Abstract
We have determined the crystal structure of the chloroperoxidase (CPO) hydroperoxo reaction intermediate (CPO compound 0) at 1.75-Å resolution. The intermediate was generated through controlled photoreduction of the CPO oxygen complex during x-ray data collection, which was monitored by recording of the crystal absorption spectra. Initially, the peroxo-anion species was formed and then protonated to yield compound 0. Quantum chemical calculations indicate that the peroxo-anion species is not stable and collapses instantaneously to compound 0. Compound 0 is present in the ferric low-spin doublet ground state and is characterized by a long O O bond length of 1.5 Å and a Fe
O bond length of 1.5 Å and a Fe O bond distance of 1.8 Å, which is also observed in the crystal structure.
O bond distance of 1.8 Å, which is also observed in the crystal structure.
Keywords: crystal structure, protein structure/function, radiolysis, reaction mechanism, QM/MM calculations
Nature devised diverse heme enzymes to metabolize oxygen by using atmospheric dioxygen or hydrogen peroxide (1, 2). Cytochrome P450 enzymes are monooxygenases that catalyze the incorporation of one atom of molecular oxygen into a range of organic substrates. P450 enzymes are unique in their ability to hydroxylate nonactivated hydrocarbon compounds and are involved in many biological processes, including the biosynthesis of steroids, lipids, fat-soluble vitamins, and antibiotics (3). Their role in the degradation of drugs and other xenobiotics is of great medical interest (4). Heme oxygenases use dioxygen as a substrate, whereas peroxidases and catalases use hydrogen peroxide (2). Chloroperoxidase (CPO) is a glycoprotein secreted from the fungus Caldariomyces fumago, which catalyzes hydrogen peroxide-dependent halogenation reactions (5). In addition to catalyzing chlorination, bromination, and iodination reactions, CPO functions as a catalase and a peroxidase and catalyzes P450-like oxygen insertion reactions. This wide-ranging activity makes CPO one of the most versatile of all heme enzymes (6, 7). The structure of CPO has unique aspects but it shares features with P450 enzymes, e.g., a cysteine as proximal heme ligand, and contains a polar distal pocket in common with peroxidases. But in contrast to other peroxidases, the distal residue of CPO is not a histidine but a glutamic acid (Glu-183), and the arginine that is highly conserved in peroxidases active site pockets is not present in CPO (8). CPO is considered to be a crucial model for the reaction intermediates of P450 enzymes. The short-lived intermediates of CPO have been characterized by a wide range of spectroscopic methods (9–15).
Despite the diverse functions of the above-mentioned heme enzymes, all are considered to share a common species, a ferric hydroperoxo intermediate (Fe-OOH−), also denoted as compound 0 (16). Characterization of this intermediate is difficult, because it is unstable and short-lived (17). Compound 0 (Fe-OOH−) was initially reported at low temperatures by stopped-flow experiments (18). This intermediate has been more recently reported to be prepared through the reduction of oxy-ferrous heme proteins by cryogenic radiolysis (19–22). During exposure to high-energy radiation, e.g., γ-radiation from 60Co sources or x-rays, electrons are released through the photoelectric effect and secondary processes, i.e., the ejection of Auger electrons. X-rays with an energy of 12 keV are commonly used for x-ray data collection at synchrotron light sources. Ninety percent of the photons at this energy interact with the sample through the photoelectric effect. A single absorbed photon leads to the generation of ≈300 electrons at this energy (23, 24).
Compound 0 (Fe-OOH−) intermediates of various heme enzymes, including P450cam (21) and HRP (22), have been generated in frozen oxy-ferrous heme solutions by cryogenic radiolysis, which has allowed the spectroscopic characterization of this species. Photoreduction can also be exploited during x-ray crystallographic structure determination to obtain otherwise short-lived reaction intermediates. One such example is the structural characterization of the P450cam (CYP101) reaction intermediates. The second electron of the P450 reaction cycle required for the reduction of the oxy-ferrous P450cam complex was generated during x-ray irradiation and allowed the structural determination of a species consistent with an iron-oxo complex. However, the compound 0 intermediate was not observed (25).
Here, we describe the crystal structure of compound 0 of CPO determined at a resolution of 1.75 Å. This cryo-trapped intermediate was generated by photoreduction of CPO oxygen complex crystals during x-ray data collection at 90 K followed by protonation. Visible absorption spectra of crystals recorded during x-ray exposure and hybrid quantum mechanical (QM)/molecular mechanical (MM) calculations confirm the formation of compound 0 and suggest a pathway for its generation.
Results and Discussion
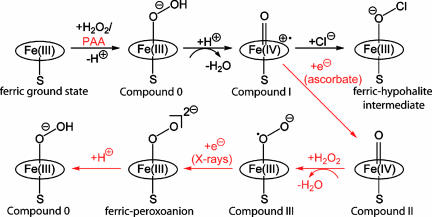
CPO crystals were soaked in a peracetic acid (PAA)/ascorbate and hydrogen peroxide-containing cryoprotectant solution at 4°C. The formed intermediate was cryo-trapped by flash-cooling of the crystals in liquid nitrogen (see Fig. 1). Unexpectedly, instead of compound II, we observed density fitting a diatomic ligand at the iron in the crystal structure. During soaking of the crystals with the cryoprotectant mixture, compound II reacted with hydrogen peroxide and yielded compound III (26, 27) (Fig. 1). Compound III corresponds to the CPO oxygen complex [Fe(II)-O2], which is isoelectronic with [Fe(III)-O2]−.
Fig. 1.
Reaction scheme of CPO. The halogenating ferric-hypohalite intermediate is formed in vivo by the reaction of CPO with hydrogen peroxide and proceeds via compound 0 and compound I. Shown in red is the experimental strategy used in this study to generate the compound 0 species, using PAA instead of the natural substrate H2O2.
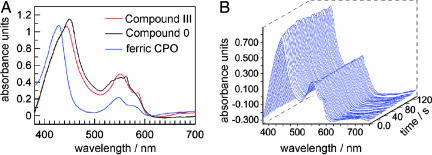
To analyze the ligand state of the crystalline CPO, we measured the UV-visible absorption spectra with a microspectrophotometer. Difficulties in interpreting these spectra can arise because of the anisotropic properties of crystals. As a consequence, the crystal absorption spectra differ depending on the orientation of the crystal with respect to the direction of the incident light. Nevertheless, in the case of CPO, both solution and crystal absorption spectra share the same features. The crystal absorption maxima could be assigned to the equivalent peaks of the solution absorption spectra. Absorption spectra recorded of the soaked CPO crystals confirmed the formation of compound III. A red shift of the Soret band is in agreement with the solution absorption spectra (Fig. 2A). The maximum of the Soret band of the compound III crystals varies between 430 and 445 nm depending on the orientation of the crystal, whereas for untreated ferric crystals, the peak of the Soret band is located between 420 and 430 nm. This finding agrees with absorption spectra measured from frozen ferric CPO solutions at 77 K, which show a Soret band peak at 424 nm (28). In comparison, a value of 432 nm for the Soret band of the compound III species was reported for CPO solution spectra recorded at 5°C (26).
Fig. 2.
Absorption spectra measured from CPO crystals, mounted in a loop and kept at 90 K. (A) Comparison of the ferric ground state (blue), compound III (red), and compound 0 (black) crystal absorption spectra. (B) Temporal evolution of absorption spectra recorded during x-ray exposure of CPO compound III crystals.
We observed that the Q bands located in the region between 500 and 600 nm were less sensitive toward changes in the orientation of the crystal. A shift of the Qα band by 5 nm to a longer wavelength (550 nm) was observed in compound III crystals compared with the ferric ground state (545 nm). Furthermore, the Qβ (583 nm) band was more pronounced in compound III crystals than in the ferric ground state, which was also consistent with the solution absorption spectra (Fig. 2A).
Because it has been observed previously that active sites of metallo-proteins are readily reduced by photoelectrons formed during x-ray exposure using synchrotron radiation (25, 29, 30, 56), we investigated whether this is also the case for the CPO compound III crystals. Indeed, when CPO compound III crystals were exposed to x-rays at the synchrotron, within seconds a new absorption peak appeared at 563 nm, and a shift of the Soret band to a longer wavelength, i.e., from 443 to 451 nm was observed (Fig. 2B), which indicated a reduction of compound III to a peroxo species (Fig. 1). Such a red shift of the Soret band is consistent with cryogenic radiolysis experiments of frozen CPO compound III solutions, where a shift of the Soret band peak from 427 to 447 nm was observed upon irradiation, although no full spectrum was reported at that time (31).
CPO crystals have the orthorhombic space group C2221, and ≈90° of data are required to obtain a complete data set. The crystals were exposed for 1 s for a 1° oscillation. Consequently, a crystal needed to be exposed for ≈90 s to obtain a complete data set. In addition to displaying rapidly changing optical spectra upon x-ray exposure, after ≈1 min of irradiation, the Soret band (451 nm) started to decay slowly. Hence, the electron density map calculated from data collected from a single crystal will show a mixture of different states: compound III, the reduced peroxo-intermediate, possibly a hydroperoxo intermediate, and the species formed after the decay of the (hydro)peroxo intermediate. Therefore, we used a composite data collection strategy using several crystals to obtain a data set of the pure accumulated peroxo species. Data were merged to yield “low dose,” “medium dose,” and “high dose” data sets at 1.75-Å resolution (as described in Materials and Methods). The Fobs − Fcalc electron density difference maps calculated from the three merged data sets show elongated densities above the heme iron. The densities observed for the low- and medium-dose data are similar and fit a diatomic ligand. However, because compound III was reduced very rapidly, even the low-dose data set contained a mixture of compound III and the (hydro)peroxo intermediate. Hence we could not determine the compound III structure. In contrast, for the high-dose data set the electron density close to the iron becomes weaker, indicating a decay of the (hydro)peroxo species at longer exposure times. Thus the medium-dose data set was the best description of the (hydro)peroxo state and was used for structure determination.
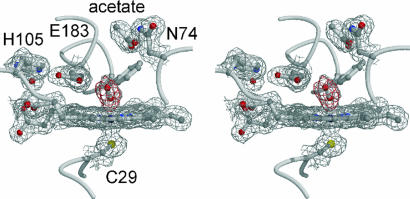
The formation of the (hydro)peroxo intermediate does not induce major conformational changes in the protein, with the exception of a twisting of the aromatic side chain of Phe-186 by 0.5 Å away from the distal oxygen compared with the ferric resting state. But this movement is more pronounced in the cyclopentanedione and acetate complexed CPO structures (32). An acetate molecule is bound in the wide substrate access channel and forms a hydrogen bond with the side chain of Asn-74 (Fig. 3). Acetate is present at a concentration of ≈150 mM in the soaking solution, because it occurs in high concentrations in PAA stock solutions. The acetate binding site has been identified previously as a binding site for polar compounds, i.e., iodide, ethylene glycol, and DMSO, all of which interact with Asn-74 in an otherwise mainly nonpolar environment (32).
Fig. 3.
Stereoview of the active site of CPO compound 0. The structure was refined against the medium-dose data. The final 1.75-Å resolution sigmaA-weighted 2m FO − DFC map (1σ contour level) is shown in gray and the mFO − DFC omit map (+3σ contour level) is superimposed in red.
The distal ligand has an O O bond length of 1.49 Å, which is obtained after refinement against the medium dose data with only weak restraints being applied to the O
O bond length of 1.49 Å, which is obtained after refinement against the medium dose data with only weak restraints being applied to the O O bond length (Fig. 3). The ligand was modeled with an occupancy of 100% and has B factors of 18.5 Å2 for the proximal oxygen and 21.5 Å2 for the distal oxygen atom. In comparison, the B factor is 14.3 Å2 for the iron. The experimentally observed bond length value of 1.49 Å is slightly but not significantly longer than the 1.46-Å bond length obtained after refinement with the low-dose data. The long O
O bond length (Fig. 3). The ligand was modeled with an occupancy of 100% and has B factors of 18.5 Å2 for the proximal oxygen and 21.5 Å2 for the distal oxygen atom. In comparison, the B factor is 14.3 Å2 for the iron. The experimentally observed bond length value of 1.49 Å is slightly but not significantly longer than the 1.46-Å bond length obtained after refinement with the low-dose data. The long O O bond length indicates that reduction of compound III has occurred, because a shorter bond length would be expected for this species. For instance, an O
O bond length indicates that reduction of compound III has occurred, because a shorter bond length would be expected for this species. For instance, an O O bond length of 1.3 Å was reported for the compound III crystal structure of HRP (29). The long O
O bond length of 1.3 Å was reported for the compound III crystal structure of HRP (29). The long O O bond observed here is strong evidence for the generation of compound 0, formed through protonation of the peroxo-anion species. This experimental value is in agreement with theoretical calculations, which gave values of 1.46 Å (33) and 1.52 Å (34) for the compound 0 intermediates of heme-thiolate model compounds. In contrast, a distinctly shorter bond length of 1.32 Å was calculated for the corresponding peroxo-anion species (33, 35). The bond length between the proximal oxygen and iron is 1.9 Å, and the bond length between iron and sulfur is 2.4 Å. The hydroperoxo group binds in an end-on fashion with a Fe–O–O angle of 131° (Fig. 3).
O bond observed here is strong evidence for the generation of compound 0, formed through protonation of the peroxo-anion species. This experimental value is in agreement with theoretical calculations, which gave values of 1.46 Å (33) and 1.52 Å (34) for the compound 0 intermediates of heme-thiolate model compounds. In contrast, a distinctly shorter bond length of 1.32 Å was calculated for the corresponding peroxo-anion species (33, 35). The bond length between the proximal oxygen and iron is 1.9 Å, and the bond length between iron and sulfur is 2.4 Å. The hydroperoxo group binds in an end-on fashion with a Fe–O–O angle of 131° (Fig. 3).
The following conclusions can be drawn from the structure for the mechanism of compound I formation. The distance of the distal oxygen atom to the carboxyl group of Glu-183 is 2.7 Å, which permits protonation of compound 0 by the protonated carboxyl group of Glu-183 and facilitates the formation of compound I (Fig. 1). In contrast, the distance between Glu-183 and the proximal oxygen is too long (3.5 Å) for a hydrogen bond, preventing proton transfer to the proximal oxygen that would reform the natural substrate hydrogen peroxide.
In this study we obtained compound 0 through reduction of the CPO oxygen complex and subsequent protonation of the formed peroxo-anion intermediate mimicking the cytochrome P450 reaction pathway. The peroxo-anion species is a powerful base (36, 37) and either could be directly protonated by Glu-183 or could abstract a proton from a water molecule in the active site. However, independent of how compound 0 was generated, the second proton that is required for compound I formation would not be available under our reaction conditions at pH 6, and therefore the enzyme could be trapped as compound 0 intermediate.
The long O O bond length observed in the crystal structure strongly supports compound 0 formation as discussed above. However, the UV-visible absorption spectra recorded from the crystals are not conclusive, because the spectra of both the peroxo-anion and compound 0 species are very similar. Only small differences were observed between both intermediates for P450cam (CYP101) (38). Hence we performed QM/MM calculations to gather further support for compound 0 formation.
O bond length observed in the crystal structure strongly supports compound 0 formation as discussed above. However, the UV-visible absorption spectra recorded from the crystals are not conclusive, because the spectra of both the peroxo-anion and compound 0 species are very similar. Only small differences were observed between both intermediates for P450cam (CYP101) (38). Hence we performed QM/MM calculations to gather further support for compound 0 formation.
QM/MM Energy Profile from the Ferric-Hydroperoxo to the Ferric-Peroxo Complexes.
We wanted to ascertain whether the crystal structure indeed corresponds to compound 0 (Fe-OOH−), or the ferric-peroxo complex (Fe-O22−), or alternatively if the two species could coexist in respective energy minima. We therefore analyzed these intermediates with QM/MM calculations. The CPO compound 0 crystal structure presented here was used as a starting point for calculations, after the addition of hydrogens and water molecules as described in supporting information (SI) Methodology.
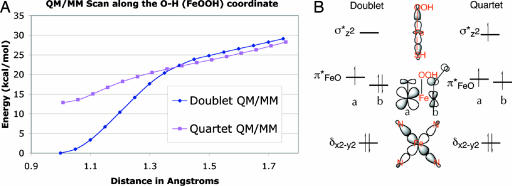
The Fe-OOH− complex had been previously investigated for cytochrome P450 and was shown to have two low lying spin states, a doublet ground state, and a quartet excited state (35). Geometry optimization for these two states led to the minimized energy structures labeled A (Fe-OOH− doublet state) and B (Fe-OOH− quartet state) in Figs. 4 and 5. Both A and B are genuine minima. Starting from these structures, we calculated the respective energy profiles in which the proton on the OOH− group is transferred back to Glu-183. The energy profile scans led to the corresponding spin states of the ferric-peroxo Fe-O22− complexes, labeled in Fig. 5 with C and D, which are associated with the A and B structures of compound 0, respectively. Importantly, neither C nor D are genuine minima, as shown in Fig. 4A. Both collapse in a barrier-free fashion to the respective compound 0 (Fe-OOH−) species A and B. The collapsing can be explained by the fact that the Fe-O22− species is a very strong base (36, 37), whereas Glu-183 is a moderately strong acid. Therefore, it is expected that the Fe-O22− species will not be stable in the CPO environment unless Glu-183 is deprotonated and will collapse instantaneously to Fe-OOH−. In conclusion, the QM/MM calculations indicate that the only existing species are the Fe-OOH− complexes in the spin states A and B, with A being predominant because it is the lowest energy species.
Fig. 4.
Characterization of the doublet and quartet states. (A) QM/MM (UB3LYP/B1:CHARMM) potential energy surfaces between the Fe-OOH− and Fe-O22- states. (B) The electronic configurations of the doublet and quartet states of Fe-OOH−.
Fig. 5.
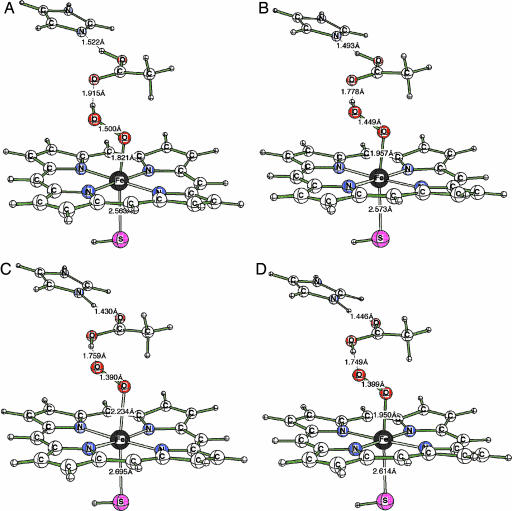
QM/MM-optimized structures corresponding to CPO compound 0 in the doublet (A) and quartet (B) spin states and of Fe-O22− species C (doublet) and D (quartet spin state) identified during the energy profile shown in Fig. 4A.
Structural information of all four species is displayed in Fig. 5. Inspection of the ferric-hydroperoxo species shows that in the doublet state (A) of Fe-OOH− the O O distance is computed to be 1.500 Å, whereas in the quartet state (B) it is 1.449 Å. By contrast, the corresponding O
O distance is computed to be 1.500 Å, whereas in the quartet state (B) it is 1.449 Å. By contrast, the corresponding O O bond distances in the ferric-peroxo structures C and D are significantly shorter (1.390 and 1.399 Å, respectively). In the initial Protein Data Bank structure used for the QM/MM calculations, the bond distance was 1.47 Å. This match in the O
O bond distances in the ferric-peroxo structures C and D are significantly shorter (1.390 and 1.399 Å, respectively). In the initial Protein Data Bank structure used for the QM/MM calculations, the bond distance was 1.47 Å. This match in the O O distances and the instability of the ferric-peroxo structures C and D led to the conclusion that the observed x-ray structure corresponds to the Fe-OOH− species, whereas the Fe-O22− species may not be a genuine minimum of the enzyme.
O distances and the instability of the ferric-peroxo structures C and D led to the conclusion that the observed x-ray structure corresponds to the Fe-OOH− species, whereas the Fe-O22− species may not be a genuine minimum of the enzyme.
The next section analyzes the computed properties of the observed Fe-OOH− species of CPO in comparison to the analogous species in other heme enzymes, e.g., P450 and HRP.
QM/MM-Computed Properties of the compound 0 Species.
SI Table 1 summarizes relative energies, group spin densities, and charges of the ferric-hydroperoxide complex in the two spin states. It is notable that the gap between the doublet and quartet for Fe-OOH− (12.9 kcal/mol) is very similar to the gap determined for P450 (14.5 kcal/mol) (39) and significantly larger than the difference in HRP (6.6 kcal/mol) (40). This finding is in agreement with EPR/electron nuclear double resonance assignment of the Fe-OOH− species in P450 (41). Thus, the fact that the doublet state is the ground state is independent of the specific protein environment. Instead it arises from the interaction of the OOH− fragment with the ferric iron of the heme. The energy difference between the doublet and quartet states is considerably larger when the proximal ligand is a cysteine rather than a histidine, consistent with the “push” effect of sulfur (6). This energy difference indicates that the observed species in the x-ray crystallographic study is the low-spin doublet state compound 0.
The assignment of the species as Fe(III)-OOH− is based on the spin density data shown in SI Table 1 and the orbital diagram in Fig. 4B. It is apparent that in the doublet ground state (A) there is a net single spin localized mainly on the iron. This spin density reflects the electronic structure of the doublet state that involves five electrons in the formally 3d-type orbitals on iron (δx2-y22 π*xz2 π*yz1; Fig. 4B). The singly occupied orbital is primarily a 3dyz atomic-orbital localized on the iron, slightly mixed with the 2py orbital of the proximal oxygen. This electronic structure results in a spin density of almost unity on Fe with a small amount on the OOH− group. Thus the assignment of the compound 0 species as Fe(III)-OOH− reflects the electronic structure and a formal electron count in the oxidation state formalism. The actual charge on iron in compound 0 is +0.5964. As is generally the case, the actual charge is not identical to the formal oxidation state.
In the quartet state, B, the orbital occupation in the d-block is δx2-y22 π*xz1 π*yz1 σ*z21, meaning that there are three unpaired electrons in the d-orbitals of iron (Fig. 4B), which is reflected by its large spin density (3.32) as listed in SI Table 1. One should also notice that there is some delocalization of spin density onto the porphyrin and onto the SH and OOH ligands. This spin density delocalization reflects the fact that the d-orbitals in Fig. 4B are delocalized, e.g., the σ*z2 orbital is delocalized over the dz2 orbital of Fe, the hybrid orbital of the thiolate and the hybrid orbital of the proximal oxygen ligated to iron. Interestingly, the quartet state has longer Fe O and shorter O
O and shorter O O bond distances compared with the doublet state (Fig. 5), which is a reflection of the electronic structures and the excitation of an electron from π*xz to σ*z2; the former orbital involves O
O bond distances compared with the doublet state (Fig. 5), which is a reflection of the electronic structures and the excitation of an electron from π*xz to σ*z2; the former orbital involves O O antibonding interaction and the latter orbital has a strong antibonding Fe
O antibonding interaction and the latter orbital has a strong antibonding Fe O interaction.
O interaction.
To summarize, the observation of compound 0 in CPO bears similarity to the situation in P450 and indicates that in both cases there should exist a heterolytic cleavage mechanism leading to the formation of compound I.
Conclusions
Compound 0 is a common intermediate of many different heme enzymes. The intermediate was formed by first generating the CPO oxygen complex, and then reducing it to the ferric-peroxoanion species through the photoelectrons released during x-ray data collection. Calculations show that the peroxoanion intermediate Fe-O22− will be instantly protonated by Glu-183 to yield compound 0, and that the ground state of this species is a doublet spin state as previously calculated for P450 and HRP.
Materials and Methods
Crystallization and CPO Compound III Formation.
Caldariomyces fumago CPO was expressed and purified as described (42). Crystals were grown by the hanging drop method at 20°C. One microliter of 15 mg/ml CPO in 5 mM sodium acetate buffer, pH 3.8 was mixed with 1 μl of reservoir solution containing 50 mM KBr, 22% PEG 3000, and 0.1 M sodium citrate, pH 3.6. Long rod-like crystals with a typical size of 400 × 50 × 20 μm3 appeared within a few days. All crystal soaks were carried out at 4°C. First, the crystals were transferred from the mother liquor into a solution containing 22% PEG 3000 in 0.1 M citrate, pH 3.6 to remove bromide, and then moved into a cryoprotectant solution containing 100 mM PAA (Fluka, St. Louis, MO), 30 mM hydrogen peroxide (present in the PAA stock solution), 15% ethylene glycol, and 22% PEG 3000 in 0.15 M sodium phosphate, pH 6.0. Crystals changed color from brown to olive green upon addition of PAA containing cryoprotectant. After incubation for ≈30 s, ascorbate containing cryoprotectant (100 mM ascorbate, 15% ethylene glycol, 22% PEG 3000, and 0.15 M sodium phosphate, pH 6.0) was added in a 1:1 ratio. Crystals turned red-brown and were flash-cooled in liquid nitrogen after incubation for 1 min in the cryoprotectant mixture.
Spectroscopy, X-Ray Data Collection, and Structure Determination.
Visible single crystal absorption spectra were recorded from cryo-cooled crystals (90 K) mounted in loops with a microspectrophotometer (4Dxray Systems AB, Uppsala, Sweden) connected with an Andor InstaSpec II Photodiode array and a MCS 500 light source (Zeiss, Thornwood, NY) (56). Diffraction data were collected at beamline X10SA (Swiss Light Source, Paul Scherrer Institute, Villigen, Switzerland) with the crystals being kept at 90 K. A composite data collection strategy was used (29, 43, 44). After indexing using one frame, the crystals were rotated to an optimal orientation to yield the most data for the first collected 10°. Diffraction data were collected from seven crystals by using exposure times of 1 s per degree. The first 10° data wedges collected from each crystal were merged to form a low-dose data set. The 10–30° wedges yielded a medium-dose data set, and the collected 60–90° wedges formed a high-dose data set. The flux of the beam at an energy of 12.3 keV with a 100-μm Al absorber was 1.7 × 1012 photons in an area of 50 × 10 μm2. During collection of the low-dose data set the crystals received a dose of 0.7 MGy, and after the medium-dose data 2.1 MGy were absorbed. At the end of the collection of the high-dose wedge, a dose of 6.4 MGy was absorbed by the crystals, which is well below the dose limit of 30 MGy (45). Dose calculations were done with RADDOSE (46).
Data were processed and scaled with the XDS software package (47). The structure was determined by molecular replacement with Amore (48) using the previously published CPO structure (Protein Data Bank ID 1cpo) (8) as a search model. The graphics programs O (49) and Coot (50) were used for manual rebuilding of the model. ArpWarp was used for identification of water molecules (51). Refinement was carried out with Refmac5 (52). The final model was refined to a R factor of 19.7% (Rfree = 21.0%) with good stereochemistry (for statistics see SI Table 2). Figures were prepared with Molscript (53) and Bobscript (54).
QM/MM Calculations.
The QM/MM method used here dissected the enzyme into two subsystems, namely the active site and the rest of the protein. The active site was described by a QM method; in the present case, density functional theory in the unrestricted Kohn-Sham formalism used the B3LYP hybrid functional. The rest of the system was treated by MM using a force field calibrated for proteins. The Chemshell software package (55) was used to perform the QM/MM calculations by integrating the TURBOMOLE package for QM with the DL-POLY program for MM using the CHARMM22 force field. More technical details are given in SI Methodologyi.
Supplementary Material
Acknowledgments
We thank Thorsten Beitlich, Robert L. Shoeman, and Clemens Schulze-Briese for help with the microspectrophotometer setup; Ehmke Pohl and Anuschka Pauluhn for support in setting up the beamline; and Ingrid Vetter for support of the crystallographic software. Diffraction data were collected at beamline X10SA (Swiss Light Source, Paul Scherrer Institute, Villigen, Switzerland). This work was supported by the Peter und Traudl Engelhorn-Stiftung (K.K.), the Deutsche Forschungsgemeinschaft Grant SFB 623 (to I.S.), National Institutes of Health Grant GM57042 (to J.T.), and an Israel Science Foundation grant (to S.S.).
Abbreviations
- CPO
chloroperoxidase
- PAA
peracetic acid
- QM
quantum mechanical
- MM
molecular mechanical.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2J5M).
This article contains supporting information online at www.pnas.org/cgi/content/full/0606285103/DC1.
References
- 1.Sono M, Roach MP, Coulter ED, Dawson JH. Chem Rev. 1996;96:2841–2888. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- 2.Matsunaga I, Shiro Y. Curr Opin Chem Biol. 2004;8:127–132. doi: 10.1016/j.cbpa.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Werck-Reichhart D, Feyereisen R. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-6-reviews3003. REVIEWS3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guengerich FP. Mol Interv. 2003;3:194–204. doi: 10.1124/mi.3.4.194. [DOI] [PubMed] [Google Scholar]
- 5.Hager LP, Morris DR, Brown FS, Eberwein H. J Biol Chem. 1966;241:1769–1777. [PubMed] [Google Scholar]
- 6.Dawson JH, Sono M. Chem Rev. 1987;87:1255–1276. [Google Scholar]
- 7.Griffin B. In: Peroxidases in Chemistry and Biology. Everse J, Everse KE, Grisham MB, editors. Vol II. Boca Raton, FL: CRC; 1991. pp. 85–137. [Google Scholar]
- 8.Sundaramoorthy M, Terner J, Poulos TL. Structure (London) 1995;3:1367–1377. doi: 10.1016/s0969-2126(01)00274-x. [DOI] [PubMed] [Google Scholar]
- 9.Rutter R, Hager LP, Dhonau H, Hendrich M, Valentine M, Debrunner P. Biochemistry. 1984;23:6809–6816. doi: 10.1021/bi00321a082. [DOI] [PubMed] [Google Scholar]
- 10.Hosten CM, Sullivan AM, Palaniappan V, Fitzgerald MM, Terner J. J Biol Chem. 1994;269:13966–13978. [PubMed] [Google Scholar]
- 11.Wagenknecht HA, Woggon WD. Chem Biol. 1997;4:367–372. doi: 10.1016/s1074-5521(97)90127-7. [DOI] [PubMed] [Google Scholar]
- 12.Egawa T, Proshlyakov DA, Miki H, Makino R, Ogura T, Kitagawa T, Ishimura Y. J Biol Inorg Chem. 2001;6:46–54. doi: 10.1007/s007750000181. [DOI] [PubMed] [Google Scholar]
- 13.Green MT, Dawson JH, Gray HB. Science. 2004;304:1653–1656. doi: 10.1126/science.1096897. [DOI] [PubMed] [Google Scholar]
- 14.Stone KL, Behan RK, Green MT. Proc Natl Acad Sci USA. 2005;102:16563–16565. doi: 10.1073/pnas.0507069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SH, Perera R, Hager LP, Dawson JH, Hoffman BM. J Am Chem Soc. 2006;128:5598–5599. doi: 10.1021/ja060776l. [DOI] [PubMed] [Google Scholar]
- 16.Makris TM, Denisov IG, Schlichting I, Sligar SG. In: Cytochrome P450. Ortiz de Montanello PR, editor. New York: Plenum; 2005. pp. 149–170. [Google Scholar]
- 17.Primus JL, Grunenwald S, Hagedoorn PL, Albrecht-Gary AM, Mandon D, Veeger C. J Am Chem Soc. 2002;124:1214–1221. doi: 10.1021/ja016907u. [DOI] [PubMed] [Google Scholar]
- 18.Baek HK, Van Wart HE. J Am Chem Soc. 1992;114:718–725. [Google Scholar]
- 19.Davydov R, Macdonald IDG, Makris TM, Sligar SG, Hoffman BM. J Am Chem Soc. 1999;121:10654–10655. [Google Scholar]
- 20.Davydov RM, Yoshida T, Ikeda-Saito M, Hoffman BM. J Am Chem Soc. 1999;121:10656–10657. [Google Scholar]
- 21.Denisov IG, Makris TM, Sligar SG. J Biol Chem. 2001;276:11648–11652. doi: 10.1074/jbc.M010219200. [DOI] [PubMed] [Google Scholar]
- 22.Denisov IG, Makris TM, Sligar SG. J Biol Chem. 2002;277:42706–42710. doi: 10.1074/jbc.M207949200. [DOI] [PubMed] [Google Scholar]
- 23.Mozumder A, Magee JL. Radiat Res. 1966;28:203–214. [PubMed] [Google Scholar]
- 24.Nave C. Radiat Phys Chem. 1995;45:483–490. [Google Scholar]
- 25.Schlichting I, Berendzen J, Chu K, Stock AM, Maves SA, Benson DE, Sweet RM, Ringe D, Petsko GA, Sligar SG. Science. 2000;287:1615–1622. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- 26.Nakajima R, Yamazaki I, Griffin BW. Biochem Biophys Res Commun. 1985;128:1–6. doi: 10.1016/0006-291x(85)91635-3. [DOI] [PubMed] [Google Scholar]
- 27.Jakopitsch C, Wanasinghe A, Jantschko W, Furtmuller PG, Obinger C. J Biol Chem. 2005;280:9037–9042. doi: 10.1074/jbc.M413317200. [DOI] [PubMed] [Google Scholar]
- 28.Hollenberg PF, Hager LP, Blumberg WE, Peisach J. J Biol Chem. 1980;255:4801–4807. [PubMed] [Google Scholar]
- 29.Berglund GI, Carlsson GH, Smith AT, Szoke H, Henriksen A, Hajdu J. Nature. 2002;417:463–468. doi: 10.1038/417463a. [DOI] [PubMed] [Google Scholar]
- 30.Adam V, Royant A, Niviere V, Molina-Heredia FP, Bourgeois D. Structure (London) 2004;12:1729–1740. doi: 10.1016/j.str.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 31.Sligar SG, Makris TM, Denisov IG. Biochem Biophys Res Commun. 2005;338:346–354. doi: 10.1016/j.bbrc.2005.08.094. [DOI] [PubMed] [Google Scholar]
- 32.Kühnel K, Blankenfeldt W, Terner J, Schlichting I. J Biol Chem. 2006;281:23990–23998. doi: 10.1074/jbc.M603166200. [DOI] [PubMed] [Google Scholar]
- 33.Silaghi-Dumitrescu R, Cooper CE. Dalton Trans. 2005:3477–3482. doi: 10.1039/b505440k. [DOI] [PubMed] [Google Scholar]
- 34.Derat E, Kumar D, Hirao H, Shaik S. J Am Chem Soc. 2006;128:473–484. doi: 10.1021/ja056328f. [DOI] [PubMed] [Google Scholar]
- 35.Shaik S, Kumar D, de Visser SP, Altun A, Thiel W. Chem Rev. 2005;105:2279–2328. doi: 10.1021/cr030722j. [DOI] [PubMed] [Google Scholar]
- 36.Harris DL, Loew GH. J Am Chem Soc. 1998;120:8941–8948. [Google Scholar]
- 37.Kamachi T, Yoshizawa K. J Am Chem Soc. 2003;125:4652–4661. doi: 10.1021/ja0208862. [DOI] [PubMed] [Google Scholar]
- 38.Denisov IG, Makris TM, Sligar SG, Schlichting I. Chem Rev. 2005;105:2253–2277. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 39.Ogliaro F, de Visser SP, Cohen S, Sharma PK, Shaik S. J Am Chem Soc. 2002;124:2806–2817. doi: 10.1021/ja0171963. [DOI] [PubMed] [Google Scholar]
- 40.Derat E, Shaik S. J Phys Chem B Condens Matter Mater Surf Interfaces Biophys. 2006;110:10526–10533. doi: 10.1021/jp055412e. [DOI] [PubMed] [Google Scholar]
- 41.Davydov R, Makris TM, Kofman V, Werst DE, Sligar SG, Hoffman BM. J Am Chem Soc. 2001;123:1403–1415. doi: 10.1021/ja003583l. [DOI] [PubMed] [Google Scholar]
- 42.Blanke SR, Yi S, Hager LP. Biotechnol Lett. 1989;11:769–774. [Google Scholar]
- 43.Fedorov R, Schlichting I, Hartmann E, Domratcheva T, Fuhrmann M, Hegemann P. Biophys J. 2003;84:2474–2482. doi: 10.1016/S0006-3495(03)75052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carugo O, Carugo KD. Trends Biochem Sci. 2005;30:213–219. doi: 10.1016/j.tibs.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 45.Owen RL, Rudino-Pinera E, Garman EF. Proc Natl Acad Sci USA. 2006;103:4912–4917. doi: 10.1073/pnas.0600973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murray JW, Rudino-Pinera E, Owen RL, Grininger M, Ravelli RB, Garman EF. J Synchrotron Radiat. 2005;12:268–275. doi: 10.1107/S0909049505003262. [DOI] [PubMed] [Google Scholar]
- 47.Kabsch W. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 48.Navaza J. Acta Crystallogr D. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- 49.Jones TA, Zou JY, Cowan SW, Kjeldgaard Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Perrakis A, Morris R, Lamzin VS. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 52.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Acta Crystallogr D. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 53.Kraulis PJ. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 54.Esnouf RM. Acta Crystallogr D. 1999;55:938–940. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]
- 55.Sherwood P, de Vries AH, Guest MF, Schreckenbach G, Catlow CRA, French SA, Sokol AA, Bromley ST, Thiel W, Turner AJ, et al. J Mol Struct Theochem. 2003;632:1–28. [Google Scholar]
- 56.Beitlich T, Kühnel K, Schulze-Briese C, Shoeman RL, Schlichting I. J Synchrotron Radiat. 2007;14:11–23. doi: 10.1107/S0909049506049806. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.