Abstract
Pseudomonas aeruginosa, an important cause of opportunistic infections in humans, delivers bacterial cytotoxins by type III secretion directly into the host cell cytoplasm, resulting in disruption of host cell signaling and host innate immunity. However, little is known about the fate of the toxins themselves following injection into the host cytosol. Here, we show by both in vitro and in vivo studies that the host ubiquitin ligase Cbl-b interacts with the type III–secreted effector exotoxin T (ExoT) and plays a key role in vivo in limiting bacterial dissemination mediated by ExoT. We demonstrate that, following polyubiquitination, ExoT undergoes regulated proteasomal degradation in the host cell cytosol. ExoT interacts with the E3 ubiquitin ligase Cbl-b and Crk, the substrate for the ExoT ADP ribosyltransferase (ADPRT) domain. The efficiency of degradation is dependent upon the activity of the ADPRT domain. In mouse models of acute pneumonia and systemic infection, Cbl-b is specifically required to limit the dissemination of ExoT-producing bacteria whereas c-Cbl plays no detectable role. To the best of our knowledge, this represents the first identification of a mammalian gene product that is specifically required for in vivo resistance to disease mediated by a type III–secreted effector.
Introduction
Pseudomonas aeruginosa is an important opportunistic human pathogen and is one of the leading causes of nosocomial infections, particularly in the settings of epithelial cell injury and/or immunocompromise (1). It can also chronically colonize the lungs of patients with cystic fibrosis, leading to persistent infections and lung damage (2). A key virulence determinant of P. aeruginosa is the type III secretion system, a highly conserved contact-dependent secretion system that delivers bacterial effectors directly into the host cell cytosol (1). The type III secretion system has been shown to contribute to P. aeruginosa virulence in both animal and plant models of disease (1). Additionally, several studies have shown a correlation between the presence of a functional type III secretion system and poor clinical outcomes in patients with acute P. aeruginosa infections (3, 4).
Exotoxins U, Y, S, and T are the 4 type III–secreted proteins that have been identified in P. aeruginosa, though almost no strain encodes all 4 of them (5). Exotoxin S (ExoS) and exotoxin T (ExoT) are 76% identical and encode an N terminal GTPase-activating protein (GAP) domain and a C terminal ADP ribosyltransferase (ADPRT) domain. The GAP domains of ExoS and ExoT both target the small GTPases Rho, Rac, and Cdc 42 (6, 7). In contrast, the substrate specificity of the ExoS and ExoT C terminal ADPRT domains are distinct. The ExoS ADPRT domain targets cytoskeletal regulators, such as Ras, Ral, several Rabs, Cdc42, ERM family proteins, and ExoS itself (6, 8). The ExoT ADPRT domain interacts with Crk family proteins and phosphoglycerate kinase (9, 10). Together, ExoS and ExoT modulate many processes that involve the actin cytoskeleton. These modulations include disruption of cell-cell and cell-substrate contacts that are key to maintaining integrity of the mucosal barrier, inhibition of wound repair by affecting cell migration and cell proliferation, disruption of signaling pathways, particularly those that regulate the innate immune response, and modulation of bacterial internalization by epithelial cells (6). ExoT is thought to be particularly important in the pathogenesis of P. aeruginosa infections since all strains encode this type III effector and isogenic mutants lacking ExoT showed reduced virulence in several models of disease, notably in the context of dissemination of bacteria to distant sites (5, 11–13).
The innate immune response plays a critical role in controlling acute P. aeruginosa (14). Recent reports have begun to identify the ubiquitin-proteasome system as an essential regulator of host response to pathogen attack (15–17). This proteolytic system selectively terminates protein function by targeted protein degradation following ubiquitination (18). Most cells express a single E1 or ubiquitin–activating enzyme, a limited number of E2s or ubiquitin–conjugating enzymes, and many E3 ubiquitin ligases, which carry out the key roles of selecting the specific target and regulating the site and number of ubiquitin residues added (19). Proteins modified by the attachment of a polyubiquitin chain consisting of at least 4 ubiquitins covalently linked at lysine 48 are targeted for degradation by the 26S proteasome (19).
In this paper, we provide what we believe is the first example of an animal host gene that is required for the in vivo resistance to disease mediated by a type III–secreted effector. Specifically, we show that the adaptor molecule and E3 ubiquitin ligase Cbl-b modulates the stability of ExoT by targeted proteasomal degradation and limits the dissemination of P. aeruginosa in murine models of infection. These results demonstrate a previously unappreciated role for Cbl-b in early host defense response.
Results
ExoT undergoes proteasome-dependent degradation in host cells.
The major identified substrates of ExoT ADPRT domain are Crk I and II, splice variants of the same gene (9, 10). Through their SH2 and SH3 domains, these scaffolding proteins associate with many proteins, including the Cbl family of E3 ubiquitin ligases (20). Recent studies have demonstrated that bacterial proteins can undergo ubiquitination and proteasomal degradation in the host cytosol (21–23). We therefore sought to determined whether ExoT was subject to degradation in the host cell cytosol.
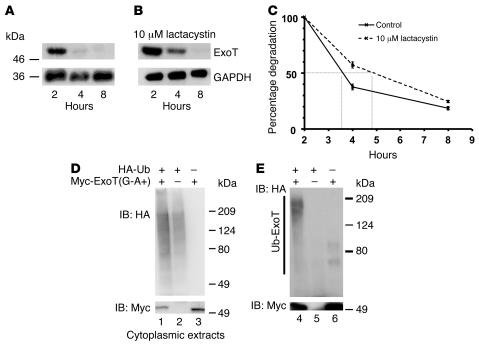
The half-life of translocated ExoT was determined in a pulse-chase experiment. Since the presence of an active GAP domain causes rapid cell rounding and cell loss (24), we performed studies in a strain encoding an inactive GAP domain, hereafter referred to as ExoT(G-A+). Bacteria were cocultivated with HeLa cells for 1.5 hours (time 0), gentamicin and chloramphenicol were added (to kill extracellular bacteria and block bacterial protein synthesis, respectively), and host cell lysates prepared at 2, 4, and 8 hours after infection (hpi) were immunoblotted with anti-ExoT Abs. Translocated ExoT was easily detectable at 2 hours after translocation (Figure 1A). At 4 hpi, there was a dramatic drop in the amount of ExoT, and it became undetectable by 8 hpi. The loss of translocated ExoT was not due to cell loss or unequal loading of cytoplasmic lysates, since the levels of the control GAPDH remained constant throughout the time course of the experiment.
Figure 1. ExoT is degraded by proteasomes and is polyubiquitinated.
(A–C) HeLa cells were cocultivated with PA103ΔexoU/exoT(G-A+) for 1.5 hours in the (A) presence or (B) absence of 10 μM lactacystin. Extracellular bacteria were then killed by the addition of amikacin. Protein synthesis of internalized bacteria was inhibited by the addition of chloramphenicol. At the indicated times, cytoplasmic extracts were prepared, and translocated ExoT was assayed by immunoblotting with anti-ExoT antiserum (upper panels, A and B). To control for loading efficiency, the blots were probed with anti-GAPDH mAbs (lower panels, A and B). (C) The rate of degradation of ExoT was quantified by normalizing the amount of ExoT at each time to the corresponding amount of GAPDH. This ratio was plotted as a percentage of the ratio at time 0 (2 hpi). Shown are the means ± SD. In some cases, the error bars are too small to be seen. (D and E) HeLa cells were transfected with the indicated plasmids for 18 hours; 10 μM lactacystin was added, and the cells were further incubated for 4 hours. (D) To assess overall levels of transfected proteins, whole-cell lysates were immunoblotted with anti-HA (upper panel) or anti-Myc (for ExoT) (lower panel). (E) ExoT was immunoprecipitated using anti-ExoT antiserum and immunoblotted with anti-HA (upper panel, ubiquitinated ExoT) or anti-Myc (lower panel, total ExoT). Polyubiquitinated ExoT, indicated by the slower migrating forms of ExoT, is only visible in cells cotransfected with the ExoT and HA-Ub vectors (lane 4, upper portion of E).
We evaluated the role of proteasomes in ExoT degradation using specific inhibitors. Addition of proteasomal inhibitors clasto-lactacystin β-lactone (data not shown), MG132 (data not shown), or lactacystin partially protected ExoT(G-A+) from degradation (Figure 1B) and increased its half-life by approximately 2-fold (Figure 1C) without affecting the stability of GAPDH (Figure 1B). The incomplete protection afforded by lactacystin may have resulted from loss of potency at the later times. Alternatively, ExoT may be subject to degradation by proteasome-independent pathways at these times. However, inhibitors of the lysosomes (E-64 and ammonium chloride), serine and cysteine proteases and cathepsin B (leupeptin), acid proteases (pepstatin), calpain (N-acetyl-L-leucyl-L-leucyl-L-methioninal or LLM), and caspases (N-benzyloxycarbonyl-Val-Ala-Asp(O-Me) fluoromethyl ketone or Z-VAD-FMK) did not have any effect on the rate of degradation of ExoT (data not shown).
ExoT is ubiquitinated in HeLa cells.
Most substrates destined for proteasome degradation are modified by polyubiquitination (18). In preliminary experiments, we were unable to detect ubiquitination of translocated ExoT in HeLa cells (data not shown). We increased the sensitivity of our assay by transiently transfecting HeLa cells with HA-tagged ubiquitin [pcDNA3-(HA-Ub)4] and Myc-tagged ExoT [pCruzMyc-ExoT(G-A+)] and by performing the experiments in the presence of lactacystin. In control experiments, cells were transfected with either pCruzMyc-ExoT(G-A+) and pcDNA3 or pCruzMyc and pcDNA3-(HA-Ub)4. Eighteen hours after transfection, lactacystin was added to inhibit proteasomal activity; cytoplasmic lysates were prepared 4 hours later. Myc-ExoT(G-A+) was immunoprecipitated using polyclonal serum against ExoT and immunoblotted with anti-Myc (to detect total ExoT) and anti-HA (to detect ubiquitinated ExoT). Total cell lysates were immunoblotted with anti-HA or anti-Myc to quantify transfected protein levels (Figure 1D). Following immunoprecipitation, the anti-HA Abs readily detected a laddering pattern characteristic of polyubiquitinated proteins in samples transfected with both Myc-ExoT(G-A+) and HA-Ub but not in cells transfected with vector only (Figure 1E). The faint bands, approximately 70 kDa and approximately 100 kDa, seen in lanes 5 and 6 (Figure 1E) represent nonspecific background bands. Transfection with a vector in which ExoT was cloned in reverse orientation did not show any laddering pattern in the presence of HA-Ub (data not shown). These results indicate that transfected ExoT is polyubiquitinated and support a role for proteasomes in the degradation of ExoT.
ExoT interacts with Crk and with the ubiquitin ligase Cbl-b.
Cbl family proteins have a multidomain organization, including a RING domain with ubiquitin ligase activity (25). The ubiquitin ligase activity of Cbl can result in mono-, multi-, or polyubiquitination of the target with functionally diverse outcomes, including regulation of receptor tyrosine kinase endocytosis, modulation of the actin cytoskeleton, and inhibition of lymphocyte signaling in the adaptive immune response. Phosphorylation of Cbl at tyrosine residues 700 and 774 promotes binding to SH2 domain–containing proteins, including Crk (26). Since the ExoT-ADPRT domain interacts with Crk and Crk interacts with Cbl, we explored the possibility that one or more isoforms of Cbl may be involved in the proteasomal degradation of ExoT.
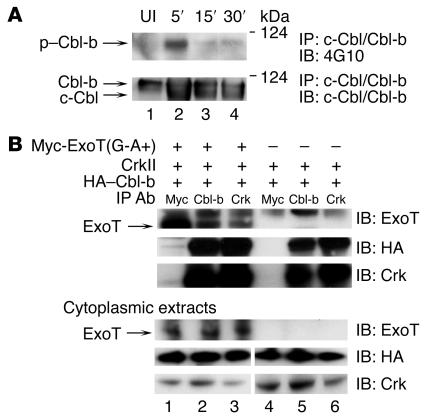
We first determined whether P. aeruginosa infection induced phosphorylation of the widely expressed isoforms c-Cbl and Cbl-b. HeLa cells were infected with bacteria expressing ExoT(G-A+), and host cell lysates were immunoprecipitated with an Ab that recognizes both c-Cbl and Cbl-b (Figure 2A). Rapid phosphorylation of Cbl-b, but not of c-Cbl, was observed within 5 minutes after the addition of the bacteria to HeLa cells (Figure 2A). Cbl-b phosphorylation was also observed upon addition of ExoT-deficient bacteria or bacteria producing ExoT(G+A+) or ExoT(G-A-) (data not shown). While Cbl-b phosphorylation was reproducibly observed within 5 minutes, the rate of dephosphorylation varied among experiments and was independent of ExoT (data not shown). Taken together, these findings suggest that P. aeruginosa rapidly induces phosphorylation of only the Cbl-b isoform independent of and likely prior to translocation of ExoT.
Figure 2. ExoT, Crk, and Cbl-b exist in a complex, and P. aeruginosa induces phosphorylation of Cbl-b.
(A) HeLa cells were coincubated with PA103ΔexoU/exoT(G-A+) for 5, 15, or 30 minutes. Cytoplasmic extracts were immunoprecipitated with anti–c-Cbl/Cbl-b Abs, H-454. Upper panel: lysates were immunoblotted with 4G10 to detect tyrosine-phosphorylated proteins. P. aeruginosa induced tyrosine phosphorylation of the upper band, Cbl-b, but not the lower band, c-Cbl. Lower panel: lysates were immunoblotted with anti–c-Cbl/Cbl-b Abs to detect total Cbl proteins. UI, uninfected. (B) HeLa cells were cotransfected with the indicated plasmids [Myc-ExoT(G-A+) and/or CrkII and HA–Cbl-b] for 18 hours and further incubated for 4 hours with 10 μM lactacystin. Cytoplasmic extracts were prepared, and proteins were immunoprecipitated with anti-Myc, anti–Cbl-b, or anti-Crk Abs. Coimmunoprecipitated ExoT was visualized by immunoblot using anti-ExoT Abs (upper panel). ExoT was coimmunoprecipitated with Cbl-b (lane 2) or CrkII (lane 3). Upper band visible in all the immunoprecipitations represents nonspecific binding to the ExoT Abs. Coimmunoprecipitated Cbl-b and Crk were visualized using anti-HA and anti-Crk Abs (bottom 2 panels). To measure expression of the transfected proteins, the cytoplasmic extracts were immunoblotted using anti-Myc, anti-HA, or anti-Crk Abs (lower 3 panels).
Because of these findings, we tested to determine whether ExoT, Crk, and Cbl-b exist in a complex that can be detected by coimmunoprecipitation. As endogenous levels of Crk and Cbl-b are very low, we cotransfected HeLa cells with Myc-ExoT(G-A+), HA–Cbl-b, and CrkII. Cellular lysates were immunoprecipitated with Abs to Myc (for ExoT), Cbl-b, or Crk and immunoblotted with anti-ExoT. ExoT coimmunoprecipitated with Cbl-b or Crk (Figure 2B). This association was dependent upon transfected ExoT. Immunoblot analyses of lysates of the transfected cells demonstrated that the amount of transfected proteins was similar in all experiments (Figure 2B). Crk coimmunoprecipitated with Cbl-b even in the absence of transfected ExoT (Figure 2B), consistent with previously published data demonstrating that Crk and Cbl-b can form constitutive as well as signal-induced complexes (20). While we were able to efficiently coimmunoprecipitate ExoT with Abs against Cbl-b and Crk, the reverse coimmunoprecipitation with anti-Myc Abs yielded relatively low amounts of Cbl-b and Crk (Figure 2B). Under similar conditions, c-Cbl and ExoT did not coimmunoprecipitate (data not shown). Although these results were obtained under conditions of overexpression, they suggest that ExoT specifically interacts with Crk and Cbl-b in the host cytosol.
Degradation of ExoT is Cbl-b dependent.
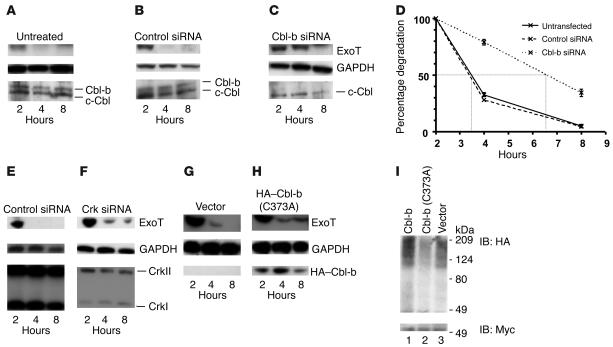
To determine whether Cbl-b modulated ExoT degradation, HeLa cells were depleted of Cbl-b by siRNA and cocultivated with bacteria expressing ExoT(G-A+); stability of the translocated protein was determined. ExoT(G-A+) was rapidly degraded and was undetectable by 4 hpi in the untransfected (Figure 3A) or control siRNA–transfected cells (Figure 3B) while GAPDH levels remained constant (Figure 3, A and B). Depletion of Cbl-b mediated by siRNA specifically resulted in decreased levels of Cbl-b without affecting the levels of c-Cbl (Figure 3C) or GAPDH. Concomitant with the Cbl-b depletion, the half-life of ExoT(G-A+) increased almost 3-fold (Figure 3D), and the protein was now readily detectable at 4 hours after translocation (Figure 3C). A faint band was also seen at 8 hours after translocation. Depletion of c-Cbl did not have any effect on the rate of degradation of ExoT (data not shown).
Figure 3. Role of Crk and Cbl-b in the ubiquitination and degradation of ExoT.
(A–E) HeLa cells were pretreated with (A) no siRNA, (B and E) control siRNA, (C) Cbl-b siRNA, or (F) Crk siRNA for 30 hours prior to infection with PA103ΔexoU/exoT(G-A+). Translocation assays were performed as described for Figure 1. Cytoplasmic extracts were analyzed by immunoblotting using Abs against ExoT (A–C, E, and F; top panels) and loading control GAPDH (A–C, E, and F; middle panels). siRNA knockdown efficiency was determined by immunoblotting against Cbl-b (A–C; bottom panels) or Crk (E and F; bottom panels). (D) Rates of degradation in the presence or absence of Cbl-b siRNA were quantified as described for Figure 1C and plotted as means ± SD. Error bars are too small to be seen at some points. (G–H) HeLa cells were pretreated with (G) vector control pCEFL or (H) pCEFL–HA–Cbl-b (C373A) for 24 hours. Translocation assays were performed as described for Figure 1. Cytoplasmic extracts were analyzed by immunoblotting using Abs against ExoT (top panels) and GAPDH (middle panels). Blots were probed with anti-HA Abs (bottom panels) to verify expression of transfected protein. (I) HeLa cells were transfected with pCruzMycB-ExoT(G-A+), pcDNA3-(HA-Ub)4 and pCEFL, pCEFL–HA–Cbl-b, or pCEFL–HA–Cbl-b (C373A) for 18 hours. Ubiquitination of ExoT was assessed as described for Figure 1D. ExoT was immunoprecipitated using anti-ExoT antiserum under conditions in which ExoT and Cbl-b do not coimmunoprecipitate (in the presence of 0.5% SDS) and immunoblotted with anti-HA (top panel, ubiquitinated ExoT) or anti-Myc (bottom panel, total ExoT).
We investigated the possibility that Crk was required for Cbl-b–mediated degradation of ExoT. CrkI and CrkII were depleted by siRNA (Figure 3F), and the stability of ExoT was measured in the absence of Crk. Crk levels did not change under conditions in which ExoT was degraded (Figure 3E). Crk depletion prolonged the half-life of ExoT(G-A+) without altering the levels of GAPDH (Figure 3F). Together, these results unambiguously demonstrate a role for Cbl-b in the degradation of ExoT and suggest that, by binding to Crk, ExoT is brought in close proximity to Cbl-b. However, it is possible that both Crk and Cbl-b target ExoT for degradation by independent pathways.
Cbl-b ubiquitin ligase activity is required for ExoT ubiquitination and degradation.
Cbl proteins contain a RING domain through which they interact with E2 ubiquitin–conjugating enzymes (27) and which is required for their ubiquitin ligase activity. To directly assess the role of ubiquitin ligase activity in the degradation of ExoT, we employed the Cbl-b (C373A) point mutant, in which a single amino acid change abolishes the ligase activity, resulting in a dominant-negative isoform of Cbl-b (27). HeLa cells were transfected with HA–Cbl-b (C373A) (27) or vector control and cocultivated with P. aeruginosa producing ExoT(G-A+); the half-life of translocated ExoT(G-A+) was then quantified. In cells transfected with vector only, ExoT(G-A+) rapidly disappeared while GAPDH levels remained constant (Figure 3G). However, in the presence of HA–Cbl-b (C373A) (Figure 3H), the half-life of ExoT(G-A+) was significantly increased without any alterations in the levels of GAPDH. Overexpression of Cbl-b increased the rate of degradation of ExoT(G-A+) whereas overexpression of the c-Cbl ubiquitin ligase mutant, c-Cbl (C351A), had no effect (data not shown).
To assess the requirement of Cbl-b ubiquitin ligase activity on the ubiquitination of ExoT, we cotransfected HeLa cells with Myc-ExoT(G-A+) and HA-Ub along with Cbl-b, Cbl-b (C373A), or vector alone and assayed for the amount of ubiquitinated ExoT following immunoprecipitation of ExoT (Figure 3I). Immunoblot analyses of lysates of the transfected cells demonstrated that the amount of transfected proteins was similar in all experiments (data not shown). We observed a reduction in the amount of ubiquitinated ExoT in the presence of Cbl-b (C373A) compared with Cbl-b or vector (Figure 3I) transfected cells. We observed a modest increase in ubiquitination of ExoT in samples transfected with Cbl-b (Figure 3I) compared with vector-transfected samples expressing endogenous Cbl-b only (Figure 3I). Together, these data confirm that the ubiquitin ligase activity of Cbl-b is required for the ubiquitination and degradation of ExoT.
The ADPRT domain of ExoT is necessary and sufficient to modulate its degradation.
The above results support a model in which Cbl-b is recruited to ExoT through its binding to Crk. To confirm the requirement for the ExoT-ADPRT domain in this process and to determine whether the binding of a specific substrate or substrate-binding partner to the ADPRT domain contributed to ExoT degradation, we compared the stability of ExoT with the closely related effector ExoS. While ExoT is highly homologous to ExoS, small differences in their ADPRT domains are sufficient for conferring distinct, nonoverlapping substrate specificities (8). Since PA103 does not encode exoS, we introduced exoS(G-A+) onto a plasmid into PA103ΔexoUΔexoT and measured its half-life in pulse-chase experiments using ExoT antiserum (which recognizes ExoS). In contrast with ExoT, translocated ExoS showed minimal degradation over an 8-hour time period (compare Supplemental Figure 1, A and B, top panels; supplemental material available online with this article; doi:10.1172/JCI28792DS1). Consistent with results presented for Figure 1, plasmid-expressed ExoT in PA103ΔexoUΔexoT was also rapidly degraded (data not shown).
To confirm that the ADPRT domain of ExoT was the key determinant of its stability, we fused the inactive GAP domain of ExoT to the wild-type ADPRT domain of ExoS [G-(ExoT)/A+(ExoS)] and expressed it in PA103ΔexoUΔexoT on a plasmid. As shown in Supplemental Figure 1C, the chimeric protein had a half-life similar to that of ExoS, indicating that the ADPRT domain of ExoS was sufficient to protect this translocated protein from degradation. This experiment also ruled out the possibility that the GAP domain of ExoS may protect it from host cell–mediated degradation.
The rate of degradation of ExoT in host cells is influenced by the presence of an active ADPRT domain.
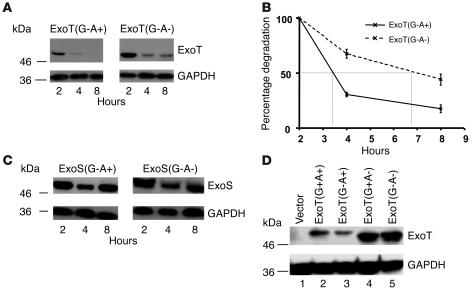
To determine whether the ADPRT activity of ExoT affected its stability, we compared the half-life of ExoT(G-A+) with an ADPRT-inactive form of the protein, ExoT(G-A-). Surprisingly, we found that the amount of ExoT(G-A+) at each time point was less than the amount of ExoT(G-A-) (Figure 4A); indeed, the presence of a catalytically active ADPRT domain decreased the apparent intracellular half-life of ExoT over 3-fold as compared with the inactive ADPRT domain (Figure 4, A and B). In contrast, the levels of GAPDH remained constant throughout the time course of the experiment (Figure 4A). Addition of lactacystin or depletion of Cbl-b or Crk by siRNA had a minor protective effect on ExoT(G-A-) stability, indicating that this form of ExoT was also degraded by the same pathway as ExoT(G-A+), though not as efficiently (data not shown). ExoT(G-A+) was not inherently more susceptible to degradation, since under conditions where bacterial protein synthesis was inhibited by chloramphenicol, the rates of degradation of ExoT(G-A+) and ExoT(G-A-) were indistinguishable in bacterial lysates (data not shown). In contrast, the stability of ExoS was unaffected by the activity of its ADPRT domain (Figure 4C) upon translocation. Both ExoS(G-A+) and ExoS(G-A-) were eventually degraded with similar kinetics, becoming undetectable by 24 hpi (data not shown).
Figure 4. The stability of ExoT in the host cytosol is modulated by the activity of its ADPRT domain.
(A) HeLa cells were cocultivated with PA103ΔexoU/exoT(G-A+) or PA103ΔexoU/exoT(G-A-) for 1.5 hours. Translocation assays were performed as described for Figure 1. Translocated ExoT was quantified by immunoblot analyses using antiserum against ExoT (upper panels). As a control, blots were probed with anti-GAPDH mAbs (lower panels). (B) The rates of degradation were quantified as described for Figure 1C. The ratio obtained at each time point was plotted as a percentage of the ratio at time 0 (2 hpi). Shown are means ± SD. In some cases, error bars are too small to be seen. (C) HeLa cells were cocultivated with PA103ΔexoUΔexoT + pUCP20ExoS(G-A+) or PA103ΔexoU/ΔexoT + pUCP20ExoS(G-A-), and translocation assays were performed as previously described. (D) HeLa cells were transfected with p-IRES–ExoT or the indicated ExoT mutants. Eighteen hours after transfection, cells were lysed and steady-state levels of ExoT quantified by immunoblotting with anti-ExoT (upper panel). Gels were immunoblotted with anti-GAPDH as a control (lower panel). As reported earlier, ADPRT-deficient variants of ExoT migrate with a slower mobility than ADPRT+ forms of ExoT (24).
To determine whether the ADPRT domain was sufficient to target ExoT for degradation and to rule out the contribution of the GAP domain, we compared the stability of wild-type ExoT(G+A+) with that of ExoT(G+A-), ExoT(G-A+), and ExoT(G-A-). Since the presence of an active GAP domain causes rapid cell rounding and cell loss during the longer time courses of pulse-chase experiments, we compared the steady state levels of transfected ExoT. As shown in Figure 4D, the levels of ExoT were reproducibly lower for proteins encoding a catalytically active ADPRT domain, ExoT(G+A+) or ExoT(G-A+), as compared with mutants of ExoT encoding an inactive ADPRT domain, ExoT(G+A-) or ExoT(G-A-) (Figure 4D). The levels of the control protein GAPDH were constant throughout the time period (Figure 4D).
Cbl-b is important for limiting ExoT-mediated P.
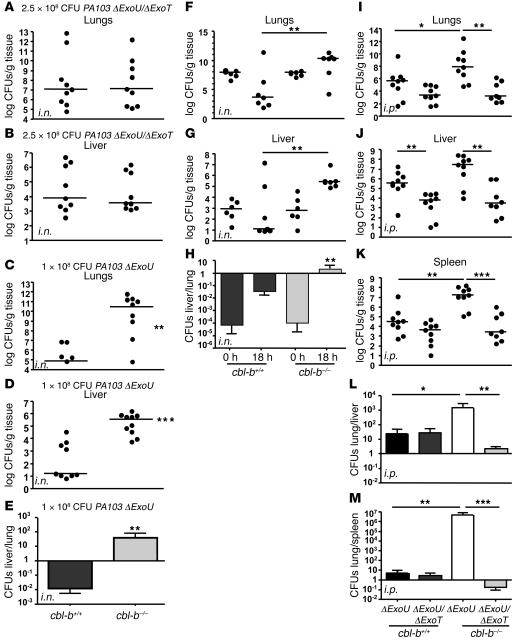
aeruginosa disease. To evaluate the role of Cbl-b in ExoT-mediated P. aeruginosa disease in vivo, we infected wild-type C57BL/6 mice (cbl-b+/+) and Cbl-b–knockout mice (cbl-b–/–) (28) i.n. or i.p. with either PA103ΔexoU or PA103ΔexoUΔexoT and quantified bacterial burden in the lungs and liver (i.n.) and in the lungs, liver, and spleen (i.p.) 18 hpi (11). Consistent with previously published observations (5, 11–13), a higher dose of the ExoT mutant (PA103ΔexoUΔexoT; 2.5 × 108 CFUs) compared to the ExoT-producing bacteria (PA103ΔexoU; 1 × 108 CFUs) was required to reproducibly induce pneumonia (data not shown). Therefore, we limited our comparisons to i.n. infections with the same strain in cbl-b+/+and cbl-b–/– mice.
In the mouse pneumonia model, at 18 hpi, the bacterial burdens in the lungs and liver of PA103ΔexoUΔexoT–infected cbl-b+/+ and cbl-b–/– mice were similar to each other (Figure 5, A and B). A comparison of the medians of the bacterial counts in the lungs (cbl-b+/+ = 1.2 × 107; cbl-b–/– = 1.3 × 107) and liver (cbl-b+/+ = 8.0 × 103; cbl-b–/– = 3.7 × 103) indicated that the observed differences seen were not statistically significant (2-tailed Mann-Whitney U test). In striking contrast, there was a 5-log difference in the number of PA103ΔexoU bacteria, which produces ExoT, recovered from the lungs of cbl-b–/–mice as compared with that from cbl-b+/+mice (Figure 5C; median CFUs for cbl-b+/+ = 7.7 × 104; cbl-b–/– = 2.9 × 1010; P = 0.0041 by the 2-tailed Mann-Whitney U test), implying that the cbl-b–/–mice were more susceptible to infection by PA103ΔexoU.
Figure 5. Cbl-b limits P. aeruginosa infection in an ExoT-dependent manner.
(A–E) C57BL/6 (cbl-b+/+) or C57BL/6 (cbl-b–/–) were infected i.n. with 2.5 × 108 of PA103ΔexoUΔexoT (A and B) or 1 × 108 of PA103ΔexoU (C and D). A higher dose of PA103ΔexoUΔexoT compared with PA103ΔexoU was necessary in order to quantify lung or liver CFUs at 18 hours. At 18 hpi, mice were sacrificed and bacterial CFUs were enumerated from the lung (A and C) and liver (B and D) tissues by plating serial dilutions of tissue homogenates on LB agar plates. (E) The average ± SEM of the liver/lung ratio of bacterial counts in cbl-b+/+ and cbl-b–/–mice was determined for each mouse. (F–G) cbl-b+/+ or cbl-b–/– mice were infected i.n. with 1 × 108 CFUs of PA103ΔexoU. Mice were sacrificed at the indicated times after infection, and bacterial CFUs in the lungs (F) and liver (G) were enumerated. Liver/lung ratio of bacterial counts was determined for each mouse and plotted as an average for cbl-b+/+ and cbl-b–/– mice (H). (I–M) cbl-b+/+ or cbl-b–/– were infected i.p. with 1 × 107 of PA103ΔexoUΔexoT or PA103ΔexoU. At 18 hpi, mice were sacrificed and bacterial CFUs were enumerated from the lungs (I), liver (J), and spleen (K). The lung/liver (L) or lung/spleen (M) ratio of bacterial counts was determined for each mouse and plotted as an average ± SEM for cbl-b+/+ and cbl-b–/–mice. Statistical analyses were performed by the 2-tailed Mann-Whitney U test. *P < 0.05; **P < 0.005; ***P < 0.001.
In parallel with the increased bacterial burden in the lungs of cbl-b–/– mice, we observed larger bacterial loads in the livers of cbl-b–/–mice (Figure 5D; median CFUs for cbl-b+/+ = 16.4; cbl-b–/– = 3.45 × 105; P < 0.0001 by the 2-tailed Mann-Whitney U test). While the absolute number of bacteria recovered from the lungs and livers of mice was variable, increased bacterial dissemination to the liver was consistently overserved in the cbl-b–/–mice as compared to the cbl-b+/+mice. Indeed, a comparison of the ratio of bacterial counts in the liver to lung in each individual mouse indicated that there was greater dissemination of bacteria to distant sites in the cbl-b–/– as compared with the cbl-b+/+ mice (Figure 5E). We also observed increased ascites fluid in these cbl-b–/– mice, consistent with the increased dissemination (data not shown). The number of bacteria recovered from the lungs and livers of c-cbl–/–mice was not significantly different from that of wild-type mice infected with PA103ΔexoU (data not shown), indicating that c-Cbl does not play a role in limiting infection.
To determine whether increased bacterial burden observed in the lungs of c-cbl–/– mice was a result of differences in colonization or a consequence of failure of the host to control the infection, we quantified the lung CFUs at various times after infection with PA103ΔU. Whereas at the early times (0, 4, and 8 hpi), there was no difference in the lung or liver bacterial load between the cbl-b+/+ and cbl-b–/– mice, there was a dramatic increase in the bacterial load in the cbl-b–/–mice between 12 and 18 hpi (data not shown and Figure 5, F and G). Additionally, while the liver/lung ratio of bacterial counts in each mouse at time 0 were roughly the same for both mouse strains, the ratios at 18 hpi indicated that the bacteria disseminated to the liver much more efficiently in the cbl-b–/– than in the cbl-b+/+ mice (Figure 5H). Together, these results establish that initial colonization and replication rates of ExoT-producing P. aeruginosa are similar in cbl-b+/+ and cbl-b–/– mice. However, the cbl-b–/– mice were unable to limit replication of ExoT-producing bacteria as early as 12–18 hpi. These results imply a previously unappreciated role for Cbl-b in limiting bacterial infection.
We confirmed the role of Cbl-b in ExoT-mediated virulence of P. aeruginosa in systemic infection following i.p. injection of bacteria. For these experiments, it was possible to infect mice with the same inoculum of ExoT-producing or ExoT-deficient bacteria. At 18 hpi, the wild-type mice consistently had higher counts of PA103ΔexoU as compared with PA103ΔexoUΔexoT in the lungs (Figure 5I; median CFUs PA103ΔexoU = 5 × 105; PA103ΔexoUΔexoT = 2.5 × 103), liver (Figure 5J; median CFUs PA103ΔexoU = 3.7 × 105; PA103ΔexoUΔexoT = 6.5 × 104), and spleen (Figure 5K; median CFUs PA103ΔexoU = 3.1 × 105; PA103ΔexoUΔexoT = 4.6 × 104) although the difference was significant only in the liver (Figure 5J; P = 0.0019). Remarkably, in the absence of Cbl-b (cbl-b–/–), PA103ΔexoU and PA103ΔexoUΔexoT bacterial counts in the lungs (Figure 5I; median CFUs, PA103ΔexoU = 8.2 × 107; PA103ΔexoUΔexoT = 1.6 × 103 P = 0.0025), liver (Figure 5J; median CFUs PA103ΔexoU = 2.8 × 107; PA103ΔexoUΔexoT = 3.1 × 103 P = 0.0016), and spleen (Figure 5K; median CFUs PA103ΔexoU = 1.7 × 108; PA103ΔexoUΔexoT = 2.6 × 104; P = 0.0010) were all significantly different. A comparison of the ratios of bacterial counts in the lungs/liver and lungs/spleen indicated that in the absence of Cbl-b and presence of ExoT, bacteria disseminated more efficiently from the liver and the spleen to the lungs (Figure 5, L and M). Together, these results demonstrate that Cbl-b modulates ExoT-mediated dissemination in vivo and suggest that Cbl-b is important in modulating the early defense response to ExoT-mediated damage by P. aeruginosa.
Discussion
In this paper, we identify what we believe to be a novel role for the E3 ubiquitin ligase and adaptor molecule Cbl-b in the early defense response and in bacterial pathogenesis. We specifically show that ExoT, a type III–secreted effector and critical virulence factor of P. aeruginosa, is ubiquitinated and undergoes rapid proteasomal degradation in a Cbl-b–dependent manner whereas the closely related effector ExoS is not rapidly degraded. We further demonstrate that the E3 ubiquitin ligase activity of Cbl-b is required for ExoT ubiquitination and degradation and that the rate of degradation is regulated by ExoT ADPRT activity. We find that Cbl-b is tyrosine phosphorylated upon addition of P. aeruginosa and is found in a complex with ExoT and its substrate Crk. Crk appears to be required for the interaction of ExoT and Cbl-b that leads to rapid degradation of ExoT. Our data demonstrate that, in vitro, ExoT has a prolonged half-life in the absence of Cbl-b or Cbl-b ubiquitin ligase activity. These observations highlight Cbl-b as one of the few identified examples of a mammalian gene product that is specifically required to degrade a type III effector (22, 29).
Using 2 different mouse models, acute pneumonia and acute peritonitis, we demonstrate the in vivo relevance of these findings. In both models, Cbl-b was specifically required to limit the dissemination of ExoT-producing bacteria whereas c-Cbl had no effect. We observed higher bacterial loads as well as increased dissemination to distant organs and increased ascites fluid in cbl-b–/– mice infected with PA103ΔexoU compared with wild-type mice. Our findings reinforce the role for ExoT in dissemination of P. aeruginosa in a mouse model of acute pneumonia and demonstrate, for what we believe is the first time, a role for ExoT in the i.p. infection model. The loss of Cbl-b did not appear to have an impact on colonization or spread of ExoT-deficient P. aeruginosa, eliminating the possibility that cbl-b–/– mice are simply more susceptible to microbial infections.
Using cbl-b–/–mice, we provide the first evidence, to our knowledge, that this multifaceted protein is involved in the early defense response against infectious agents: the defect in limiting dissemination of ExoT-producing bacteria was observed as early as 12–18 hpi. These findings are distinct from previous studies that implicated a role for Cbl-b in humoral immunity, specifically in limiting T cell receptor stimulation in response to antigen (25, 28, 30). As Cbl-b is widely expressed (25), it might be predicted to play multiple roles in host defense. For example, by enhancing the degradation of a bacterial toxin, it may help to preserve the integrity of the epithelial or endothelial barrier. Alternatively, it may be required in neutrophil- or macrophage-mediated responses. Experiments to test these possibilities are in progress.
Our data support a model in which ExoT, Crk, and Cbl-b form a complex that leads to ubiquitination of ExoT by Cbl-b. Phosphorylation of Cbl-b occurs rapidly upon addition of P. aeruginosa, within 5 minutes, prior to the translocation of ExoT (which is not detectable under the conditions of our experiments prior to 15 minutes; unpublished data). Phosphorylated Cbl-b binds to Crk SH2 domain. Once translocated, ExoT binds to the Crk–Cbl-b complex. Cbl-b is not a known substrate of ExoT (9, 10) and therefore would not be predicted to bind ExoT directly. As Crk itself does not have any enzymatic activity, we propose that Crk, functioning as a molecular scaffold, recruits Cbl-b to ExoT. Indeed, siRNA depletion of Crk alone is sufficient to increase ExoT stability. In the absence of Cbl-b or Cbl-b ubiquitin ligase activity, ExoT has a longer half-life, thus leading to increased ExoT-mediated damage to host cells and presumably increased bacterial dissemination in vivo.
The formation and activity of the ExoT–Crk–Cbl-b complex may be subject to extensive regulation, at least in response to P. aeruginosa infection. We found that addition of P. aeruginosa increased the phosphorylation of both Crk and Cbl-b, which may have complicated and opposing biological consequences (unpublished data). Additionally, we found that ExoT-ADPRT activity enhances degradation. Changes in the amount of tyrosine phosphorylated Cbl-b or Crk may modulate the interaction of Cbl-b and ExoT. We have preliminary data indicating that ADP-ribosylation by ExoT decreases tyrosine phosphorylation of CrkII (unpublished data). ADP ribosylation of Crk itself also alters the affinity of Crk for some SH2-binding proteins (10). Whether this affects Cbl-b binding to Crk is currently unknown. It is also possible that ExoT(G-A+) and ExoT(G-A-) bind to Crk and/or Cbl-b with different affinities, resulting in their differential susceptibility to degradation.
Our findings that cbl-b–/– mice but not c-cbl–/– mice are uniquely susceptible to ExoT-producing P. aeruginosa, that only Cbl-b is phosphorylated by P. aeruginosa, and that Cbl-b is uniquely involved in ExoT degradation provides extensive new information to support the notion that c-Cbl and Cbl-b, though highly homologous, can have functionally distinct roles. These findings are consistent with reports that c-Cbl and Cbl-b perform different roles in regulating T cell signaling (25, 27, 28, 30, 31). c-Cbl primarily regulates the T cell receptor complex whereas Cbl-b controls T cell signaling through the Rho family guanine exchange factor VAV and Wiskott-Aldrich syndrome protein (WASP), which eventually leads to cytoskeletal remodeling. Understanding how ExoT discriminates between these 2 proteins may shed fundamental insights into their function. One possibility is that bacterial binding and entry lead to Crk and specifically Cbl-b recruitment, consistent with its proposed involvement in regulating components of the actin cytoskeleton during T cell signaling. Indeed, we have preliminary evidence that Cbl-b plays a role in P. aeruginosa entry into cultured epithelial cells (unpublished observations).
Alternatively, the interaction between ExoT and Cbl-b may be a mechanism by which P. aeruginosa temporally regulates the expression of the toxin in the host cell. Indeed, previous studies have demonstrated that, while type III–secreted effectors are necessary for the establishment of an initial acute infection, these functions are lost under conditions of chronic persistence (3, 4, 32, 33). Thus, modulation of ExoT stability by Cbl-b–mediated degradation may facilitate a transition from an acute infection to a chronic or persistent infection.
The differential susceptibility of the closely related toxins ExoT and ExoS to proteasomal degradation adds them to the list of bacterial toxins whose activity is modulated by host-mediated proteasomal degradation (21–23, 34, 35). Interestingly, unlike the Salmonella enterica proteins SopE and SptP, whose N terminal secretion and translocation domains modulate the temporal regulation of their activities through ubiquitination and differential proteasomal degradation (21), the rate of degradation of ExoS and ExoT is regulated by their ADPRT-domain binding partners.
The results described in this paper describe a novel role for Cbl-b in bacterial pathogenesis and suggest that Cbl-b is important in host response to infection. By identifying a host protein that specifically limits bacterial dissemination mediated by a single effector, they highlight how the host develops a fine-tuned mechanism to downregulate or inactivate bacterial toxins. This new information could help in developing strategies directed toward enhancing host defense and/or limiting excessive host response to improve outcome in P. aeruginosa lung infections.
Methods
Plasmids, bacterial strains, and media.
Plasmids and bacterial strains used in this study are described in Supplemental Table 1. To avoid ExoU-mediated cytotoxicity, all experiments were performed in PA103ΔexoU, a mutant that encodes and secretes only ExoT. Bacterial strains were routinely cultured in Luria-Bertani (LB) medium, with antibiotics as needed for cloning purposes.
Translocation assays.
Translocation assays were performed as previously described (24). The half-life of translocated proteins was determined by a pulse-chase experiment. HeLa cells were transferred to MEM at the end of the translocation period (1.5 hours). Amikacin (400 μg/ml; to kill extracellular bacteria) and chloramphenicol (150 μg/ml; to inhibit protein synthesis in all remaining bacteria) were added (time 0). The cells were further incubated at 37°C and 5% CO2. When required, lactacystin, MG-132, clasto-lactacystin β-lactone (10 μM), E-64 (100 μM), N-acetyl-L-leucyl-L-leucyl-L-methioninal or LLM, leupeptin (1 mM), NH4Cl (20 mM; Sigma-Aldrich), or Z-VAD-FMK (100 μM; Sigma-Aldrich) were added during and after translocation. At various times after addition of bacteria, cytoplasmic extracts were prepared by lysing cells in PBS containing 1% Triton X-100 and Complete protease inhibitor cocktail (Roche Diagnostics) for 10 minutes at room temperature. Following centrifugation at 20,800 g for 5 minutes, the supernatants were transferred to a fresh tube containing 4× sample buffer. Blots were probed with anti-ExoT Abs to detect translocated ExoT and with anti-GAPDH mAbs (Chemicon International) as a control for loading efficiency.
Transfection of HeLa cells.
For DNA transfections, HeLa cells were plated to achieve approximately 50%–60% confluence. DNA was transfected using Effectene (Qiagen). For siRNA transfections, HeLa cells were seeded at approximately 1.2 × 105 cells in a 6-well dish. Cells were treated with c-Cbl, Cbl-b, Crk, or control siRNA (Santa Cruz Biotechnology Inc.) for 30 hours. The efficiency of protein depletion was determined by immunoblotting using anti–c-Cbl and anti–Cbl-b Abs (H-454; Santa Cruz Biotechnology Inc.) or anti-Crk Abs (BD Biosciences — Transduction Laboratories). As a control, immunoblots were also performed against GAPDH.
Immunoprecipitation and coimmunoprecipitation.
HeLa cells were transfected with appropriate plasmids for 18 hours. The cells were treated with 10 μM of lactacystin for 4 hours and lysed in lysis buffer (50 mM Tris HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 1 mM EDTA, 1 mM PMSF, 10 mM N-ethylmaleimide, 1 mM sodium orthovanadate, 1 mM okadaic acid, 10 μM lactacystin, and Complete protease inhibitors; Roche Diagnostics). After centrifugation at 20,800 g for 5 minutes to remove cell debris, the supernatants were transferred to fresh tubes. To detect ubiquitinated ExoT, lysis buffer contained 0.5% SDS, used to remove coimmunoprecipitating proteins. Supernatants containing anti-ExoT polyclonal serum preconjugated to Protein G Sepharose TM 4 Fast Flow (GE HealthCare) for 45 minutes at 4°C with gentle rocking. Immunoprecipitates were recovered by centrifugation, washed 3 times in lysis buffer and twice in 50 mM Tris HCl pH7.5, eluted by boiling in ×4 SDS sample buffer, and subjected to SDS-PAGE. Immunoblots were performed with anti-Myc mAbs (Santa Cruz Biotechnology Inc.) to detect total ExoT or anti-HA mAbs (Covance) to detect ubiquitinated ExoT.
To detect phosphorylation of Cbl proteins, HeLa cells were cocultured with bacteria and samples were collected at different times and lysed as described above (without lactacystin). Lysates were immunoprecipitated with anti-c-Cbl/Cbl-b Abs, H-454 (Santa Cruz Biotechnology Inc.). Immunoblots were probed with H454 to detect total c-Cbl/Cbl-b and with anti-phosphotyrosine Abs (4G10; Upstate USA Inc.) to detect phosphorylated Cbl species.
For coimmunoprecipitations, lysates were immunoprecipitated with anti-Myc mAbs, anti–Cbl-b Abs (Santa Cruz Biotechnology Inc.), or anti-CrkII Abs (BD Biosciences — Transduction Laboratories) and immunoblotted with anti-ExoT Abs. Total expression of the transfected proteins was determining by performing immunoblots on cytoplasmic extracts using anti-ExoT, anti-HA, or anti-Crk Abs.
Animal experiments.
Eight- to twelve-week-old C57BL/6 (cbl-b +/+) or Cbl-b–knockout (cbl-b–/– backcrossed at least 7 generations into C57BL/6) mice (28) were infected i.n. with approximately 1 × 108 PA10ΔexoU or 2.5 × 108 PA103ΔexoUΔexoT or PBS or i.p. with 1 × 107 of each bacterial strain. Animals were sacrificed 18 hpi by an overdose of pentobarbital sodium (200 mg/kg i.p.). Lungs, liver, and spleen were harvested and homogenized in PBS, and bacterial counts were determined by plating serial dilutions on LB plates. Statistical analyses were performed using the 2-tailed Mann-Whitney U test (P < 0.05 was considered significant). All experiments used in this study were approved by the University of California, San Francisco Committee on Animal Research.
Supplementary Material
Acknowledgments
We thank M. Matthay, M. Looney, A. Ma, E. Brown, and members of the Engel lab for advice and encouragement. We thank B. Mayer, S. Lipkowitz, T. Ohta, J. Olson, and H. Gu for reagents. This work was supported by grants from the NIH (J.N. Engel and L. Dragone), the Howard Hughes Medical Research Institute (A. Weiss), and the Rosalind Russell Medical Research Center for Arthritis (A. Weiss).
Footnotes
Nonstandard abbreviations used: ADPRT, ADP ribosyltransferase; ExoT, exotoxin T; GAP, GTPase-activating protein; hpi, hours after infection; LB, Luria-Bertani.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:419–427 (2007). doi:10.1172/JCI28792
Lynne Garrity-Ryan’s present address is: Paratek Pharmaceuticals, Boston, Massachusetts, USA.
References
- 1. Engel, J.N. 2003. Molecular pathogenesis of acute Pseudomonas aeruginosa infections. InSevere infections caused by Pseudomonas aeruginosa. A. Hauser and J. Rello, editors. Kluwer Academic/Plenum Press. New York, New York, USA. 201–230. [Google Scholar]
- 2.Lau G.W., Hassett D.J., Britigan B.E. Modulation of lung epithelial functions by Pseudomonas aeruginosa. . Trends Microbiol. 2005;13:389–397. doi: 10.1016/j.tim.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 3.Roy-Burman A., et al. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. . J. Infect. Dis. 2001;183:1767–1774. doi: 10.1086/320737. [DOI] [PubMed] [Google Scholar]
- 4.Hauser A.R., et al. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. . Crit. Care Med. 2002;30:521–528. doi: 10.1097/00003246-200203000-00005. [DOI] [PubMed] [Google Scholar]
- 5.Shaver C.M., Hauser A.R. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. . Infect. Immun. 2004;72:6969–6977. doi: 10.1128/IAI.72.12.6969-6977.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbieri J.T., Sun J. Pseudomonas aeruginosa ExoS and ExoT. . Rev. Physiol. Biochem. Pharmacol. 2004;152:79–92. doi: 10.1007/s10254-004-0031-7. [DOI] [PubMed] [Google Scholar]
- 7.Kazmierczak B., Engel J. Pseudomonas aeruginosa ExoT acts in vivo as a GTPase activating protein for RhoA, Rac1, and Cdc42. . Infect. Immun. 2002;70:2198–2205. doi: 10.1128/IAI.70.4.2198-2205.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun J., Maresso A.W., Kim J.J., Barbieri J.T. How bacterial ADP-ribosylating toxins recognize substrates. Nat. Struct. Mol. Biol. 2004;11:868–876. doi: 10.1038/nsmb818. [DOI] [PubMed] [Google Scholar]
- 9.Sun J., Barbieri J.T. Pseudomonas aeruginosa ExoT ADP-ribosylates CT10 regulator of kinase (Crk) proteins. . J. Biol. Chem. 2003;278:32794–32800. doi: 10.1074/jbc.M304290200. [DOI] [PubMed] [Google Scholar]
- 10.Deng Q., Sun J., Barbieri J.T. Uncoupling Crk-signal transduction by Pseudomonas ExoT. . J. Biol. Chem. 2005;280:35953–35960. doi: 10.1074/jbc.M504901200. [DOI] [PubMed] [Google Scholar]
- 11.Garrity-Ryan L., et al. The arginine finger domain of ExoT is required for actin cytoskeleton disruption and inhibition of internalization of Pseudomonas aeruginosa by epithelial cells and macrophages. . Infect. Immun. 2000;68:7100–7113. doi: 10.1128/iai.68.12.7100-7113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vance R.E., Rietsch A., Mekalanos J.J. Role of the type III secreted exoenzymes S, T, and Y in systemic spread of Pseudomonas aeruginosa PAO1 in vivo. . Infect. Immun. 2005;73:1706–1713. doi: 10.1128/IAI.73.3.1706-1713.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee V.T., Smith R.S., Tummler B., Lory S. Activities of Pseudomonas aeruginosa effectors secreted by the Type III secretion system in vitro and during infection. . Infect. Immun. 2005;73:1695–1705. doi: 10.1128/IAI.73.3.1695-1705.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sadikot R.T., Blackwell T.S., Christman J.W., Prince A.S. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. . Am. J. Respir. Crit. Care Med. 2005;171:1209–1223. doi: 10.1164/rccm.200408-1044SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyer L., Lemichez E. Targeting of host-cell ubiquitin and ubiquitin-like pathways by bacterial factors. Nat. Rev. Microbiol. 2004;2:779–788. doi: 10.1038/nrmicro1005. [DOI] [PubMed] [Google Scholar]
- 16.Perrin A.J., Jiang X., Birmingham C.L., So N.S., Brumell J.H. Recognition of bacteria in the cytosol of Mammalian cells by the ubiquitin system. Curr. Biol. 2004;14:806–811. doi: 10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 17.Khan S., et al. Immunoproteasomes largely replace constitutive proteasomes during an antiviral and antibacterial immune response in the liver. J. Immunol. 2001;167:6859–6868. doi: 10.4049/jimmunol.167.12.6859. [DOI] [PubMed] [Google Scholar]
- 18.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 19.Weissman A.M. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 20.Feller S.M. Crk family adaptors-signalling complex formation and biological roles. Oncogene. 2001;20:6348–6371. doi: 10.1038/sj.onc.1204779. [DOI] [PubMed] [Google Scholar]
- 21.Kubori T., Galan J.E. Temporal regulation of Salmonella virulence effector function by proteasome-dependent protein degradation. . Cell. 2003;115:333–342. doi: 10.1016/s0092-8674(03)00849-3. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y., Higashide W., Dai S., Sherman D.M., Zhou D. Recognition and ubiquitination of Salmonella type III effector SopA by a ubiquitin E3 ligase, HsRMA1. . J. Biol. Chem. 2005;280:38682–38688. doi: 10.1074/jbc.M506309200. [DOI] [PubMed] [Google Scholar]
- 23.Schnupf P., Portnoy D.A., Decatur A.L. Phosphorylation, ubiquitination and degradation of listeriolysin O in mammalian cells: role of the PEST-like sequence. Cell. Microbiol. 2006;8:353–364. doi: 10.1111/j.1462-5822.2005.00631.x. [DOI] [PubMed] [Google Scholar]
- 24.Garrity-Ryan L., et al. The ADP ribosyltransferase domain of Pseudomonas aeruginosa ExoT contributes to its biological activities. . Infect. Immun. 2004;72:546–558. doi: 10.1128/IAI.72.1.546-558.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duan L., Reddi A.L., Ghosh A., Dimri M., Band H. The Cbl family and other ubiquitin ligases: destructive forces in control of antigen receptor signaling. Immunity. 2004;21:7–17. doi: 10.1016/j.immuni.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 26.Standaert M.L., Sajan M.P., Miura A., Bandyopadhyay G., Farese R.V. Requirements for pYXXM motifs in Cbl for binding to the p85 subunit of phosphatidylinositol 3-kinase and Crk, and activation of atypical protein kinase C and glucose transport during insulin action in 3T3/L1 adipocytes. Biochemistry. 2004;43:15494–15502. doi: 10.1021/bi049222q. [DOI] [PubMed] [Google Scholar]
- 27.Davies G.C., et al. Cbl-b interacts with ubiquitinated proteins; differential functions of the UBA domains of c-Cbl and Cbl-b. Oncogene. 2004;23:7104–7115. doi: 10.1038/sj.onc.1207952. [DOI] [PubMed] [Google Scholar]
- 28.Chiang Y.J., et al. Cbl-b regulates the CD28 dependence of T-cell activation. Nature. 2000;403:216–220. doi: 10.1038/35003235. [DOI] [PubMed] [Google Scholar]
- 29.Weinrauch Y., Drujan D., Shapiro S.D., Weiss J., Zychlinsky A. Neutrophil elastase targets virulence factors of enterobacteria. Nature. 2002;417:91–94. doi: 10.1038/417091a. [DOI] [PubMed] [Google Scholar]
- 30.Naramura M., Kole H.K., Hu R.J., Gu H. Altered thymic positive selection and intracellular signals in Cbl-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 1998;95:15547–15552. doi: 10.1073/pnas.95.26.15547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J., Chiang Y.J., Hodes R.J., Siraganian R.P. Inactivation of c-Cbl or Cbl-b differentially affects signaling from the high affinity IgE receptor. J. Immunol. 2004;173:1811–1818. doi: 10.4049/jimmunol.173.3.1811. [DOI] [PubMed] [Google Scholar]
- 32.Goodman A.L., et al. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. . Dev. Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 33.Jain M., et al. Type III secretion phenotypes of Pseudomonas aeruginosa strains change during infection of individuals with cystic fibrosis. . J. Clin. Microbiol. 2004;42:5229–5237. doi: 10.1128/JCM.42.11.5229-5237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcus S.L., Knodler L.A., Finlay B.B. Salmonella enterica serovar Typhimurium effector SigD/SopB is membrane-associated and ubiquitinated inside host cells. . Cell. Microbiol. 2002;4:435–446. doi: 10.1046/j.1462-5822.2002.00202.x. [DOI] [PubMed] [Google Scholar]
- 35.Stirling F.R., Cuzick A., Kelly S.M., Oxley D., Evans T.J.2006Eukaryotic localization, activation and ubiquitinylation of a bacterial type III secreted toxin. Cell. Microbiol. 81294 – 1309 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.