Abstract
Malignant rhabdoid tumor (MRT) is an aggressive, highly lethal cancer of young children. Tumors occur in various locations, including kidney, brain, and soft tissues. Despite intensive therapy, 80% of affected children die, often within 1 year of diagnosis. The majority of MRT samples and cell lines have sustained biallelic inactivating mutations of the hSNF5 (integrase interactor 1) gene, suggesting that hSNF5 may act as a tumor suppressor. We sought to examine the role of Snf5 in development and cancer in a murine model. Here we report that Snf5 is widely expressed during embryogenesis with focal areas of high-level expression in the mandibular portion of the first branchial arch and central nervous system. Homozygous knockout of Snf5 results in embryonic lethality by embryonic day 7, whereas heterozygous mice are born at the expected frequency and appear normal. However, beginning as early as 5 weeks of age, heterozygous mice develop tumors consistent with MRT. The majority of tumors arise in soft tissues derived from the first branchial arch. Our findings constitute persuasive genetic evidence that Snf5, a core member of the Swi/Snf chromatin-remodeling complex, functions as a tumor suppressor gene, and, moreover, Snf5 heterozygotes provide a murine model of this lethal pediatric cancer.
Chromatin structure plays a critical role in eukaryotic gene transcription (1). Multiprotein complexes modify and remodel nucleosomes and thereby exert regulatory effects on gene expression. These complexes can be assigned to two major classes, those that covalently modify core histones and others that use the energy of ATP hydrolysis to enzymatically alter nucleosome conformation or location. Examples of the first class include the histone acetylase and deacetylase complexes, which add and remove acetyl groups from the four core histones and generally, but not always, result in relaxation and constriction of chromatin respectively. The second class consists of large, multisubunit ATPase-containing chromatin-remodeling machines. These complexes use either a Snf2 or ISWI-related ATPase partnered with varying subunits to achieve alteration in nucleosome position or spacing. Notably, these complexes play roles in both gene activation and silencing.
The most intensively studied remodeling machine, Swi/Snf, was originally identified in yeast as a 2-MDa complex that is highly conserved in all eukaryotes (2–5). The individual components had previously been identified in screens for genes controlling mating type switching (Swi) and the ability to use sucrose as a food source (sucrose nonfermenting or SNF; refs. 6 and 7). Evidence that Swi/Snf was involved in chromatin remodeling emerged when genetic suppressors of Swi/Snf mutations were found to encode histones and other chromatin components (8, 9). Unlike the closely related RSC chromatin-remodeling complex, which is present in 10-fold excess of Swi/Snf and is required for cell viability, Swi/Snf is nonessential and is required for the expression of only ≈5% of genes in yeast (10, 11). Swi/Snf-mediated control of transcriptional regulation is exerted at the level of individual promoters rather than larger chromosomal domains and is involved in both transcriptional activation and repression (11). Although data are emerging for the targets of transcriptional control in yeast, the individual genes and classes of genes that are controlled by Swi/Snf in mammalian cells remain unknown.
Clues to the function of Snf5, a core subunit of Swi/Snf complexes, have arisen from diverse areas of investigation. Early on, Snf5 was shown to be a transcriptional activator and to antagonize repression mediated by nucleosomes at the SUC2 locus in yeast (8). In humans, hSNF5 was independently isolated in a yeast two-hybrid screen performed to identify binding targets of the integrase of the HIV (12). hSNF5 was shown to specifically bind to HIV integrase and stimulate integration of HIV into the genome. Human Snf5 was thus given the name integrase interactor 1. Deletion of Snf5 in yeast abolishes the lethal phenotype induced by expression of HIV-1 integrase (13). Snf5 has also been found to interact with other DNA-binding proteins. The Drosophila Trithorax (Trx) gene and the human counterpart MLL (ALL-1) activate the expression of homeotic genes by establishing areas of open chromatin. Both Trx and MLL physically interact with Snf5 (14). The Epstein–Barr virus transcriptional transactivator Epstein–Barr virus-encoded nuclear antigen-2, when phosphorylated, also binds to Snf5 (15). Furthermore, c-myc was shown to interact with Snf5 and to require the Swi/Snf complex for its transactivation function (16).
Further insight into potential functions of hSNF5 was gained with the discovery of biallelic inactivating mutations of hSNF5 in the majority of malignant rhabdoid tumors (MRTs; refs. 17 and 18). According to one report, approximately 14% of children with MRTs have constitutional mutations in one allele and inactivation of the second allele in their tumor (18). In several instances, patients with constitutional mutations have developed two independent primary tumors (19–21). Moreover, families have been identified in which members share a constitutional hSNF5 mutation and develop tumors, a condition termed the rhabdoid predisposition syndrome (19, 22). These findings implicated an ATPase-containing chromatin remodeling complex in the genesis of cancer. In this study, we sought to characterize the in vivo requirements of Snf5/integrase interactor 1 in mammalian development and to investigate its role in oncogenesis. We report that Snf5 is widely expressed in the embryo, with several areas of particularly high expression. Absence of Snf5 results in early embryonic lethality, and haploinsufficiency of Snf5 predisposes mice to the development of MRTs.
Materials and Methods
Cloning of Murine Snf5.
The dBEST murine databank was searched using the human SNF5 sequence. Five overlapping expressed sequence tags were identified that covered the entire ORF. Image clone ID 355345 (GenBank accession no. W48248) contained a 1640-bp insert spanning the entire ORF of murine Snf5. The clone was used to screen a 129-strain murine genomic DNA library, and two nonoverlapping phages were identified. A phage containing exons 1–5 was used to map intron/exon boundaries and subsequently used for generation of the targeting construct.
Northern Analysis.
A murine embryo RNA blot (CLONTECH no. 7763-1) was hybridized with a randomly primed probe generated from the entire ORF of murine Snf5, using the manufacturers hybridization solution, ExpressHyb (CLONTECH). Hybridization was carried out for 1 h at 68°C, and the final wash was in 0.1× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7), 0.1% SDS at 5°C according to the manufacturer's instructions.
In Situ Hybridization.
Whole-mount RNA in situ hybridization was carried out by using a 440-bp PvuII fragment (bases 54–493 relative to the initiation codon) as previously described (23).
Generation of Snf5+/− Mice.
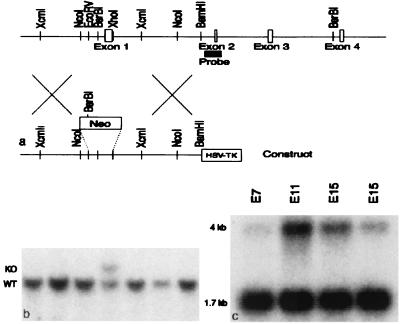
An 8.8-kb region of homology beginning 4.1 kb upstream of exon1 was used to generate the targeting construct in the pSP72 vector. EcoRV and XhoI were used to remove a 1.2-kb fragment containing exon I, which was replaced with a neomycin resistance cassette. An HSV-TK cassette was inserted at the BamHI site at the end of the 3′ homology region. This construct was electroporated into TL1 ES cells (strain 129), and clones were selected in G418 and gancyclovir. Three of 145 clones were correctly targeted and were injected into C57/BL6 blastocysts. Chimeric offspring were bred to C57/BL6 females, and two clones gave rise to germline transmission. The colony has been maintained on a mixed 129 and C57/BL6 background. Tail DNA was analyzed by Southern blotting.
Anti-Snf5 Immunohistochemistry.
An affinity-purified goat polyclonal antibody raised against an N-terminal peptide of Snf5 (identical human and murine sequence) was obtained from Santa Cruz Biotechnology (catalog no. sc-9749). Tumor tissue was fixed overnight in 10% buffered formalin and embedded in paraffin. Sections (4 μm) were cut and deparaffinized, and antigen unmasking was carried out by heating in 10 mM sodium citrate at 95°C for 5 min. Immunoperoxidase staining was then performed with a kit (Santa Cruz Biotechnology no. sc-2053) according to the manufacturer's protocol, with the primary antibody used at a concentration of 5 μg/ml and a developing time of 5 min. Sections were counterstained in hematoxylin for 14 s.
Results
Murine Snf5 Is Highly Conserved and Widely Expressed During Development.
By using the reported sequence of human SNF5 (12), we first searched a murine expressed sequence tag database (dBEST), identified murine Snf5, and determined the entire ORF. The deduced amino acid sequence of murine Snf5 is highly conserved relative to the human protein (Fig. 1). Murine and human Snf5 are identical at 384 of 385 amino acids, displaying a single conserved amino acid change. Northern analysis revealed the presence of Snf5 RNA at all developmental times examined, on embryonic days (E) 7, 11, 15, and 17 (Fig. 3c). Whole-mount RNA in situ hybridization was performed on E8, E9.5, and E11.5 (Fig. 2 a–d). On E8, while overall expression is widespread, it is particularly high in the neural folds, including the head fold. Expression within the heart primordium is weak (Fig. 2a). On E9.5, diffuse expression persists, with intense expression within the neuroepithelium. A high level of expression is also seen in the newly forming first branchial arch, which will give rise to the mandible and surrounding tissues of the face, and in the developing hindlimb bud. Expression is low to absent within the heart (Fig. 2b). On E11.5, expression is widespread and continues more intensely in the neuroepithelium, first branchial arch, and hindlimb (Fig. 2 c and d).
Figure 1.
Alignment of murine and human Snf5.
Figure 3.
Homologous recombination knockout construct and embryonic expression of Snf5. (a) The murine Snf5 genomic locus and targeting construct. (b) Southern blot of embryonic stem cell DNA cut with BsrBI and XhoI and hybridized with the probe. The wild-type band is 11.3 kb, and the targeted band is 12.5 kb. (c) Northern blot of murine whole-embryo RNA hybridized with a probe covering the entire ORF of the cDNA of murine Snf5. Transcripts of 1.7 and 4 kb are detected.
Figure 2.
Snf5 expression during development. Wild-type embryos were harvested at E8 (a), E9.5 (b), and E11.5 (c and d). In situ hybridization was carried out with antisense (a, b, and d) or sense (c) probes to Snf5. Expression is detected throughout the embryos but is particularly intense in the headfolds (black arrows), neural folds (black arrowheads), first branchial arch (white arrow), and hindlimb bud (white arrowheads).
Homozygous Inactivation of Snf5 Results in Early Embryonic Lethality.
To inactivate the Snf5 gene, we used a targeting construct in which exon 1, which contains the initiation codon, was replaced by a neomycin resistance cassette (Fig. 3a). Two independent lines of targeted mice were generated. Heterozygous mice appeared grossly normal. No homozygous mutant pups were born as a result of the interbreeding of heterozygotes. Moreover, no Snf5−/− embryos were present in multiple litters examined at E7.5, indicating that the absence of Snf5 results in embryonic lethality before the onset of gastrulation and organogenesis.
Mice Haploinsufficient for Snf5 Develop Malignant Rhabdoid Tumors.
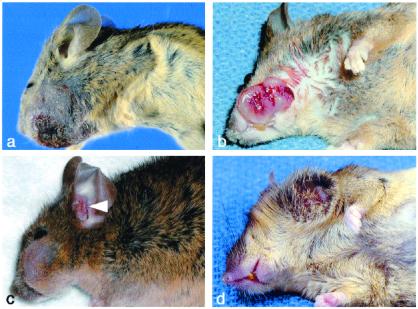
To assess the role of Snf5 as a potential tumor suppressor, we followed cohorts of Snf5+/− and littermate control animals for up to 11.5 months. Snf5+/− mice were born at the expected frequency and were normal in appearance, growth, and fertility (not shown). Commencing as early as 5 weeks of age, tumors appeared in a relatively small fraction of mice. To date, in the cohort of mice older than 8 months, 8 of 125 Snf5+/− animals have developed frank tumor masses, whereas none of the 124 controls have (P < 0.005; Table 1). At the time of this analysis, the colony has a median age of 9 months (range: 2–11.5 months). In seven of eight cases the tumor originated in the soft tissues of the head in structures derived from the first branchial arch (Fig. 4). One of these seven mice presented with neurologic symptoms and pronounced cranial enlargement. Necropsy revealed a tumor arising in the soft tissues of the head that had invaded the skull and caused secondary hemorrhage. The remaining tumor arose on the anterior chest wall. The tumors behave in an aggressive manner, displaying rapid growth, epidermal ulceration, and hemorrhage (Figs. 4 and 5a).
Table 1.
Tumor occurrence
| Mouse no. | Age at death, months | Tumor location: metastases |
|---|---|---|
| 32 | 6.4 | Inferolateral face |
| 229 | 5.4 | External ear; lymph nodes |
| 130 | 5.9 | Inferolateral face; lymph nodes |
| 5 | 7.4 | Lateral face; lung, ear |
| 53 | 8.1 | Lateral face/neck |
| 108 | 7.5 | Anterior chest wall; lymph nodes |
| 341 | 4.9 | Posterolateral face; widespread emboli |
| 396 | 1.2 | Lateral face with intracranial extension |
Figure 4.
Gross appearance of tumors. (a) Mouse 32. (b) Mouse 130. (c) Mouse 5. Two tumors are present: one on the lateral face and a metachronous/metastatic tumor in the external ear canal (arrowhead). (d) Mouse 53.
Figure 5.
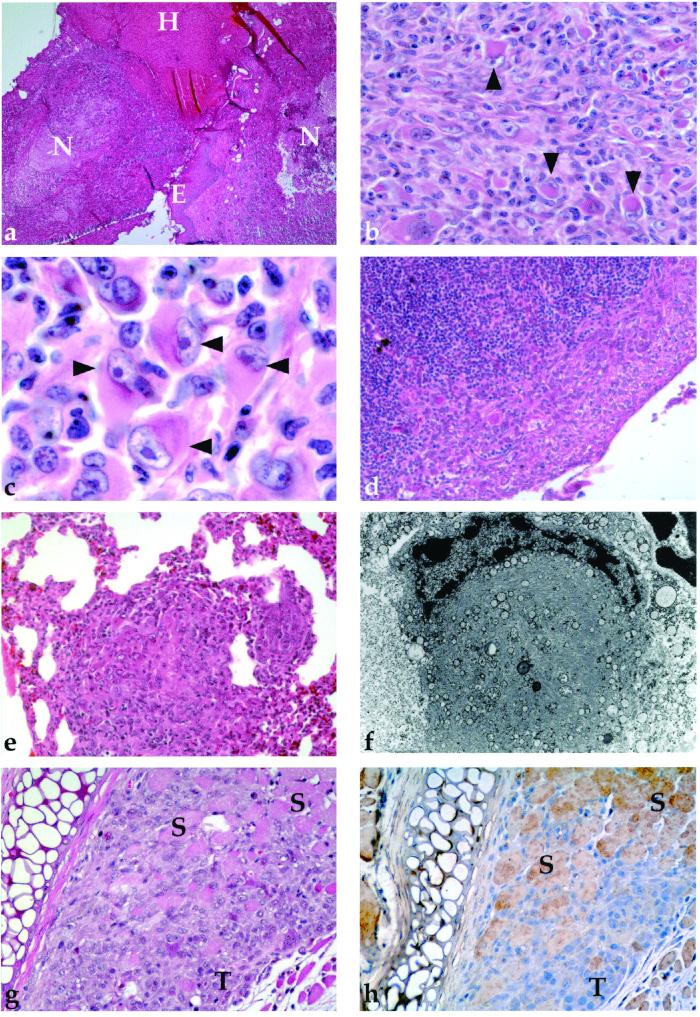
Microscopic appearance of tumors. (a) Low-power view demonstrating areas of necrosis (N), hemorrhage (H), and epithelial invasion (E) (×40). (b and c) Medium-power and high-power views revealing classic rhabdoid cells (arrows) with large eccentric nuclei, prominent nucleoli, and a prominent eosinophilic cytoplasm (×400, ×1,000). (d) Lymph node with subcapsular metastasis (×200). (e) Lung with metastatic tumor (×200). (f) EM of rhabdoid cell demonstrating large cytoplasm containing whorls of intermediate filaments. (g and h) Corresponding sections demonstrating H&E and immunohistochemical staining for Snf5 in tumor infiltrating skeletal muscle. Entrapped eosinophilic muscle fibers (S) are positive for Snf5, whereas the tumor cells (T) are negative (×200).
On routine histological examination all tumors involved subcutaneous tissue and skeletal muscle and presented a consistent appearance. Each was composed of atypical spindle cells admixed with variable numbers (5–20%) of cells with prominent hyaline cytoplasmic (“rhabdoid”) inclusions, vesicular nuclei, and large nucleoli (Fig. 5 b and c). Metastatic disease in regional lymph nodes or lung was present in five of eight cases (Fig. 5 d and e), with one case having widespread tumor emboli in the heart and systemic blood vessels. Tumor cells do not express keratin, epithelial membrane antigen, desmin, or S100 protein, as assessed by immunohistochemistry (not shown).
Ultrastructural examination revealed paranuclear whorls of intermediate filaments in the rhabdoid cells (Fig. 5f). Immunohistochemical staining with an anti-Snf5 antibody demonstrated that the tumors were negative for Snf5 expression (Fig. 5 g and h). The murine tumors described herein are entirely comparable to human MRTs and appear similarly aggressive. To our knowledge, such tumors have not previously been reported to occur in mice.
Discussion
Abrogation of Snf5 in yeast results in slowed growth, altered chromatin structure at target promoters, and inactivation of several metabolic pathways (8). Further studies in yeast revealed that inactivation of the Swi/Snf complex results in altered expression of dozens of transcripts constituting ≈5% of all genes (10, 11). Combined with the large number of genes likely to be regulated by Snf5, our data demonstrating widespread embryonic expression and early embryonic lethality suggest that Snf5 is required for a fundamental growth process in the mammalian embryo.
In conjunction with reports of mutations of Snf5 in children with MRTs, the occurrence of tumors in Snf5+/− mice and the absence of Snf5 protein in tumor cells provides persuasive evidence that Snf5 functions as a tumor suppressor. Further studies are required to determine the mechanism of inactivation of the remaining Snf5 allele. The striking similarity of histopathology in tumors from humans and Snf5+/− mice is particularly noteworthy and suggests that the mechanism by which Snf5 acts as a tumor suppressor is evolutionarily conserved. As in children who have inherited a constitutional mutation in one allele of hSnf5, the latency of tumor development in Snf5+/− mice is likely related to a requirement for spontaneous inactivation of the remaining Snf5 allele within a susceptible cell type.
Extensive debate has occurred regarding the nature and classification of MRTs (24). The first cases were described as arising in the kidneys of infants and containing cells that resemble rhabdomyoblasts (“rhabdoid cells”), despite the lack of evidence of true skeletal muscle differentiation (25). Tumors with an identical histologic and ultrastructural appearance also occur in extrarenal sites, including soft tissues, brain, meninges, and visceral organs. Significant disagreement has arisen among pathologists over whether extrarenal tumors are truly MRTs or, rather, histologic variants of other tumor types (24). Part of the difficulty stems from the fact that non-MRT tumors can occasionally display rhabdoid cytomorphology. Consequently, despite the use of a combination of histologic, immunohistochemical, and ultrastructural features, errors in the diagnosis of MRT are not infrequent (24, 26). Now, however, the occurrence of Snf5 mutations in both renal and extrarenal MRTs, in concert with our data, indicates that this formerly controversial and difficult to define tumor type may now be more reproducibly and meaningfully defined by its genotype.
Given that Snf5 protein is known to directly bind to and activate oncogenic transcription factors such as c-myc and MLL (14,16), Snf5 now emerges as a critical regulator of oncogenesis. CBP/p300, MOZ, and MLL are members of complexes that alter DNA structure through covalent modification of histones, and each has been shown to be involved in oncogenesis. Although the retinoblastoma protein also has been reported to interact with hBRM, a Snf2-related ATPase-containing subunit of the SWI/SNF complex, a direct role for hBRM in oncogenesis remains speculative (27). Thus, Snf5, a member of an ATPase-containing chromatin-remodeling complex, is directly implicated in cancer formation. Given the close resemblance between the murine and human tumors and their shared genetic basis, Snf5+/− mice are likely to be an excellent model of the human MRT. These mice can be used both to test novel therapies for this lethal pediatric cancer as well as to investigate the molecular basis of tumor suppression by a chromatin remodeling complex.
Acknowledgments
We thank E. S. Meade and M. M. Leroux for expert technical assistance, M. Fleming for expert guidance in systematic necropsy, and Y. Fujiwara for expert assistance in maintenance of the animal colony. C.W.M.R. is supported by a Physician-Scientist Award from the Howard Hughes Medical Institute. S.H.O. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- MRT
malignant rhabdoid tumor
- Snf
SNF, sucrose nonfermenting
- E
embryonic day
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250492697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250492697
References
- 1.Kingston R E, Narlikar G J. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 2.Peterson C L, Dingwall A, Scott M P. Proc Natl Acad Sci USA. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cote J, Quinn J, Workman J L, Peterson C L. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 4.Cairns B R, Kim Y-J, Sayre M H, Laurent B C, Kornberg R D. Proc Natl Acad Sci USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sudarsanam P, Winston F. Trends Genet. 2000;16:345–351. doi: 10.1016/s0168-9525(00)02060-6. [DOI] [PubMed] [Google Scholar]
- 6.Neigeborn L, Carlson M. Genetics. 1984;108:741–753. doi: 10.1093/genetics/112.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stern M, Jensen R E, Herskowitz I. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 8.Hirschhorn J N, Brown S A, Clark C D, Winston F. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 9.Winston F, Carlson M. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 10.Holstege F C P, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 11.Sudarsanam P, Iyer V R, Brown P O, Winston F. Proc Natl Acad Sci USA. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 13.Parissi V, Caumont A, Richard de Soultrait V, Dupont C-H, Pichuantes S, Litvak S. Gene. 2000;247:129–136. doi: 10.1016/s0378-1119(00)00108-6. [DOI] [PubMed] [Google Scholar]
- 14.Rozenblatt-Rosen O, Rozovskaia T, Burakov D, Sedkov Y, Tillib S, Blechman J, Nakamura T, Croce C M, Mazo A, Canaani E. Proc Natl Acad Sci USA. 1998;95:4152–4157. doi: 10.1073/pnas.95.8.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu D Y, Kalpana G V, Goff S P, Schubach W H. J Virol. 1996;70:6020–6028. doi: 10.1128/jvi.70.9.6020-6028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng S W, Davies K P, Yung E, Beltran R J, Yu J, Kalpana G V. Nat Genet. 1999;22:102–105. doi: 10.1038/8811. [DOI] [PubMed] [Google Scholar]
- 17.Versteege I, Sevenet N, Lange J, Rousseau-Merck M-F, Ambros P, Handgretinger R, Aurias A, Delattre O. Nature (London) 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 18.Biegel J A, Zhou J-Y, Rorke L B, Stenstrom C, Wainwright L M, Fogelgren B. Cancer Res. 1999;59:74–79. [PubMed] [Google Scholar]
- 19.Sevenet N, Sheridan E, Amram D, Schneider P, Handgretinger R, Delattre O. Am J Hum Genet. 1999;65:1342–1348. doi: 10.1086/302639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salva J, Chen T T-Y, Schneider N R, Timmons C F, Delattre O, Tomlinson G E. J Natl Cancer Inst. 2000;92:648–650. doi: 10.1093/jnci/92.8.648. [DOI] [PubMed] [Google Scholar]
- 21.Biegel J A, Fogelgren B, Wainwright L M, Zhou J-Y, Bevan H, Rorke L B. Genes Chromosomes Cancer. 2000;28:31–37. doi: 10.1002/(sici)1098-2264(200005)28:1<31::aid-gcc4>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 22.Taylor M D, Gokgoz N, Andrulis I L, Mainprize T G, Drake J M, Rutka J T. Am J Hum Genet. 2000;66:1403–1406. doi: 10.1086/302833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkinson D G. In Situ Hybridization. Oxford: IRL Press; 1992. [Google Scholar]
- 24.Ogino S, Ro J Y, Redline R W. Adv Anat Pathol. 2000;7:181–190. doi: 10.1097/00125480-200007030-00007. [DOI] [PubMed] [Google Scholar]
- 25.Weeks D A, Beckwith J B, Mierau G W, Luckey D W. Am J Surg Pathol. 1989;13:439–458. [PubMed] [Google Scholar]
- 26.Biegel J A, Fogelgren B, Zhou J Y, James C D, Janss A J, Allen J C, Zagzag D, Raffel C, Rorke L B. Clin Cancer Res. 2000;6:2759–2763. [PubMed] [Google Scholar]
- 27.Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. Proc Natl Acad Sci USA. 1997;94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]