Abstract
Thiamin diphosphate, a key coenzyme in sugar metabolism, is comprised of the thiazolium and 4′-aminopyrimidine aromatic rings, but only recently has participation of the 4′-aminopyrimidine moiety in catalysis gained wider acceptance. We report the use of electronic spectroscopy to identify the various tautomeric forms of the 4′-aminopyrimidine ring on four thiamin diphosphate enzymes, all of which decarboxylate pyruvate: the E1 component of human pyruvate dehydrogenase complex, the E1 subunit of Escherichia coli pyruvate dehydrogenase complex, yeast pyruvate decarboxylase, and pyruvate oxidase from Lactobacillus plantarum. It is shown that, according to circular dichroism spectroscopy, both the 1′,4′-iminopyrimidine and the 4′-aminopyrimidine tautomers coexist on the E1 component of human pyruvate dehydrogenase complex and pyruvate oxidase. Because both tautomers are seen simultaneously, these two enzymes provide excellent evidence for nonidentical active centers (asymmetry) in solution in these multimeric enzymes. Asymmetry of active centers can also be induced upon addition of acetylphosphinate, an excellent electrostatic pyruvate mimic, which participates in an enzyme-catalyzed addition to form a stable adduct, resembling the common predecarboxylation thiamin-bound intermediate, which exists in its 1′,4′-iminopyrimidine form. The identification of the 1′,4′-iminopyrimidine tautomer on four enzymes is almost certainly applicable to all thiamin diphosphate enzymes: this tautomer is the intramolecular trigger to generate the reactive ylide/carbene at the thiazolium C2 position in the first fundamental step of thiamin catalysis.
Keywords: 1′,4′-iminothiamin diphospate; 2-oxoacid decarboxylases; active site asymmetry
The hypothesis that the 4′-aminopyrimidine (AP) ring of the coenzyme thiamin diphosphate (ThDP) (1, 2) actively participates in the reaction of enzymes that use it was suggested some years ago (3, 4). The hypothesis gained wider acceptance with the observation of two highly conserved features noted in the first structures of ThDP-dependent enzymes (5–7): (i) a distance shorter than 3.5 Å between the N4′ atom of the AP and the C2 atom of the thiazolium ring, creating the possibility for intramolecular proton abstraction to generate the C2 carbanion/ylide, identified by Breslow (8) as the nucleophile that initiates the sequence of reactions involving multiple ThDP-bound covalent intermediates [exemplified with yeast pyruvate decarboxylase (YPDC) (EC 4.1.1.1) in Scheme 1] and (ii) a glutamate residue within short hydrogen bonding distance of the N1′ atom, poised to catalyze the tautomerization shown on the left side of Scheme 1. The tautomerization reaction requires three forms of the AP ring of which two are neutral, the AP and 1′,4′-iminopyrimidine (IP), but these forms must interconvert via the positively charged, N1′-protonated 4′-aminopyrimidinium ion (APH+). Our goal is to establish which form of the AP ring is present on each intermediate on the pathway in Scheme 1, and indeed in all ThDP enzymes. We here report two examples of ThDP enzymes on which both the AP and IP tautomers of ThDP are present simultaneously. The results provide strong solution evidence for ThDP enzymes to (i) stabilize the unstable IP tautomer that can generate the C2-carbanion/ylide and (ii) have asymmetry of active centers even in the absence of substrate.
Scheme 1.
Mechanism of YPDC.
We have selected four ThDP enzymes, all of which carry out the reaction in Scheme 1 through decarboxylation, to assess the state of tautomerization both in the absence and presence of a pyruvate analogue. We use CD spectroscopy, which offers remarkably rich insight to ThDP mechanisms because all enzyme-bound intermediates are in a chiral environment by virtue of the “V” conformation universally enforced and conserved on all ThDP enzymes, whether or not these intermediates possess a chiral center (9). The data enable us to assign the tautomeric form (AP, IP) and APH+ of the AP ring in each ThDP-bound intermediate on the reaction pathway for this rare coenzyme possessing two aromatic rings, both of which are involved in catalysis. With this understanding, we can address several current hypotheses regarding active center interactions in ThDP enzymes, such as alternating active sites (10) and communication between ThDP cofactors via the “proton wire” mechanism (11).
Results
Human Pyruvate Dehydrogenase E1 Component (PDHc-E1h).
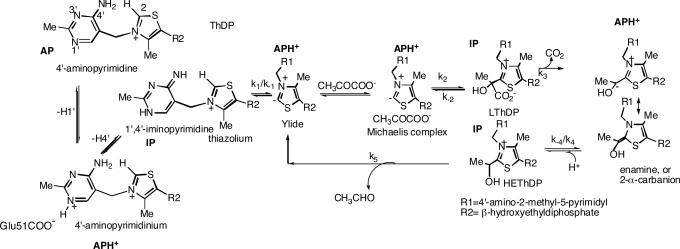
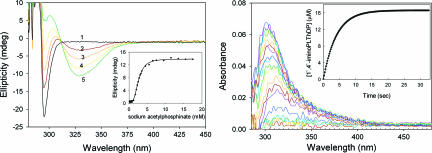
The assignments to different tautomeric forms of the AP ring of ThDP rest on an understanding of the origins of two CD bands pertaining to enzyme-bound species: a positive one in the 300- to 310-nm range and a negative one at 320–330 nm. For the assignments of the two CD bands, PDHc-E1h [EC 1.2.4.1; two active centers in a α2β2 heterotetrameric E1 (12)] provides the clearest evidence (Fig. 1Left). Both the positive (305 nm) and negative (330 nm) (13) bands are related to or derived from ThDP bound to PDHc-E1h, as confirmed by the observation that addition of ThDP to the apo-PDHc-E1h (protein free of ThDP) results in the simultaneous generation of both bands (Fig. 1 Left). Addition of thiamin 2-thiothiazolone diphosphate, a potent “transition state analogue,” to apo-PDHc-E1h produces a positive band (325 nm), a band that persists upon addition of even a large excess of ThDP (Fig. 1 Right), also affirming that the positive CD band at 305 nm and the negative one at 330 nm pertain to ThDP on the PDHc-E1h. In prior studies including chemical model systems, we assigned the electronic transition responsible for the positive CD band at 300–310 nm to the 1′,4′-imino tautomer of the coenzyme (14–17), IP. Concomitantly, we used both enzyme-bound and chemical model observations to show that the negative band at 320–330 nm (18, 19) corresponds to a charge-transfer transition between the thiazolium and the AP rings of ThDP (16). Because, upon ThDP binding to PDHc-E1h, both CD bands are clearly visible, we conclude that the two active centers are occupied by different tautomeric forms of ThDP, i.e., IP and AP.
Fig. 1.
Near-UV CD spectra of PDHc-E1h. (Left) Spectrum 1 shows PDHc-E1h (concentration of active centers = 25 μM) in 10 mM KH2PO4 (pH 7.0) with 0.20 mM ThDP and 1 mM MgCl2. Spectrum 2 shows PDHc-E1h in the absence of ThDP. (Center) Near-UV CD spectra of PDHc-E1h titrated with AcP−. PDHc-E1h (concentration of active centers = 19.6 μM) in 10 mM KH2PO4 (pH 7.0) containing 0.20 mM ThDP and 1 mM MgCl2 was titrated with AcP− (1–150 μM). Inset shows that saturation is reached for a 1:1 molar ratio of [AcP−]/[PDHc-E1h active centers]. (Right) The PDHc-E1h (1.0 mg/ml, concentration of active centers = 13.0 μM) was diluted in 50 mM KH2PO4 (pH 7.0) containing 1 mM MgCl2 and then thiamin 2-thiothiazolone diphosphate (1–50 μM) was added. Upon addition of thiamin 2-thiothiazolone diphosphate the positive CD band at 325 nm developed and reached saturation with Kd,ThTTDP = 0.59 μM.
Upon incremental addition of the substrate analogue sodium acetylphosphinate (CH3C( O)P(H)OO− or AcP−), the positive band at 305 nm increases while the negative band decreases then disappears (Fig. 1 Center). We interpret this to mean that AcP− is used as a substrate analogue by PDHc-E1h, and it is converted by the enzyme to the 1′,4′-iminophosphinolactyl-ThDP, a stable analogue of C2α-lactylThDP (LThDP) as in Scheme 2. This conclusion is supported by studies with sodium methyl acetylphosphonate (NaMAP), a somewhat bulkier pyruvate analogue but similar to AcP−, on both the PDHc-E1h and the Escherichia coli PDHc E1 subunit (PDHc-E1ec) (data not shown) by a variety of experiments (15, 16). Addition of either phosphonolactylThDP (a preformed adduct of ThDP and NaMAP) or NaMAP itself to PDHc-E1ec generates the same positive CD band at 305 nm. From the accumulated evidence we concluded that ThDP-bound intermediates with a tetrahedral substituent at the C2 thiazolium position, such as LThDP or C2α-hydroxyethylThDP (HEThDP) (Scheme 1), exist in the IP tautomeric form (16).
O)P(H)OO− or AcP−), the positive band at 305 nm increases while the negative band decreases then disappears (Fig. 1 Center). We interpret this to mean that AcP− is used as a substrate analogue by PDHc-E1h, and it is converted by the enzyme to the 1′,4′-iminophosphinolactyl-ThDP, a stable analogue of C2α-lactylThDP (LThDP) as in Scheme 2. This conclusion is supported by studies with sodium methyl acetylphosphonate (NaMAP), a somewhat bulkier pyruvate analogue but similar to AcP−, on both the PDHc-E1h and the Escherichia coli PDHc E1 subunit (PDHc-E1ec) (data not shown) by a variety of experiments (15, 16). Addition of either phosphonolactylThDP (a preformed adduct of ThDP and NaMAP) or NaMAP itself to PDHc-E1ec generates the same positive CD band at 305 nm. From the accumulated evidence we concluded that ThDP-bound intermediates with a tetrahedral substituent at the C2 thiazolium position, such as LThDP or C2α-hydroxyethylThDP (HEThDP) (Scheme 1), exist in the IP tautomeric form (16).
Scheme 2.
Formation of 1′,4′-iminophosphinolactyl-ThDP, a stable LThDP analogue.
Pyruvate Oxidase (POX).
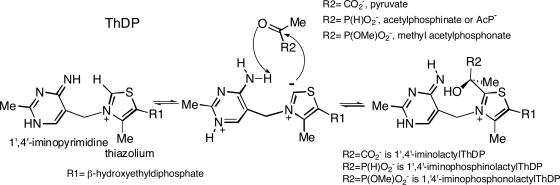
POX from Lactobacillus plantarum (EC 1.2.3.3) (6, 20) is a α4 homotetramer, with each subunit containing one tightly bound FAD, ThDP, and Mg+2. The CD spectrum of POX displays two bands from FAD, a positive one at 393 nm and a negative one at 469 nm, corresponding to the well known bands observed in the UV-VIS spectrum (Fig. 2).
Fig. 2.
Near-UV CD spectra of POX. Spectrum 1 shows POX (concentration of active centers = 2.0 mg/ml or 30 μM) at 30°C in 50 mM KH2PO4 (pH 6.0) containing 0.1 mM ThDP, 1 mM MgCl2, and 6% glycerol. Spectra 2, 3, and 4 were recorded after addition of 5, 10, and 40 μM AcP−, respectively, at 30°C and show the presence of 1′,4′-iminophosphinolactyl-ThDP at 308 nm and the Michaelis complex at 336 nm. Inset shows the enlarged spectra in the 290- to 380-nm range upon addition of acetylphosphinate.
There appear two additional bands in the spectrum (Fig. 2), a positive one at 305 nm and a negative one at 328 nm, corresponding to ThDP as assigned above. The active centers of POX are occupied by different tautomers of ThDP, IP, and AP, according to the CD data, as observed with PDHC-E1h above. Upon addition of AcP−, two changes are revealed: (i) the positive 305-nm CD band shifts to 308 nm, signaling a new species, which we assign to the 1′,4′-iminophosphinolactyl-ThDP; (ii) the negative CD band also shifts from 328 to 336 nm, but, in this case, there is saturation of a distinct new negative band, which we assign to a Michaelis complex type species, POX.ThDP(APH+).AcP−. This band was also evident, and assigned to a Michaelis complex, when NaMAP was added to YPDC (16). The experiments with POX enable us to identify four enzyme-bound species, AP and IP with only ThDP bound, and two new species formed with the pyruvate analogue bound, one covalently bound to ThDP (represented as an IP) and the other as a Michaelis complex, POX.ThDP(APH+).AcP−.
YPDC.
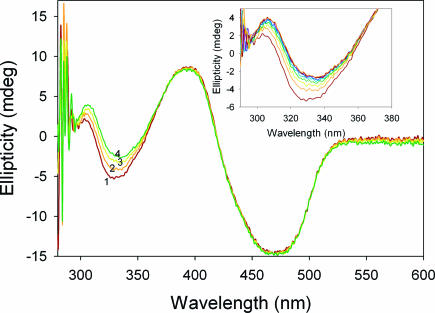
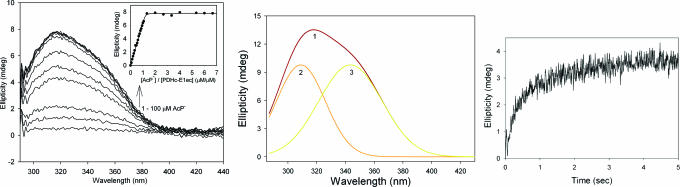
The recombinant YPDC is a α4 homotetramer (7, 21), and, as was the case with PDHc-E1ec, with ThDP bound, YPDC is fully occupied by APH+ in all active centers, deduced from the absence of a CD band for AP or IP. Incremental addition of AcP− to YPDC results in simultaneous appearance of the positive CD band at 302 nm and a negative one at 328 nm (Fig. 3Left), indicating occupancy of the active centers by the 1′,4′-iminophosphinolactyl-ThDP in one half of the active centers and by the Michaelis complex [YPDC.ThDP(APH+).AcP−] in the second half of what is termed a functional dimer (22). Hence, once substrate or analogue AcP− is bound, both IP and AP tautomers are in evidence. Further evidence for the assignments on YPDC is gleaned from kinetic data on the rate of formation of the two UV-VIS bands (corresponding to the CD bands): (i) the band at 328 nm attributed to the Michaelis complex is fully formed within the 2.5 ms required for recording the first spectrum; and (ii) the rate constant for formation of the positive CD band at 302 nm related to formation of 1′,4′-iminophosphinolactyl-ThDP is 0.25 s−1 (Fig. 3 Right). We could assign the tautomeric form of ThDP present to each species and also determine the time course for its formation.
Fig. 3.
Formation of 1′,4′-iminophosphinolactyl-ThDP on YPDC. (Left) Near-UV CD spectra of YPDC. Spectrum 1 shows YPDC (concentration of active centers is 2.5 mg/ml or 42 μM) in 50 mM KH2PO4 (pH 6.0) containing 0.1 mM ThDP and 2.0 mM MgCl2. Spectra 2, 3, 4, and 5 were recorded after addition of 2, 2.5, 3.5, and 17.5 mM sodium acetylphosphinate, respectively, at 30°C. The spectra show presence of 1′,4′-iminophosphinolactyl-ThDP at 302 nm and of the Michaelis complex at 328 nm. (Inset) Dependence of 1′,4′-iminophosphinolactyl-ThDP formation at 302 nm on concentration of AcP− after subtraction of the spectrum of YPDC in the absence of AcP−. Strong cooperativity (Hill coefficient ≈ 3.17) probably reflects the presence of an allosteric substrate activation site in addition to the active centers in the functional dimer (1, 2). (Right) Photodiode array stopped-flow UV-VIS difference spectra showing the formation of 1′,4′-iminophosphinolactyl-ThDP at 302 nm in YPDC. YPDC (2 mg/ml) in 50 mM phosphate (pH 6.0) containing 0.1 mM ThDP and 2.0 mM MgCl2 in one syringe was mixed with an equal volume of 40 mM AcP− in the same buffer in the second syringe. The reaction was monitored for 33 s, and the spectra were recorded at 2.5-ms interval at 30°C. Inset shows the rate of formation of 1′,4′-iminophosphinolactyl-ThDP at 302 nm (kobs = 0.25 s−1).
E. coli Pyruvate Dehydrogenase Complex E1 Subunit.
With PDHc-E1ec [EC 1.2.4.1; α2 homodimer with two active centers (23)] we see no evidence for either bound AP or IP with saturating ThDP; hence, we conclude that it is in the APH+ state in both active centers (Scheme 1). Addition of AcP− to PDHc-E1ec produces only a single broad positive CD band (Fig. 4Left and Center), which corresponds to the 1′,4′-iminophosphinolactyl-ThDP, similar to results reported with phosphonolactylThDP (16, 24). Stopped-flow kinetic studies helped to assign the band to a single species (Fig. 4 Right). Upon saturation with AcP−, all active centers appear to be filled with the 1′,4′-iminophosphinolactyl-ThDP, i.e., the IP form. With only ThDP bound, the difference between PDHc-E1h and PDHc-E1ec is truly striking; both the IP and AP forms can be seen on the former. We reported earlier that the negative band at 330 nm could be induced on the PDHc-E1ec as a Michaelis complex with pyruvate once one center was occupied by HEThDP (16) (Scheme 1).
Fig. 4.
Formation of 1′,4′-iminophosphinolactyl-ThDP on PDHc-E1ec. (Left) Difference CD spectra of the PDHc-E1ec titrated with AcP−. PDHc-E1ec (concentration of active centers = 15 μM) in 10 mM KH2PO4 (pH 7.0) containing 2 mM MgCl2 and 0.20 mM ThDP was titrated with AcP− (1–100 μM). Difference spectra were obtained upon subtraction of the spectrum of PDHc-E1ec in the absence of AcPI−. Inset shows that the CD maximum was reached at a 1:1 molar ratio of [AcP−]/[PDHc-E1ec active centers]. (Center) The CD curve resulting from addition of saturating AcP− (70 μM) to PDHc-E1ec (2.43 mg/ml; 0.1 mM ThDP/1 mM MgCl2/20 mM potassium phosphate, pH 7.0) was resolved into peaks 2 and 3 by using Peak Fit v 4.12 from Systat Software (San Jose, CA). The λmax for peaks 1, 2, and 3 are 318, 309, and 343 nm, respectively, and the rate constants measured at these wavelengths were identical within experimental error. (Right) Stopped-flow CD determination of the rate of 1′,4′-iminophosphinolactyl-ThDP formation at 30°C. PDHc-E1ec [5 mg/ml or 50 μM active centers dissolved in 20 mM KH2PO4 (pH 7.0) also containing 0.1 mM ThDP and 1 mM MgCl2] in one syringe was mixed with 50 μM AcP− in the same buffer. The reaction was monitored for 5 s, 2,000 data points were collected at 2.5-ms intervals, and the trace was fitted to a double exponential equation with rate constants of 4.44 ± 0.34 s−1 and 0.593 ± 0.064 s−1.
Discussion
Evidence for Different ThDP Tautomers in the Absence of Substrate.
The most significant observation is that both the AP and IP ThDP tautomers are present on PDHc-E1h and POX but not on YPDC or PDHc-E1ec. On PDHc-E1h and POX the evidence reveals the presence of the IP tautomer ready to initiate the intramolecular proton transfer to produce the ylide before arrival of substrate. While not assigning the CD band at 305 nm, similar observations were reported earlier on two other mammalian PDHc-E1 (19, 25). The ability of PDHc-E1h and POX to stabilize the 1′,4′-imino tautomer once again points to the exceptional active center environment of ThDP enzymes already demonstrated for YPDC (26) and PDHc-E1ec (27).
Addition of Substrate Analogue.
Addition of substrate analogue acetylphosphinate, AcP−, an excellent electrostatic and steric pyruvate mimic (the compound acetylphosphinate had been reported to be an efficient inhibitor of PDHc-E1ec in ref. 28), to any of the four enzymes leads to highly efficient synthesis of 1′,4′-iminophosphinolactyl-ThDP, a stable analog of LThDP, as reflected by the rate constant for formation of the positive CD band in the 300- to 310-nm range (see Fig. 4 Right for PDHc-E1ec; k = 4.44 s−1). Such a stable LThDP analogue derived from NaMAP (rather than AcP−) was recently identified by high-resolution x-ray studies on both PDHc-E1ec (24) and POX (20), as was LThDP itself on POX (20).
Asymmetry of Active Centers with ThDP Present.
An asymmetry of active centers is revealed by results on POX and PDH-E1h in the absence of AcP−, showing the simultaneous presence of both ThDP tautomers. This constitutes the first spectroscopic evidence for active center asymmetry in ThDP enzymes in solution (10). Such asymmetry of active centers was suggested for bacterial (29, 30) and mammalian E1 components (12) on the basis of x-ray crystal structures. Earlier, on the basis of kinetic and CD evidence, an alternating-sites mechanism was proposed for mammalian E1 component (31), very recently receiving direct kinetic support on PDHc-E1h (32). In summary, there is now kinetic, crystallographic, and spectroscopic evidence supporting an alternating-site mechanism in ThDP enzymes. The “proton wire mechanism” proposed by Perham and coworkers (11) provides one possible explanation for asymmetry. Our results on POX and PDHc-E1h suggest that the proton wire connects an IP to an AP tautomer in the two active sites.
Asymmetry of Active Centers with Substrate Analogue Bound.
With added AcP−, the four enzymes comprise two distinct groups: PDHc-E1ec and PDHc-E1h (two active centers) revealed the presence of only the 1′,4′-iminophosphinolactyl-ThDP, whereas YPDC and POX (four active centers) revealed the additional presence of a Michaelis complex. Hence, one active center is filled with either IP or the 1′,4′-iminophosphinolactyl-ThDP, and the other active center is filled with either AP (in the absence of AcP− on POX and PDHc-E1h) or the Michaelis complex analogue POX.ThDP(APH+).AcP− or YPDC.ThDP(APH+).AcP−. We suggest that this could be a reflection of the alternating active center mechanism in homotetramers, already identified on YPDC (23) and benzoylformate decarboxylase (33) (also a homotetrameric ThDP enzyme).
State of 4′-Aminopyrimidine Present on all Intermediates on ThDP Enzymes.
The current “complete” picture for YPDC is shown in Scheme 1, identifying the tautomeric/protonation state of each intermediate. There remain two questions on the pathway: the route to the Michaelis complex and the form of the AP at the enamine/C2α-carbanion stage. Regarding the latter issue, unpublished experiments from Rutgers (S.C., A.B., and F.J.) signal that, when the enamine is present, there is no evidence for the IP tautomer or for AP; hence, we suggest APH+ next to the structure. Given the intramolecular proton transfer from the thiazolium C2H to the 4′-imino nitrogen, the most logical step for forming the Michaelis complex appears to be from the ylide/C2-carbanion, a deduction with significant mechanistic implications (34). For example, the best probability of observing both the 1′,4′-imino tautomer and perhaps even the ylide/C2-carbanion is predicted to be with POX or YPDC, the IP with a substrate analogue added.
By analogy, we suggest that for all ThDP enzymes those intermediates with sp3-hybridized C2α substituents, such as the LThDP, HEThDP, and their analogues, will exist in the IP form. By contrast, those intermediates with a sp2-hybridized C2α substituents, such as the enamine/C2α-carbanion present on virtually all ThDP enzymes, the hydroyethylideneThDP radical formed from one-electron oxidation of this enamine/C2α-carbanion on pyruvate:ferredoxin oxidoreductase (35), or the 2-acyl-ThDP in the 2-oxoacid dehydrogenases and POX, will be in the AP or APH+ form. Our spectroscopic method provides a useful complement to the numerous crystal structures of ThDP enzymes being reported with bound intermediates (2, 20), where unequivocal structural assignment is often limited by the resolution.
Finally, we note the remaining questions suggested by the results. The method here exploited can identify two of three forms of enzyme-bound ThDP at equilibrium in Scheme 1: AP and IP, but not APH+. When evidence is lacking for the AP and IP forms, presumably, the APH+ form occupies all active centers. Yet, even when AP and IP forms are present, some active sites could remain occupied by APH+, which we cannot detect. What are possible occupancies of the homodimeric active sites where both AP and IP are in evidence? The choices are IP2, AP2, and AP.IP, suggesting that where both are detected almost certainly one center is occupied with IP and the other is occupied with AP. The position of protolytic equilibrium [APH+]/{[AP] + [IP]} is dictated by the pKa for bound APH+; therefore, shifting this pKa from one enzyme to the next (mammalian vs. bacterial E1 component) could account for our ability to observe the CD signals for the AP and IP tautomers in some cases but not in others.
Materials and Methods
Bacterial Strains and Plasmids.
E. coli strain JRG3456 and plasmid pGS878 with the aceE1 gene encoding the PDHc-E1ec subunit of E. coli PDHc were from J. Guest (University of Sheffield, Sheffield, U.K.). The construction of plasmid pET22b(+)-pdcI encoding YPDC with the C-terminal His6 tag was described previously (36). A coexpression vector for PDHc-E1h (pET-28b-PDHA1/PDHB) harboring coding sequences of both human PDHc-E1α and PDHc-E1β subunits was constructed as described previously (37).
Overexpression and Purification of Proteins.
E. coli strain JRG3456 transformed with pGS878 was used for overexpression of the aceE gene encoding the PDHc-E1ec subunit of E. coli PDHc according to ref. 9, and purification of PDHc-E1 was according to ref. 38. YPDC with the C-terminal His6 tag was overexpressed and purified according to ref. 36. Recombinant human PDHc-E1h was overexpressed in E. coli BL21 cells and purified by using Ni-NTA-agarose chromatography as described previously (9). POX from L. plantarum was a kind gift from Roche Molecular Biochemicals (Pennzberg, Germany).
Synthesis of acetylphosphinate was carried out according to the procedure reported in ref. 39. The acetylphosphinate is a stoichiometric active center titrant for both PDHc-E1 enzymes (40) and POX; hence, our method could be used for this purpose on all such enzymes. CD spectra were recorded with an Aviv Model 202 CD or Chirascan spectrometer (30°C; Applied Photophysics, Leatherhead, England). Stopped-flow CD spectra were recorded on a π* −180 CDF spectrometer, and stopped-flow photodiode UV-VIS spectra were recorded on a SX.18MV array reaction analyzer, both from Applied Photophysics.
Acknowledgments
Research at Rutgers was funded by National Institutes of Health Grants GM-050380 and GM-062330.
Abbreviations
- ThDP
thiamin diphosphate
- YPDC
yeast pyruvate decarboxylase
- POX
pyruvate oxidase
- PDHc-E1h
human pyruvate dehydrogenase E1 subunit
- PDHc-E1ec
E. coli PDHc E1 subunit
- AP
4′-aminopyrimidine
- IP
1′,4′-iminopyrimidine
- APH+
N1′-protonated 4′-aminopyrimidinium ion
- LThDP
C2α-lactylThDP
- HEThDP
C2α-hydroxyethylThDP
- NaMAP
sodium methyl acetylphosphonate
- AcP−
sodium acetylphosphinate.
Footnotes
The authors declare no conflict of interest.
References
- 1.Jordan F. Nat Prod Rep. 2003;20:184–201. doi: 10.1039/b111348h. [DOI] [PubMed] [Google Scholar]
- 2.Jordan F, Nemeria NS. Bioorg Chem. 2005;33:190–215. doi: 10.1016/j.bioorg.2005.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schellenberger A. Biochim Biophys Acta. 1998;1385:177–186. doi: 10.1016/s0167-4838(98)00067-3. [DOI] [PubMed] [Google Scholar]
- 4.Jordan F, Mariam YH. J Am Chem Soc. 1978;100:2534–2540. [Google Scholar]
- 5.Lindquist Y, Schneider G, Ermler U, Sundstroem M. EMBO J. 1992;11:3273–3279. doi: 10.1002/j.1460-2075.1992.tb05301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller YA, Schulz GE. Science. 1993;259:965–967. doi: 10.1126/science.8438155. [DOI] [PubMed] [Google Scholar]
- 7.Dyda F, Furey W, Swaminathan S, Sax M, Farrenkopf B, Jordan F. Biochemistry. 1993;32:6165–6170. doi: 10.1021/bi00075a008. [DOI] [PubMed] [Google Scholar]
- 8.Breslow R. J Am Chem Soc. 1958;80:3719–3726. [Google Scholar]
- 9.Nemeria N, Yan Y, Zhang Z, Brown AM, Arjunan P, Furey W, Guest JR, Jordan F. J Biol Chem. 2001;276:45969–45978. doi: 10.1074/jbc.M104116200. [DOI] [PubMed] [Google Scholar]
- 10.Jordan F, Nemeria NS, Sergienko E. Acc Chem Res. 2005;38:755–763. doi: 10.1021/ar040244e. [DOI] [PubMed] [Google Scholar]
- 11.Frank RAW, Titman CM, Pratap JV, Luisi BF, Perham RN. Science. 2004;306:872–876. doi: 10.1126/science.1101030. [DOI] [PubMed] [Google Scholar]
- 12.Ciszak EM, Korotchkina LG, Dominiak PM, Sidhu S, Patel MS. J Biol Chem. 2003;278:21240–21246. doi: 10.1074/jbc.M300339200. [DOI] [PubMed] [Google Scholar]
- 13.Korotchkina LG, Ali MS, Patel MS. Arch Biochem Biophys. 1999;369:277–287. doi: 10.1006/abbi.1999.1364. [DOI] [PubMed] [Google Scholar]
- 14.Jordan F, Zhang Z, Sergienko E. Bioorg Chem. 2002;30:188–198. doi: 10.1006/bioo.2002.1249. [DOI] [PubMed] [Google Scholar]
- 15.Jordan F, Nemeria NS, Zhang S, Yan Y, Arjunan P, Furey W. J Am Chem Soc. 2003;127:12732–12738. doi: 10.1021/ja0346126. [DOI] [PubMed] [Google Scholar]
- 16.Nemeria NS, Baykal A, Joseph E, Zhang S, Yan Y, Furey W, Jordan F. Biochemistry. 2004;43:6565–6575. doi: 10.1021/bi049549r. [DOI] [PubMed] [Google Scholar]
- 17.Baykal A, Kakalis L, Jordan F. Biochemistry. 2006;45:7522–7528. doi: 10.1021/bi060395k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kochetov GA, Usmanov RA. Biochem Biophys Res Commun. 1970;41:1134–1140. doi: 10.1016/0006-291x(70)90203-2. [DOI] [PubMed] [Google Scholar]
- 19.Khailova LS, Korotchkina LG, Severin SE. Ann NY Acad Sci. 1989;573:36–54. doi: 10.1111/j.1749-6632.1989.tb14985.x. [DOI] [PubMed] [Google Scholar]
- 20.Wille G, Meyer D, Steinmetz A, Hinze E, Golbik R, Tittmann K. Nat Chem Biol. 2006;2:324–328. doi: 10.1038/nchembio788. [DOI] [PubMed] [Google Scholar]
- 21.Arjunan P, Umland T, Dyda F, Swaminathan S, Furey W, Sax M, Farrenkopf B, Gao Y, Zhang D, Jordan F. J Mol Biol. 1996;256:590–600. doi: 10.1006/jmbi.1996.0111. [DOI] [PubMed] [Google Scholar]
- 22.Sergienko EA, Jordan F. Biochemistry. 2001;40:7382–7403. doi: 10.1021/bi002857e. [DOI] [PubMed] [Google Scholar]
- 23.Arjunan P, Nemeria N, Brunskill A, Chandra-sekhar K, Sax M, Yan Y, Jordan F, Guest JR, Furey W. Biochemistry. 2002;41:5213–5221. doi: 10.1021/bi0118557. [DOI] [PubMed] [Google Scholar]
- 24.Arjunan P, Sax M, Brunskill A, Chandrasekhar N, Nemeria NS, Zhang S, Jordan F, Furey W. J Biol Chem. 2006;281:15296–15303. doi: 10.1074/jbc.M600656200. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Bisswanger H. Biol Chem. 2005;386:11–18. doi: 10.1515/BC.2005.002. [DOI] [PubMed] [Google Scholar]
- 26.Jordan F, Li H, Brown A. Biochemistry. 1999;38:6369–6373. doi: 10.1021/bi990373g. [DOI] [PubMed] [Google Scholar]
- 27.Zhang S, Zhou L, Nemeria N, Yan Y, Zhang Z, Zou Y, Jordan F. Biochemistry. 2005;44:2237–2243. doi: 10.1021/bi047696j. [DOI] [PubMed] [Google Scholar]
- 28.Schönbrunn-Hanebeck E, Laber B, Amrhein N. Biochemistry. 1990;29:4880–4885. doi: 10.1021/bi00472a018. [DOI] [PubMed] [Google Scholar]
- 29.Nakai T, Nakagawa N, Maoka N, Masui R, Kuramitsu S, Kamiya N. J Mol Biol. 2004;337:1011–1033. doi: 10.1016/j.jmb.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 30.Frank RAW, Pratap JV, Pei XY, Perham RN, Luisi BF. Structure (London) 2005;13:1119–1130. doi: 10.1016/j.str.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 31.Khailova LS, Korotchkina LG, Severin SE. In: Biochemistry and Physiology of ThDP Enzymes. Bisswanger H, Ullrich J, editors. Weinheim, Germany: VCH; 1990. [Google Scholar]
- 32.Seifert F, Golbik R., Brauer J, Lilie H, Schroder-Tittmann K, Hinze E, Korotchkina LG, Patel MS, Tittmann K. Biochemistry. 2006;45:12775–12785. doi: 10.1021/bi061582l. [DOI] [PubMed] [Google Scholar]
- 33.Sergienko EA, Wang J, Polovnikova L, Hasson MS, McLeish MJ, Kenyon GL, Jordan F. Biochemistry. 2000;39:13862–13869. doi: 10.1021/bi001214w. [DOI] [PubMed] [Google Scholar]
- 34.Kern D, Kern G, Neef H, Tittmann M, Killen-berg-Jabs M, Wikner C, Schneider G, Hübner G. Science. 1997;275:67–70. doi: 10.1126/science.275.5296.67. [DOI] [PubMed] [Google Scholar]
- 35.Mansoorabadi SO, Seravalli J, Furdui C, Krymov V, Gerfen GJ, Begley TP, Melnick J, Ragsdale SW, Reed GH. Biochemistry. 2006;45:7122–7131. doi: 10.1021/bi0602516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu M, Sergienko EA, Guo F, Wang J, Tittmann K, Hübner G, Furey W, Jordan F. Biochemistry. 2001;40:7355–7368. doi: 10.1021/bi002855u. [DOI] [PubMed] [Google Scholar]
- 37.Korotchkina LG, Sidhu S, Patel MS. J Biol Chem. 2006;281:9688–9696. doi: 10.1074/jbc.M511481200. [DOI] [PubMed] [Google Scholar]
- 38.Nemeria N, Volkov A, Brown AM, Yi J, Zipper L, Guest JR, Jordan F. Biochemistry. 1998;37:911–922. doi: 10.1021/bi9722251. [DOI] [PubMed] [Google Scholar]
- 39.Baillie AC, Wright BJ, Wright K. 4,339,443. US Patent. 1982
- 40.Nemeria N, Korotchkina L, Chakraborty S, Patel MS, Jordan F. Bioorg Chem. 2006;34:362–379. doi: 10.1016/j.bioorg.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]