably, that poly(A) polymerases have the same highly conserved protein template residues [E211, D212, and R215 in the E. coli poly(A) polymerase] as CCA-adding enzymes (SI Fig. 5). To ask whether the E. coli poly(A) polymerase could be transformed into a poly(G) polymerase by altering the protein template, we designed a double mutant (N212E215) and two triple mutants (N211N212E215 and S211N212E215) intended to reverse the polarity of the hydrogen bonds to the Watson–Crick edges of the incoming nucleotides (Fig. 2A). We assayed poly(G), poly(A), poly(U), and poly(C) polymerase activity using two different unlabeled substrates, oligo(A)15 and tRNA lacking 3′-terminal A (tRNA-NCC) (SI Fig. 10). The mutant enzymes behaved much as predicted: S211N212E215, N212E215, and (more weakly) N211N212E215 added 5–70 G residues to oligo(A)15 and discriminated against ATP, CTP, and UTP (SI Fig. 10A).
Wild-type poly(A) polymerase discriminates against GTP but does incorporate UTP and even more weakly CTP; the mutant poly(G) polymerases incorporate all four nucleotides into tRNA-NCC, albeit with very different efficiencies (SI Fig. 10B). Thus, as previously observed (24), poly(A) polymerase is surprisingly nonspecific: The enzyme can add an A residue to immature tRNA-NCC in vivo (19), augmenting tRNA repair by the CCA-adding enzyme, but it can also add a poly(A) tail to mature tRNA in vitro (25) (SI Fig. 10B). The latter potentially damaging activity may be avoided in vivo because (i) mature tRNAs in both prokaryotes and eukaryotes appear to be almost fully aminoacylated except under conditions of nutritional deprivation (26, 27) and (ii) eubacterial poly(A) polymerase preferentially tails readily denaturable mutant tRNAs, flagging them for subsequent degradation (28). Most revealingly, nucleotide specificity is compromised only with the tRNA-NCC substrate, not with the oligo(A)15 primer (compare A and B in SI Fig. 10). We interpret this as further evidence that the RNA and nucleotide binding sites communicate. CCA-adding enzymes would be finely tuned to exploit communication between these sites to achieve position-dependent nucleotide specificity; poly(A) polymerases would retain the potential for communication because it as an intrinsic property of the class II NTR protein structure.
Transforming the CCA-Adding Enzyme into a dCdCdA-Adding Enzyme.
The BstCCA cocrystal structures with CTP and ATP (14) suggest that a single hydrogen bond between the 2′ hydroxyl of the incoming NTP and the guanidinium group of the highly conserved R111 enables the enzyme to discriminate rNTPs from dNTPs (Fig. 1). To test this prediction, we assayed a variety of point mutants in R111 of the BstCCA enzyme. The R111E mutant was inactive with both rNTPs and dNTPs; R111A and R111M were mildly impaired for CCA addition but could not use dNTPs; and R111I and R111K added both CCA and dCdCdA efficiently (SI Table 2). A variety of other point mutations in the immediate vicinity of the nucleotide binding site failed to affect the relative specificity for rNTPs and dNTPs (data not shown).
Intriguingly, R111I added CTP (C74 and C75) much better than ATP (A76) (SI Fig. 11 and SI Table 3) although R111I and wild type both add C74 and C75 at comparable rates (data not shown), and neither R111I nor R111K causes misincorporation of UTP, GTP, dUTP, or dGTP. These data suggest that the 2′ hydroxyls of CTP and ATP may be oriented differently in the mutant, and perhaps also in wild type. Consistent with this scenario, superposition of the structures of the BstCCA enzyme with CTP and ATP (14) indicates that the nucleobases are similarly oriented relative to the protein template, but the ribose moieties differ (data not shown). Taken together, these data echo the speculation above that the (U,G)-adding enzyme may undergo a conformational change before GTP addition (SI Table 1 and SI Fig. 6).
Using steady-state kinetics, we next asked whether these effects were mediated through Km, kcat, or both (SI Fig. 11 and SI Table 3). The data indicate that the nonconservative R111I mutation uses dCTP and dATP some 8- to 11-fold more efficiently than the conservative R111K mutation and is only 3-fold compromised for use of CTP although 100-fold compromised for use of ATP. The efficiency of dNTP incorporation by R111I suggests that recognition of the incoming nucleotide may be affected by the sugar pucker of the incoming dNTP (C2′-endo) compared with an incoming rNTP (C3′-endo). In fact, R111K, R111I, and R111M have no dramatic effects on use of CTP, but R111I and R111M both increase Km and decrease kcat for ATP by ≈10-fold. This suggests that R111, in addition to discriminating against dNTPs, is required to properly position ATP but not CTP for binding and catalysis. Once again, this would be consistent with the notion that a conformational change is required for 3′-terminal purine addition at position 76.
The Mechanism of CCA Addition by Class I and Class II Enzymes May Be Similar.
Consistent with a common origin for class II eubacterial poly(A) polymerases and CCA-adding enzymes (6), simple point mutations are sufficient to transform a CCA-adding enzyme into a poly(C,A) polymerase and to enable eubacterial CCA-, CC-, and A-adding enzymes, as well as poly(A) polymerase, to use of UTP and GTP instead of CTP and ATP. Most importantly, mutations in helix J can affect nucleotide specificity, although helix J directly contacts the 3′ end of the tRNA and not the incoming nucleotide (Fig. 4A and SI Fig. 8C). Taken together, the data suggest that specificity is modulated by a network of hydrogen bonds connecting the 3′ end of the tRNA substrate to the nucleotide binding site on the same face of helices G and J. For example, one hydrogen bonding pathway may lead from phosphate 74 through R194 and D154, which then affects the nucleobase directly or indirectly through E153 and R157; another pathway may lead from phosphate 74 to E198, then bifurcate through R160 and R163 to the triphosphate moiety of the ribose ring (Fig. 4A). A short, weakly conserved β-turn nearby may also interact with the 3′ end of the tRNA (20, 21) as the larger, highly conserved β-turn does in class I enzymes (13, 22). Indeed, the intricate network of hydrogen bonds seen in class II enzymes (Fig. 4A and SI Fig. 8C) is reminiscent, both structurally and functionally, of the ribonucleoprotein template seen in archaeal class I CCA-adding enzymes where the growing 3′ end of the tRNA forms part of the template for the incoming nucleotide (13). Thus, the mechanism of CCA addition by class I and class II enzymes may be more similar than previously appreciated (13, 14, 21, 22, 29), and intimate collaboration between RNA and protein may help to explain the remarkable evolutionary diversity of the NTR superfamily (2–8).
Methods
Structure-based redesign of the BstCCA enzyme to reverse the polarity of hydrogen bonds to the Watson–Crick edges of CTP and ATP (Fig. 2A) was carried out on a napkin. In wild type, the N6 amino group of adenine or N4 of cytosine donates a hydrogen bond to D154. D154 was mutated to the isosteric N154, enabling the enzyme to donate a hydrogen bond to the carbonyl groups that occupy equivalent positions in guanine or uracil. Also in wild type, N1 of adenine or N3 of cytosine accepts a hydrogen bond from the ND1 of R157. The equivalent positions of guanine and uracil are hydrogen bond donors, but the only three amino acids with hydrogen bond acceptors at the δ position are E, Q, and H. We ruled out H because CE1 could interfere with hydrogen bonding of guanine N2 to other groups. Q was ruled out because an amide can be either an acceptor or a donor, potentially compromising specificity for U or G. This left the mutation R157E as the only logical choice. In wild type, R157 donates a hydrogen bond to the O2 carbonyl of cytosine. Uracil also presents an O2 acceptor at this position, but the equivalent N2 amino group of guanine is a donor. Confronted with these conflicting requirements, we chose not to introduce additional mutations, hoping that N154 and D157 alone would suffice for specificity and that intervening water molecule(s) would alleviate or resolve the conflicting demands of O2 and N2; note that R is 52 Å3 or approximately three water molecules larger than E. Finally, we considered the immediate neighbors of the mutated residues N154 and D157. D154N should have no effect on the protein because this residue interacts solely with the incoming NTP; however, R157E creates an electrostatic conflict with E157 adjacent to E153. In both the CTP and ATP complexes, R157 forms a salt bridge with E153, which in turn forms a salt bridge with R150. R157 also interacts with a protein backbone carbonyl belonging to R110 in the CTP complex and R150 in the ATP complex. We therefore decided to replace E153 with a donor that could hydrogen-bond to E157, preferably through a short side chain because the E157 carboxylate was expected to be much closer to the polypeptide backbone than the wild-type amide nitrogen that participates in the salt bridge. Only two replacements, E153N and E153S, appeared to satisfy these requirements. We also hoped that intervening water molecule(s) or movement of R150 would facilitate further interactions with N153 or S153.
Other protocols (mutagenesis, protein expression, preparation of RNA substrates, and enzyme assays) have been described previously (12, 22), and details are available upon request.
Supplementary Material
Acknowledgments
We thank Tom Pavelitz and Arnold Bailey for advice and both reviewers for helpful suggestions. This work was supported by National Institutes of Health Grant GM059804 (to A.M.W.).
Abbreviations
- NTR
nucleotidyltransferase
- BstCCA
Bacillus stearothermophilus CCA-adding enzyme.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0606961104/DC1.
References
- 1.Deutscher MP. Enzymes. 1982;15:183–215. [Google Scholar]
- 2.Holm L, Sander C. Trends Biochem Sci. 1995;20:345–347. doi: 10.1016/s0968-0004(00)89071-4. [DOI] [PubMed] [Google Scholar]
- 3.Houseley J, Tollervey D. EMBO Rep. 2006;7:205–211. doi: 10.1038/sj.embor.7400612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin G, Keller W. EMBO J. 1996;15:2593–2603. [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J. Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- 6.Yue D, Maizels N, Weiner AM. RNA. 1996;2:895–908. [PMC free article] [PubMed] [Google Scholar]
- 7.Aphasizhev R, Sbicego S, Peris M, Jang SH, Aphasizheva I, Simpson AM, Rivlin A, Simpson L. Cell. 2002;108:637–648. doi: 10.1016/s0092-8674(02)00647-5. [DOI] [PubMed] [Google Scholar]
- 8.Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke BJ. RNA. 2006;12:1494–1504. doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho HD, Weiner AM. J Biol Chem. 2004;279:40130–40136. doi: 10.1074/jbc.M405518200. [DOI] [PubMed] [Google Scholar]
- 10.Yue D, Weiner AM, Maizels N. J Biol Chem. 1998;273:29693–29700. doi: 10.1074/jbc.273.45.29693. [DOI] [PubMed] [Google Scholar]
- 11.Shi PY, aizels N, Weiner AM. EMBO J. 1998;17:3197–3206. doi: 10.1093/emboj/17.11.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho HD, Verlinde CL, Weiner AM. J Biol Chem. 2005;280:9555–9566. doi: 10.1074/jbc.M412594200. [DOI] [PubMed] [Google Scholar]
- 13.Xiong Y, Steitz TA. Nature. 2004;430:640–645. doi: 10.1038/nature02711. [DOI] [PubMed] [Google Scholar]
- 14.Li F, Xiong Y, Wang J, Cho HD, Tomita K, Weiner AM, Steitz TA. Cell. 2002;111:815–824. doi: 10.1016/s0092-8674(02)01115-7. [DOI] [PubMed] [Google Scholar]
- 15.Cho HD, Oyelere AK, Strobel SA, Weiner AM. RNA. 2003;9:970–981. doi: 10.1261/rna.2110903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- 17.Tomita K, Weiner AM. Science. 2001;294:1334–1336. doi: 10.1126/science.1063816. [DOI] [PubMed] [Google Scholar]
- 18.Tomita K, Weiner AM. J Biol Chem. 2002;277:48192–48198. doi: 10.1074/jbc.M207527200. [DOI] [PubMed] [Google Scholar]
- 19.Bralley P, Chang SA, Jones GH. J Bacteriol. 2005;187:5927–5936. doi: 10.1128/JB.187.17.5927-5936.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin G, Keller W. RNA. 2004;10:899–906. doi: 10.1261/rna.5242304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomita K, Fukai S, Ishitani R, Ueda T, Takeuchi N, Vassylyev DG, Nureki O. Nature. 2004;430:700–704. doi: 10.1038/nature02712. [DOI] [PubMed] [Google Scholar]
- 22.Cho HD, Chen Y, Varani G, Weiner AM. J Biol Chem. 2006;281:9801–9811. doi: 10.1074/jbc.M512603200. [DOI] [PubMed] [Google Scholar]
- 23.Sarkar N. Annu Rev Biochem. 1997;66:173–197. doi: 10.1146/annurev.biochem.66.1.173. [DOI] [PubMed] [Google Scholar]
- 24.Yehudai-Resheff S, Schuster G. Nucleic Acids Res. 2000;28:1139–1144. doi: 10.1093/nar/28.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Betat H, Rammelt C, Martin G, Morl M. Mol Cell. 2004;15:389–398. doi: 10.1016/j.molcel.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 26.Hopper AK. Crit Rev Biochem Mol Biol. 2006;41:3–19. doi: 10.1080/10409230500405237. [DOI] [PubMed] [Google Scholar]
- 27.Nathanson L, Deutscher MP. J Biol Chem. 2000;275:31559–31562. doi: 10.1074/jbc.C000385200. [DOI] [PubMed] [Google Scholar]
- 28.Li Z, Reimers S, Pandit S, Deutscher MP. EMBO J. 2002;21:1132–1138. doi: 10.1093/emboj/21.5.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okabe M, Tomita K, Ishitani R, Ishii R, Takeuchi N, Arisaka F, Nureki O, Yokoyama S. EMBO J. 2003;22:5918–5927. doi: 10.1093/emboj/cdg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.