Summary
Retinoic acid (RA) is essential for normal vertebrate development, including the patterning of the central nervous system. During early embryogenesis, RA is made in the trunk mesoderm through the metabolism of vitamin A derived from the maternal diet, and behaves as a morphogen in the developing hindbrain where it specifies nested domains of Hox gene expression. Loss of endogenous sources of RA can be rescued by treatment with a uniform concentration of exogenous RA, indicating that domains of RA responsiveness can be shaped by mechanisms other than simple diffusion of RA from a localized posterior source. Here, we show that the cytochrome p450 enzymes of the Cyp26 class, which metabolize RA into polar derivatives, function redundantly to shape RA-dependent gene expression domains during hindbrain development. In zebrafish embryos depleted of the orthologs of the three mammalian Cyp26 genes, Cyp26a1, b1 and c1, the entire hindbrain expresses RA-responsive genes that are normally restricted to nested domains in the posterior hindbrain. Furthermore, we show that Cyp26 enzymes are essential for exogenous RA to rescue hindbrain patterning in RA-depleted embryos. We present a “gradient-free” model for hindbrain patterning in which differential RA responsiveness along the hindbrain anterior-posterior axis is shaped primarily by the dynamic expression of RA-degrading enzymes.
Keywords: retinoic acid, hindbrain, cyp26, hox, morphogen
Introduction
Retinoic acid (RA) is a known teratogen with critical roles in the patterning of the vertebrate nervous system. In the hindbrain, RA is essential for the establishment of anterior-posterior pattern, as demonstrated by embryos in which RA is depleted either dietarily, pharmacologically, or genetically (Begemann et al., 2004; Dupe and Lumsden, 2001; Maden et al., 1996; Niederreither et al., 1999). RA is produced in the anterior paraxial mesoderm by the activity of retinaldehyde dehydrogenase 2 (Aldh1a2), which oxidizes retinal to RA (Begemann et al., 2001; Gavalas, 2002; Niederreither et al., 1999). RA diffuses or is transported from the paraxial mesoderm into the adjacent central nervous system. RA directly regulates gene expression through its nuclear hormone receptor RAR and co-receptor RXR, which bind retinoic acid response elements (RAREs) in the enhancers of target genes (Bastien and Rochette-Egly, 2004). In the hindbrain, RA regulates 3′-Hox genes through direct (in the case of Hox-1 and Hox-4) or indirect (in the case of Hox-3) mechanisms (Gould et al., 1998; Hernandez et al., 2004; Marshall et al., 1994; Nolte et al., 2003; Studer et al., 1994; Zhang et al., 2000). More anterior RA-responsive genes (Hox-1) are expressed earlier and at lower RA concentrations than more posterior RA-responsive genes (Hox-4)(Dupe and Lumsden, 2001; Maves and Kimmel, 2005; Simeone et al., 1990). Based on the effects of switching the RAREs of Hox-1 and Hox-4 genes, Gould et al. proposed that Hox-1 genes are expressed at more anterior levels than Hox-4 genes because their RAREs are more sensitive to RA (Gould et al., 1998).
These data have lead to a model in which a continuous spatio-temporal gradient of RA through the hindbrain generates nested domains of RA-responsive gene expression. These are then resolved by secondary mechanisms into non-overlapping domains that correspond with the morphological segments of the hindbrain, the rhombomeres. However a number of observations suggest that an RA gradient is neither detectable nor required for normal hindbrain development. First and foremost, embryos depleted of endogenous RA can be fully rescued by a uniform concentration of exogenous RA (Begemann et al., 2004; Begemann et al., 2001; Gale et al., 1999; Grandel et al., 2002; Mic et al., 2002; Niederreither et al., 2000). Second, this rescue can be accomplished by a range of RA concentrations and over a range of developmental stages (Dupe and Lumsden, 2001; Maves and Kimmel, 2005). Third, when RA responsiveness is measured by the expression of a RARE-LacZ reporter, no gradient of expression is detected in the hindbrain. Instead, distinct boundaries of reporter expression that shift over time are detected (Rossant et al., 1991; Sirbu et al., 2005). Finally, in contrast to earlier findings (Gould et al., 1998), recent evidence has suggested that in the context of their intact enhancers, a Hox-1 RARE is equally responsive to RA as a Hox-4 RARE (Nolte et al., 2003).
These data suggest that cells in the presumptive hindbrain neuroepithelium can be patterned by RA in a manner that is independent both of concentration and duration of exposure, necessitating a new model for RA-dependent hindbrain patterning. Here, we propose such a model based on the hindbrain patterning defects caused by preventing RA metabolism by the cytochrome P450 enzymes of the Cyp26 class. The Cyp26 enzymes (Cyp26a1, Cyp26b1 and Cyp26c1) have been proposed to function in the regulation of RA-dependent gene expression through their ability to metabolize RA into hydroxylated polar derivatives (Fujii et al., 1997; White et al., 1996). In the mouse tail bud and limbs, loss of Cyp26 function leads to increased RA-dependent gene expression, spina bifida and caudal agenesis similar to the teratogenic effects of high concentrations of exogenous RA (Abu-Abed et al., 2001; Sakai et al., 2001; Yashiro et al., 2004).
With regard to hindbrain patterning, cyp26a1 is expressed during gastrulation in the anterior neurectoderm (Dobbs-McAuliffe et al., 2004; Kudoh et al., 2002). Based on this expression domain, it was hypothesized that localized regions of RA synthesis in the anterior trunk mesoderm and degradation in the anterior neural plate provide a classical “source-and-sink” mechanism for the spatial regulation of RA in the central nervous system (Kudoh et al., 2002; Swindell et al., 1999). However cyp26a1 mutants in the fish and mouse exhibit relatively subtle hindbrain patterning defects inconsistent with a global role for cyp26a1 in hindbrain patterning (Abu-Abed et al., 2001; Emoto et al., 2005; Kudoh et al., 2002; Sakai et al., 2001). The recent identification of other cyp26 genes has suggested that these may participate in shaping RA responsiveness in the hindbrain (Abu-Abed et al., 2002; Gu et al., 2005; MacLean et al., 2001; Reijntjes et al., 2005; Reijntjes et al., 2004; Sirbu et al., 2005; Tahayato et al., 2003; Taimi et al., 2004; Zhao et al., 2005). Here, we demonstrate that the zebrafish orthologs of mammalian Cyp26b1 and Cyp26c1 function redundantly with cyp26a1 to pattern the hindbrain, since embryos depleted of all three proteins exhibit a profound posterior transformation of the hindbrain. Furthermore, we demonstrate that cyp26 genes are responsible for the ability of exogenous RA to rescue embryos depleted of endogenous sources of RA. In embryos depleted of Cyp26 activity, the low RA concentrations that normally rescue the RA-depleted hindbrain are highly teratogenic. Based on our results, we present a “gradient-free” model for RA-dependent hindbrain patterning in which the spatially regulated inactivation of RA by Cyp26 enzymes is responsible for the establishment of RA-responsive gene expression domains in the hindbrain.
Materials and Methods
Cloning
Cyp26b1 was initially identified as an EST (Nelson, 1999), and we cloned the 5′end of the coding sequence with the SMART RACE kit (Clontech). cyp26c1 was identified in a Blast search of the zebrafish genome sequence using the human CYP26C1 protein sequence and then was amplified from 12 hour post-fertilization (hpf) whole zebrafish embryo cDNA.
Morpholinos, RNA in situ hybridizations and genotyping
Table 1 summarizes the sequences of the morpholinos (MOs) we used in this study, the combinatorial depletion experiments we performed and their outcomes. Experiments to test for MO efficacy are described in the supplementary information. All of the experiments described in this manuscript used cyp26b1 MO1 and cyp26c1 MO1, however cyp26b1MO2 and cyp26c1MO2 gave the same phenotypes. Unlike cyp26c1MO1, cyp26c1MO2 was toxic at higher concentrations. Our control MO was targeted to the deadend mRNA and eliminates primordial germ cells but does not affect other aspects of development (Weidinger et al., 2003). To assay for redundancy betweencyp26 genes, MOs were injected alone or together into embryos from a cyp26a1+/- intercross (Emoto et al., 2005). In order to control for non-specific effects due to MO injections, all embryos were injected with a total of 5 ng MO as determined by measuring the diameter of the injected bolus in mineral oil (see Table 1).
Table 1.
Summary of combinatorial cyp26 knock-down experiments and their outcomes.
| treatment | summary of phenotype | |
|---|---|---|
| cyp26a1+ | cyp26a1-/- | |
| 5ng control MO | none | very mild (expanded r4) |
| 2.5 ng cyp26b1 MO1 + 2.5 ng control MO | none | very mild (expanded r4) |
| 2.5 ng cyp26c1 MO1 + 2.5 ng control MO | none | medium (up to cerebellum) |
| 2.5 ng cyp26b1 MO + 2.5 ng cyp26c1 MO | none | severe (up to cerebellum) |
| 5 nM RA | none | severe (throughout brain) |
| 10 μM DEAB + 5 nM RA | none | severe (throughout brain) |
| Morpholino sequences: | |
|---|---|
| cyp26b1 MO1 (ATG) | 5’-CTCGAAGAGCATGGCTGTGAACGTC-3’ |
| cyp26b1 MO2 (exon 2-intron 2 splice) | 5’- ATTGACCTTACCTTCCTCCTTTTGC-3’ |
| cyp26c1 MO1 (exon 3-intron 3 splice) | 5’-AAACTCGGTTATCCTCACCTTGCGC-3’ |
| cyp26c1 MO2 (intron 1-exon 2 splice) | 5′-GGAACCCTGTCACAACATAACAGAG-3′ |
| control (deadend MO) | 5′-GCTGGGCATCCATGTCTCCGACCAT-3′ |
Two-color RNA in situs were performed essentially as described (Prince et al., 1998), except that Iodo-Nitrotetrazolium Violet (Sigma) was used as the red Alkaline Phosphatase substrate. Embryos were de-yolked and flat-mounted for photomicroscopy using a Zeiss Axioplan II microscope. After photographing, individual embryos were unmounted and genotyped for the cyp26a1 mutation as described (Emoto et al., 2005).
Drug treatments
Dechorionated embryos from wild-type or cyp26+/- parents were incubated in the dark in pharmacological agonists and antagonists of the RA metabolism pathway as follows: 4-(diethylamino)benzaldehyde (DEAB, an inhibitor of retinaldehyde dehydrogenases (Russo et al., 1988); Aldrich): 10 μM, beginning at 50% epiboly (5.25 hours post fertilization (hpf)(Kimmel et al., 1995)); R115866, a specific inhibitor of Cyp26 enzymes (Janssen Pharmaceutica): 10 μM, beginning at dome stage (4.33 hpf); all-trans RA (Sigma): 0.1-100 nM, beginning at 50% epiboly. In each case, the compound was diluted to 1000 times its final concentration in DMSO, and then diluted 1000-fold in embryo medium. Controls were treated with carrier alone (0.1% DMSO). For retinal treatments, embryos were injected at the 1-cell stage with 1 nl of 20 pmol/nl all-trans retinal in DMSO (Sigma), and controls were injected with 1 nl DMSO alone.
Results
cyp26b1 and cyp26c1 are expressed dynamically during hindbrain development
Mammalian genomes contain three CYP26 genes:
CYP26A1, B1 and C1. Cloning of the zebrafish cyp26a1 ortholog has been described (White et al., 1996). We cloned zebrafish homologs of cyp26b1 and cyp26c1 and examined their expression during development. Cyp26c1 has been previously described as cyp26d1 (Gu et al., 2005) and cyp26b1-like (Kawakami et al., 2005)(ZDB gene 050714-2). Based on two lines of evidence we argue that this gene is in fact the ortholog of mammalian CYP26C1. First, phylogenetic analysis of zebrafish cyp26 genes places it in the same clade as mouse and human Cyp26c1 genes with moderate bootstrap support (Suppl. Fig. 1A). Second, CYP26A1 and CYP26C1 are adjacent to one another on human Chromosome 10q23-q24, and current zebrafish genomic sequence data and radiation hybrid data places zebrafish cyp26a1 and cyp26c1/d1/b1-like in regions of zebrafish linkage groups (LGs) 12 and 17, respectively, that show synteny to human chromosome 10 (Suppl. Fig. 1B)(Woods et al., 2005). Since draft genomes for other tetrapod vertebrates also show Cyp26a1 and Cyp26c1 as adjacent genes, it is likely that they were adjacent genes in the ancestral vertebrate genome and that following the proposed genome duplication early in teleost evolution, the duplicate of cyp26c1 was lost from LG 12 and the duplicate of cyp26a1 was lost from LG17. Henceforth we refer to the gene previously named cyp26d1/cyp26b1-like as cyp26c1.
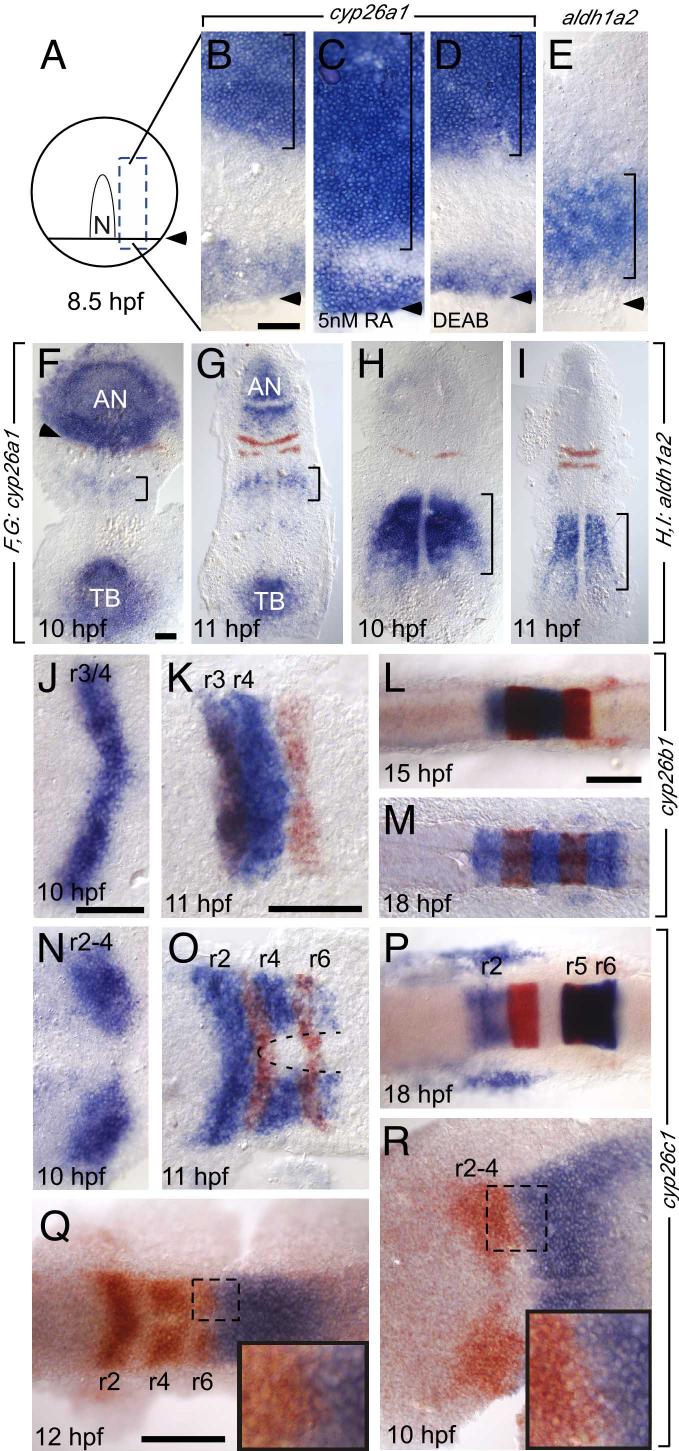
The expression patterns of zebrafish cyp26a1, cyp26b1 and cyp26c1 have been described (Dobbs-McAuliffe et al., 2004; Gu et al., 2005; Kudoh et al., 2002; Zhao et al.,2005). We focus here on their expression during hindbrain development. During gastrulation, cyp26a1 is expressed in the anterior neurectoderm (bracket in Fig. 1B) and in a narrow domain at the margin at 8.5 hours post-fertilization (hpf; arrowhead in Fig. 1B). The posterior limit of cyp26a1 expression at 8.5 hpf abuts the anterior limit of hoxb1b expression at the r3/4 boundary (Kudoh et al., 2002) but rapidly recedes anteriorly to lie at the r2/3 boundary at 10 hpf and further anterior still at 11 hpf (Fig. 1F,G). As described previously, cyp26a1 is directly RA-inducible, even at subteratogenic concentrations of RA (5 nM) which cause the ectodermal domain of expression to expand towards the margin (Fig. 1C)(Dobbs-McAuliffe et al., 2004; Kudoh et al., 2002; Loudig et al., 2000; White et al., 1996). In spite of its strong RA-inducibility, the early anterior neurectodermal expression of cyp26a1 is established independent of RA, since it is unaffected in embryos treated with 4-(diethylamino)benzaldehyde (DEAB), a specific inhibitor of retinaldehyde dehydrogenase (Fig. 1D)(Dobbs-McAuliffe et al., 2004; Sirbu et al., 2005). Throughout the hindbrain patterning period, neurectodermal cyp26a1 expression lies significantly anterior to aldh1a2 which is restricted to the anterior mesoderm (Fig. 1E,H,I)(Dobbs-McAuliffe et al., 2004; Kudoh et al., 2002; Sirbu et al., 2005; Swindell et al., 1999). Cyp26a1 is also expressed in the tailbud and in a crescent in the anterior trunk mesoderm immediately anterior to the aldh1a2-expressing domain (brackets in Fig. 1F,G).
Fig. 1.
cyp26 expression in the developing hindbrain. Whole mount in situ hybridizations during the hindbrain patterning period. All embryos are shown as dorsal views. Anterior is to the top in A-I and to the left in J-R. In situ probes are noted in brackets beside the panels, embryonic age is noted in hours post fertilization (hpf). A: schematic of an 80% epiboly (8.5 hpf) embryo. The dotted box is the region shown in the flat-mounted embryos in B-E; the arrowhead indicates the advancing margin of the epiblast. During gastrulation, cyp26a1 (B-D,F,G) is expressed in the ectoderm (bracket in B-D) anterior to the domain of RA synthesis indicated by aldh1a2 expression (bracket in E). C,D: ectodermal cyp26a1 expression expands in the presence of sub-teratogenic concentrations of RA (C), but is established independent of RA (D). F, G: cyp26a1 expression recedes anteriorly at the onset of somitogenesis. Krox20 is shown in r3 and r5. Arrowhead indicates the posterior limit of cyp26a1 expression. Bracket marks weak cyp26a1 expression in the anterior trunk mesoderm. AN; anterior neurectodermal expression; TB, tailbud expression. H,I: aldh1a2 expression during early somitogenesis. Bracket shows expression in trunk mesoderm. J-R: dynamic cyp26b1 (J-M) and cyp26c1 (N-R) expression during somitogenesis. Krox20 expression in r3 and 5 is in red in J-P. In Q and R cyp26c1 is in red while hoxd4 (Q) and vhnf1 (R) are in blue. Insets in Q, R correspond to the dotted boxes. Dotted curve in O indicates the cyp26c1-free domain in ventral r3-r6. Scale bars: 100 μM. Scale bar in B is for B-E; scale bar in F is for F-I; scale bar in J is for J,N,R; scale bar in K is for K,O; scale bar in L is for L,M,P.
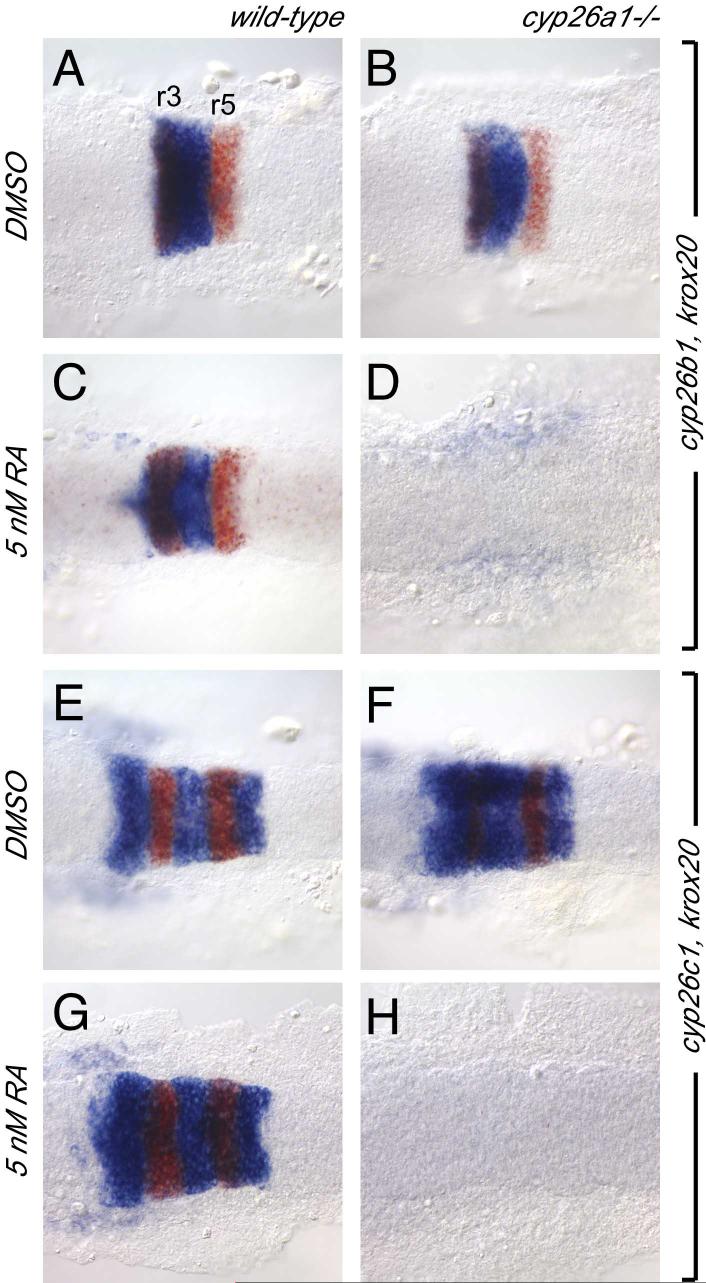
Cyp26b1 and cyp26c1 are expressed in the developing hindbrain in a dynamic, rhombomere-restricted fashion. Cyp26b1 expression is initiated in r3 and r4 beginning at tailbud stage (10 hours post fertilization (hpf) Fig. 1J). This expression slowly expands to include r2 by the 12 somite stage (15 hpf; Fig. 1L), and r5 and r6 by the 20 somite stage(19 hpf; Fig. 1M). Cyp26c1 expression overlaps with, but precedes cyp26b1 expression at each stage. cyp26c1 expression is initiated earlier, before the end of gastrulation (9 hpf) in presumptive r2 through r4 in a domain that abuts the anterior limit of vhnf1 (tcf2; ZDB gene 020104-1) expression at the presumptive r4/5 boundary (Fig. 1N,R). Expression rapidly expands posteriorly to include r6 by the 6 somite stage (12 hpf), at which time the posterior limit of cyp26c1 expression abuts the anterior limit of hoxd4 expression (Fig. 1O,Q). At the same time, cyp26c1 (but not cyp26b1) expression is down-regulated in r3. Cyp26c1 (but not cyp26b1) expression is excluded from the ventral most hindbrain above the anterior tip of the notochord during early somite stages (dotted line in Fig. 1O). Although the significance of this ventral exclusion of cyp26c1 expression for RA distribution is not known, we find that the ventral hindbrain is more sensitive to exogenous RA than more dorsal hindbrain regions (Suppl. Fig. 2). By 14 hpf, cyp26c1 expression is down-regulated in r2-r4 and is strongly up-regulated in r5 and r6 (Fig. 1P).
Unlike cyp26a1, neither cyp26b1 nor c1 are globally upregulated by exogenous RA (Suppl. Fig. 3B,E and data not shown). RA is also not required for the normal onset of their expression, since both genes are expressed in DEAB-treated embryos (Suppl. Fig. 3C,F). However we did observe effects on cyp26b1 and c1 expression at the 3 somite stage that suggest that both genes are affected indirectly by RA-dependent patterning events in the hindbrain (see legend, Suppl. Fig. 3). Briefly, in 100 nM RA r4 is expanded anteriorly and with it the r4 expression of cyp26b1 and cyp26c1, while in 10 μM DEAB r2 and r3 are expanded posteriorly and with them the r2 and r3 expression of cyp26b1 and cyp26c1. Sirbu et al (2005) showed that cyp26c1 expression in r4 is dependent on RA. We do not see clear evidence of this in the zebrafish, although the r4 domain of cyp26c1 expression is consistently reduced at the 3-somite stage in DEAB-treated embryos (Suppl. Fig. 3B).
cyp26b1 and cyp26c1 contribute to normal hindbrain patterning
We tested the function of cyp26b1 and c1 by knocking down their function using antisense morpholinos (MO). We performed all of our experiments in embryos generated by intercrossing cyp26a1 heterozygotes so that we could examine cyp26b1 and cyp26c1 function both in the presence and the absence of Cyp26a1 function (Table 1). The hindbrain phenotype of cyp26a1 mutants is subtle: r4 (marked by hoxb1a and bounded by the r3 and r5 stripes of krox20 (egr2b)) is slightly expanded in length and the anterior hindbrain (r1-r3) is slightly reduced (Fig. 2A,B, 4A,B and suppl. Fig. 4A,B) (Emoto et al., 2005). Furthermore, the posterior-most hindbrain, marked by high levels of hoxd4 expression and comprising r7 and the long unsegmented “vagal” rhombomere, r8, that lies between the segmented hindbrain and the first somite (Lumsden, 1990), is expanded in length as described previously (Emoto et al., 2005)(Fig. 2A,B; Fig. 4A,B).
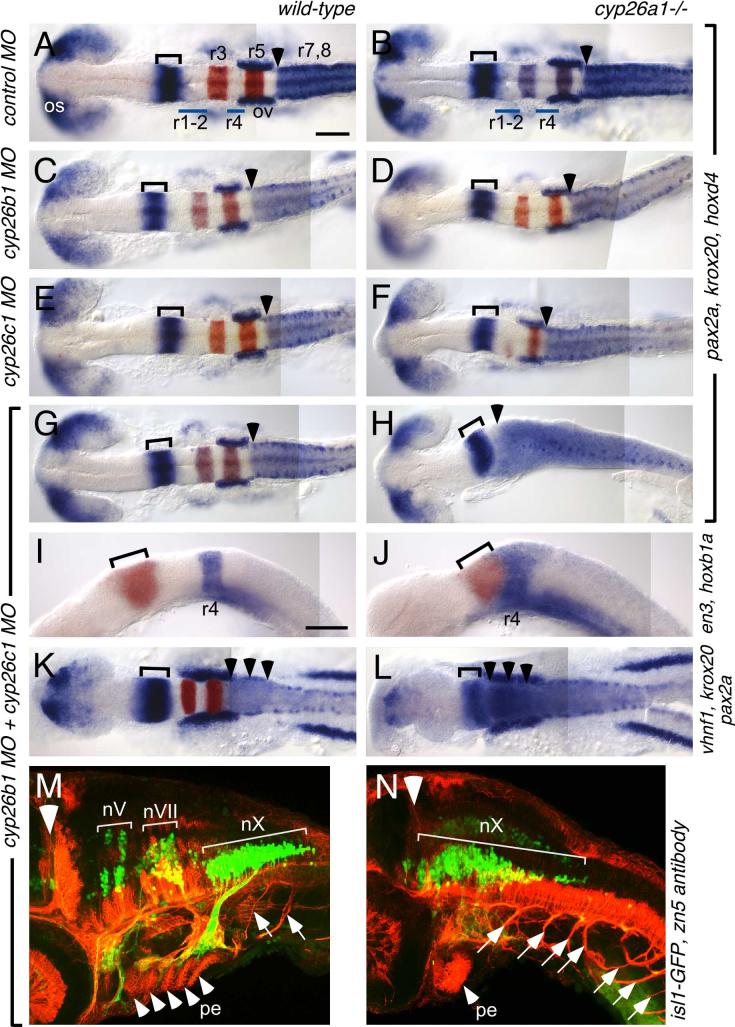
Fig. 2.
cyp26b1 and cyp26c1 function redundantly with cyp26a1 to pattern the hindbrain.
Whole-mount RNA in situ hybridizations at 18 hpf (A-J) and 13 hpf (K,L) and immunostaining at 48 hpf (M,N) in wild-type (left column) and cyp26a1-/- (right column) embryos injected with MOs as shown on the left. A-H: pax2a (blue) marks the optic stalk (os), posterior midbrain and cerebellum (bracket), and the otic vesicles (ov), while hoxd4 (also blue) marks the r7-8 territory and krox20 (red) marks r3 and r5. MO depletion of Cyp26b1 and/or c1 does not affect this pattern in wild-type embryos (C,E,G), but progressively posteriorizes the hindbrain in cyp26a1-/- embryos (D,F,H). Arrowhead marks the r6/7 boundary which is shifted to the anterior hindbrain in Cyp26-depleted embryos. I,J: en3 (red) marks the posterior midbrain and cerebellum (bracket), hoxb1a (blue) marks r4 which is shifted anteriorly in Cyp26-depleted embryos. K,L: pax2a (blue) and krox20 (red) are expressed as described above. vhnf1 is expressed in the posterior hindbrain up to the r5/6 boundary (arrowheads) and is also shifted anteriorly in Cyp26-depleted embryos. M,N: the isl1-GFP transgene (green) marks cranial motor neurons (nV: trigeminal motor neurons in r2 and r3; nVII: facial motor neurons in r4-6; nX: vagal motor neurons in r8) while the zn5 antibody (red) marks spinal motor neurons (arrows), pharyngeal arch endoderm (pe, arrowheads mark individual pharyngeal arches) and other structures. The large white arrowhead indicates the mid-hindbrain boundary. In Cyp26-depleted embryos, the motor neurons of the vagus nerve (nX) are expanded anteriorly, as are the spinal motor neurons. Scale bar: 100 μm. Scale bar in A is for A-H,K,L; scale bar in I is for I,J.
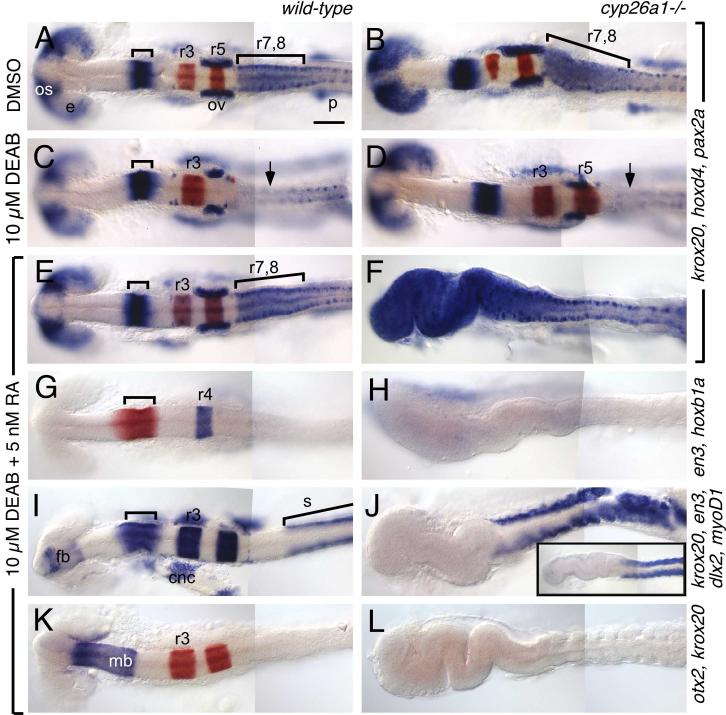
Fig. 4.
cyp26a1 protects the hindbrain from exogenous RA.
Wild-type (left column) and cyp26a1-/- (right column) embryos treated with DMSO (A,B), 10 μM DEAB (C,D) or 10 μM DEAB + 5 nM RA (E-L). RNA in situs use the markers described in Fig. 2 except for I,J, which is a mix of en3 (bracket), krox20 (r3, r5), dlx2 (cranial neural crest (cnc) and forebrain (fb)) and myoD (somites; s). os: optic stalk; e: eye; p: pronephros. Large bracket in (A) indicates the r7-8 region, which is elongated in cyp26a1 mutants (B). C: In DEAB-treated embryos, posterior rhombomeres (r5-8) are absent (arrow indicates the absence of high hoxd4 expression characteristic of r7-8). D: This phenotype is partially rescued in cyp26a1 mutants, as seen by rescue of r5 but not r7-8. E-L: The DEAB phenotype is fully rescued in wild-type embryos by treatment with 5 nM RA (E, G, I, K), while in cyp26a1 mutants this low dose of RA causes strong posteriorization of the brain (F,H,J,L). This phenotype resembles that of wild-type embryos treated with 200 nM RA (inset in J). Scale bar: 100 μm.
Depleting embryos of Cyp26b1, Cyp26c1, or both enzymes caused no brain patterning phenotype in wild-type embryos aside from a subtle shortening of the hindbrain (Fig. 2 and suppl. Fig. 4, left columns). However depleting both enzymes strongly enhanced the cyp26a1-/- hindbrain phenotype (Fig. 2 and suppl. Fig. 4, right columns). In cyp26b1 MO-injected cyp26a1-/- embryos, r4 is further expanded (Fig. 2D and suppl. Fig .4C,D) and the r6/7 boundary is shifted slightly towards r5 (arrowhead in Fig. 2C,D). Knocking down cyp26c1 caused a stronger enhancement of the cyp26a1-/- phenotype, consistent with its earlier onset of expression in the presumptive hindbrain. In these embryos, r3 is strongly reduced or absent, while r4 is expanded anteriorly so that its anterior limit lies adjacent the posterior limit of en3 expression in the presumptive cerebellum (Fig. 2F and suppl. Fig. 4F). The r6/7 boundary is again shifted anteriorly but remains posterior to a narrow r5 (arrowhead in Fig. 2F).
Embryos depleted of all three Cyp26 proteins have a strongly posteriorized hindbrain (Fig. 2G-L, suppl. Fig. 4G,H). Both r3 and r5 are eliminated (Fig. 2G,H) and r4 abuts the cerebellum (Fig. 2I,J, suppl. Fig. 4G,H). The anterior limit of vhnf1 expression, which by the 8-somite stage (13 hpf) marks the r5/6 boundary, is also shifted to abut the cerebellum (arrowheads in Fig. 2K,L). The r6/7 boundary of hoxd4 expression is similarly shifted, coming to lie within a few cell diameters of cerebellum (arrowhead in Fig. 2H). Thus three RA-responsive genes (hoxb1a, vhnf1 and hoxd4) that normally form nested expression domains in the hindbrain, are all expanded into the anterior-most hindbrain. In spite of this strong transformation of the hindbrain, the patterning of the mid- and forebrain, marked by pax2a, otx2, dlx2a and eomes appears unaffected except for a decrease in length detectable in cyp26a1 single mutants (data not shown)(Emoto et al., 2005).
We examined the neuronal organization of cyp26- depleted embryos. In control MO-injected or cyp26b1 and c1 MO-injected cyp26a1+ embryos, we observed normal patterns of cranial and spinal motor nerve differentiation (Fig. 2M). However in cyp26b1 and c1 MO-injected cyp26a1-/- embryos, the vagal (nX) neurons characteristic of r8 are expanded to the mid-hindbrain boundary (large arrowhead in Fig. 2M,N) and spinal motor roots (arrows in Fig. 2M,N) extend from hindbrain levels into a disorganized branchial region (small arrowheads in Fig. 2M,N). This occurs in spite of the fact that there are no somites to innervate at this level (data not shown). These neuronal phenotypes are consistent with our analysis of marker gene expression, in which the RA-inducible hox gene characteristic of r7-8 (hoxd4) is expanded anteriorly throughout the hindbrain region.
A pharmacological inhibitor of Cyp26 activity phenocopies Cyp26 depletion
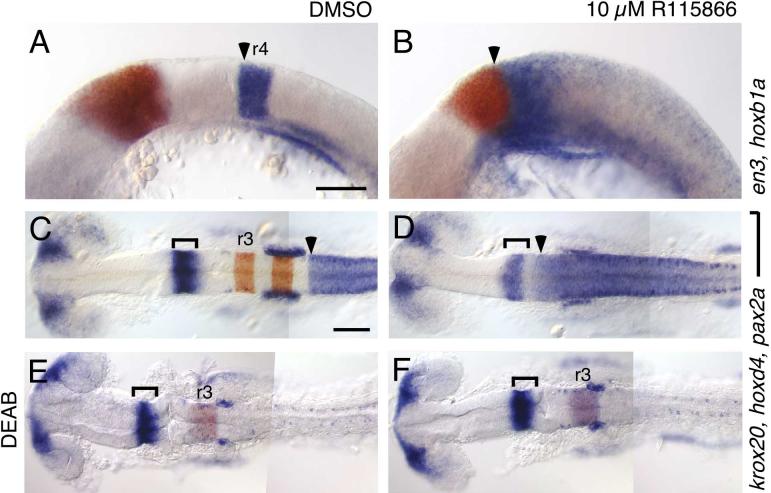
Pharmacological antagonists that inhibit RA metabolism have been developed as tools for treatment of dermatological diseases and cancer (Njar, 2002; Njar et al., 2006). The compound R115866 is a highly selective antagonist of Cyp26a1 activity in vitro, and exerts retinoidal effects in adult rats (Stoppie et al., 2000). Its effects on cyp26b1 and cyp26c1 have not been examined. We observed that treating zebrafish embryos with 10 μM R115866 caused a phenotype identical to that of embryos depleted of all three Cyp26 enzymes (compare Fig. 2J,H to Fig. 3B,D). This suggests that R115866 inhibits cyp26b1 and c1 as effectively as does knocking down their expression with MOs, and confirms our above observation that Cyp26 activity is essential for normal hindbrain patterning. The effects of R115866 treatment are completely reversed by the addition of DEAB, so that embryos treated with both drugs resemble embryos treated with DEAB alone (Fig. 3E,F). This demonstrates that, as for other phenotypes observed in cyp26a1 mutant fish and mice (Emoto et al., 2005; Niederreither et al., 2002), the posteriorized hindbrain phenotype caused by blocking all Cyp26 activity is due to accumulation of excess RA and not to the absence of bioactive Cyp26-generated RA derivatives. While such derivatives have been observed to have significant retinoidal effects in cells and inembryos and have been postulated to have functions in vivo (Idres et al., 2002; Pijnappel et al., 1993), we see no evidence for their having a role in hindbrain patterning.
Fig. 3.
a selective antagonist of cyp26 enzymes recapitulates the cyp26a1; b1;c1 phenotype.
RNA in situs with the markers described in Fig. 2. Compared to DMSO-treated controls (A,C,E), treatment with 10 μM R115866 (B,D,F) causes an anterior shift of hoxb1a (arrowhead in A,B) and hoxd4 (arrowhead in C,D) toward the presumptive cerebellum marked by en3 (red in A,B) and by pax2a (brackets in C-F). This effect of R115866 is reversed by co-treatment with 10 μM DEAB (E,F). Scale bar: 100 μm. Bar in A is for A and B; Bar in C is for C-F.
Cyp26a1 protects against RA teratogenicity.
The long-standing observation that depletion of endogenous RA can be rescued by treatment with a low concentration of exogenous RA demonstrates that an RA gradient is not strictly necessary for hindbrain patterning (Begemann et al., 2004; Begemann et al., 2001; Gale et al., 1999; Niederreither et al., 2000). However the basis of this rescue phenomenon has not been determined, and has significant implications for the mechanism of hindbrain patterning. We hypothesized that Cyp26 enzymes enable this rescue by inactivating exogenous RA in a patterned manner. We tested the roles of cyp26 genes by performing the RA rescue experiment in Cyp26-depleted embryos.
The effects of blocking RA synthesis in zebrafish with DEAB have been described (Begemann et al., 2004; Maves and Kimmel, 2005). They include loss of posterior hindbrain identities (r5-r8, Fig. 4C), expansion of anterior hindbrain identities (r2-4) and a dramatic anterior shift of paraxial and lateral plate mesoderm-derived tissues (pronephros and somites; Fig. 4C). In wild-type (cyp26a1+/+ and cyp26a1+/-) DEAB-treated embryos, this phenotype is rescued by treatment with between 0.5 and 10 nM RA; concentrations that are non-teratogenic or weakly teratogenic in wildtype embryos (Fig. 4E,G,I,K and data not shown). In the experiments described below, we used 5 nM RA as our “rescuing” concentration. Whereas in wild-type embryos 5 nM RA is non-teratogenic, it strongly posteriorizes cyp26a1-/- embryos, either in the presence or in the absence of DEAB, causing anterior expansion of r7-8 identity (Fig. 4E,F and data not shown) and loss of all brain regions anterior to r7: r3 and r5 (marked by krox20; Fig. 4E,F,I-L), r4 (marked by hoxb1a; Fig. 4G,H), the cerebellum and posterior tectum (marked by en3; Fig. 4G-J), the diencephalon and midbrain (marked by otx2; Fig. 4K,L), and the telencephalon and eyes (marked by dlx2; Fig. 4I,J and eomes; data not shown). Embryos posteriorized in this manner typically exhibited an accordion-like folding of the anterior neural tube. Exactly the same effects are observed in cyp26a1-/- embryos treated with 5 nM RA in the absence of DEAB (data not shown). This phenotype strongly resembles the effects of 40-fold higher levels of RA on wild-type embryos (inset in Fig. 4J), demonstrating that it is the ability of cyp26a1 to inactivate RA that enables RA-deficient embryos to be rescued by exogenous RA. Furthermore, these results demonstrate that cyp26a1 is able to protect embryos from the potentially teratogenic effects of low concentrations of RA. We did not see a similar sensitivity to exogenous RA in cyp26b1 and/or cyp26c1 MO-injected embryos.
We asked why under normal circumstances cyp26b1 and c1 can compensate for lack of cyp26a1 (Fig. 2), while in the presence of 5 nM RA they cannot (Fig. 4). 5 nM RA is sufficient to induce expression of cyp26a1 far posterior to its normal limit in the hindbrain (Fig. 1C). In spite of this, cyp26b1 and cyp26c1 expression is initiated at the correct anterior-posterior level and subsequent hindbrain patterning is unaffected (Fig. 5C,G and data not shown). In stark contrast, in cyp26a1-/- embryos treated with 5 nM RA, cyp26b1 and cyp26c1 are not expressed and the entire brain is strongly posteriorized (Fig. 4; Fig. 5D,H).
Fig. 5.
Exogenous RA disrupts cyp26b1 and cyp26c1 expression in cyp26a1-/- embryos but not in wild-type.
Cyp26b1 (A-D) and cyp26c1 (E-H) expression is established normally in wild-type (A,E) and cyp26a1-/- (B,F) embryos at the 6-somite stage (12 hpf). Cyp26b1 and cyp26c1 expression is also established normally in wild-type embryos treated with a sub-teratogenic concentration of RA (5 nM; C,G), but not in cyp26a1-/- embryos treated with 5 nM RA (D,H).
Cyp26a1 protects against potentially teratogenic RA precursors
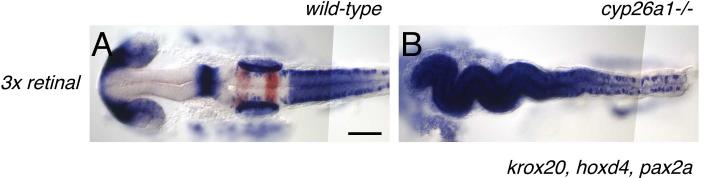
Our findings demonstrate that Cyp26a1 protects against the potentially teratogenic effects of RA. Maternally-derived RA is present at very low levels in zebrafish eggs and early embryos prior to the onset of embryonic RA synthesis, and is therefore unlikely to be a teratogenic risk (Costaridis et al., 1996). However the levels of maternally loaded retinal, the immediate precursor of RA, are higher (9 pmol/egg) (Costaridis et al., 1996; Lampert et al., 2003). We asked whether Cyp26a1 protects against teratogenicity of RA precursors. We increased retinal levels in wild-type and cyp26a1-/- eggs by injecting retinal directly into the yolk of 1-cell stage embryos. While wild-type embryos tolerate over 10-times the normal amount of retinal in the yolk (data not shown), cyp26a1-/- embryos are strongly posteriorized by only a 3 times the normal amount of retinal (27 pmol; Fig. 6). The teratogenic effects of a small increase in RA precursor in the absence of Cyp26a1 demonstrate that Cyp26a1 may normally play an important role in protecting the embryo against the potentially teratogenic effects of maternally derived RA precursors. They also suggest that the RA biosynthetic enzyme, Aldh1a2, is unable to buffer changes in the levels of its substrate. Since retinal is itself derived directly from dietary vitamin A, it may be expected to fluctuate depending on maternal diet. These observations emphasize the critical importance of a tightly regulated RA degradative pathway in nervous system patterning.
Fig. 6.
cyp26a1 protects against teratogenic effects of the RA precursor retinal.
Wild-type (A) and cyp26a1-/- (B) embryos injected with 18 pmol retinal at the 1-cell stage. Wild-type embryos are only mildly affected by approximately triple the normal levels of retinal, while cyp26 mutants are strongly posteriorized, with hoxd4 expression extending throughout the brain. Scale bar: 100 μm.
Discussion
A gradient-free model for hindbrain patterning by Retinoic Acid
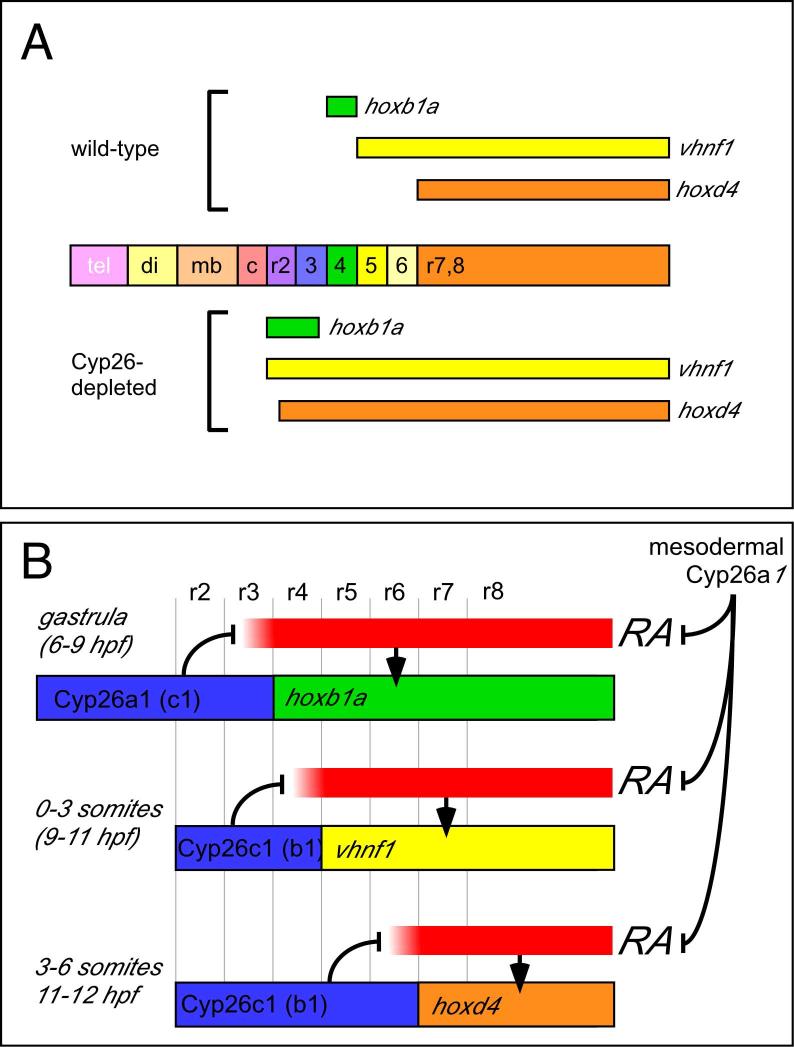
A robust model for the mechanism of hindbrain patterning must explain the following observations: 1) RA is essential for normal hindbrain development, however neither the concentration of RA, nor the localization of its synthesis are critical for this pattern; 2) RA-dependent gene expression occurs in a spatio-temporal sequence, with anterior RA-responsive genes being expressed earlier than posterior ones, however the duration of RA exposure is not critical for this temporal sequence (Maves and Kimmel, 2005). We have identified a critical role for Cyp26 RA metabolizing enzymes in establishing hindbrain pattern, since depleting them alone and in combination leads to a progressive posteriorization of the hindbrain. In fully Cyp26-depleted embryos, three RA-dependent genes that normally form nested expression domains with distinct anterior limits (hoxb1a, r3/4; vhnf1, r4/5; and hoxd4, r6/7) are all expanded up to the anteriormost hindbrain (Fig. 7A). Based on the dynamic expression of cyp26 genes in the hindbrain and on the effects of depleting embryos of Cyp26 activity, we propose a gradient-free model for RA-dependent events in hindbrain patterning, in which RA degradation by Cyp26 enzymes determines progressively more posterior limits of RA-dependent gene expression in a step-wise manner (Fig. 7B). We note that cyp26 genes are expressed similarly, although not identically, in tetrapods, predicting a similar combinatorial role for Cyp26 enzymes in mammalian hindbrain development.
Fig. 7.
A model for hindbrain patterning through regulated RA inactivation by Cyp26 enzymes.
A: RA-responsive gene expression in Cyp26-depleted embryos. Embryos depleted of all three cyp26 genes experience unpatterned RA signaling; as a result the three RA-responsive genes examined in this study, hoxb1a (green), vhnf1 (yellow), and hoxd4 (orange) are expressed throughout the transformed hindbrain. Tel: telencephalon; di; diencephalon; mb: midbrain; c: cerebellum. B: A “gradient free” model for hindbrain patterning through regulated RA inactivation. Dynamic patterns of Cyp26 expression in the hindbrain (blue bars) antagonize RA-dependent gene expression by eliminating RA(red bars) first in the anterior hindbrain (6-9 hpf), then in r2-4 (9-11 hpf), then in r2-6 (11-12 hpf). At each point, sequential RA-responsive genes (colored bars) are limited to progressively more posterior rhombomeres. At the same time, Cyp26a1-dependent RA degradation in the trunk mesoderm suppresses global RA levels (black hammers on right side).
In step 1, complete by 9 hpf, the anterior limit of hoxb1b and hoxb1a, the functional homologs of mammalian hoxa1 and hoxb1, are established by the posterior limit of cyp26a1 expression. This event establishes the r3/4 boundary (Kudoh et al., 2002), the first morphological boundary in the hindbrain (Moens et al., 1998). This function can be compensated for by cyp26c1, since the anterior limit of r4 is strongly affected only in the absence of both cyp26a1 and cyp26c1. In step 2, complete by 11 hpf, the anterior limit of the next RA-responsive gene, vhnf1, is determined by the posterior limit of cyp26c1 expression at the r4/5 boundary. This function can be partially compensated for by cyp26b1. In step 3, complete by 12 hpf, the anterior limit of the last RA-responsive gene, hoxd4, is determined by the posterior limit of cyp26c1 at the r6/7 boundary, a function that can also be compensated for by the overlapping expression of cyp26b1. Some of the mechanistic underpinnings of this model and its broader implications are discussed further below.
A similar model was previously proposed based on the correspondence between cyp26a1 and cyp26c1 expression domains and boundaries of RA-dependent reporter gene expression in the mouse (Sirbu et al., 2005). These authors predicted that cyp26a1 establishes the r2/3 boundary and that cyp26c1 subsequently establishes the r4/5 boundary. Our combinatorial functional analysis of cyp26 genes confirms this model in the general sense that Cyp26 activity determines sequential boundaries of RA-responsive gene expression in the hindbrain. However our observations demonstrate a different and broader role for cyp26 genes in hindbrain patterning, involving all three cyp26 genes functioning to establish three sequential RA-responsiveness boundaries: r3/4, r4/5 and r6/7. We do not observe a function for Cyp26 enzymes at the r2/3 boundary: r1-r3 are entirely lost while r4 identity shifts anteriorly to abut the forming cerebellum.
We find that cyp26a1 and cyp26c1 are both required to establish the anterior limit of hoxb1a expression at the r3/4 boundary, and that all three genes are required to establish the r4/5 and r6/7 boundaries. This degree of redundancy is unexpected given the lack of overlap between cyp26 expression domains in the hindbrain. The posterior limit of cyp26a1 expression lies in the anterior hindbrain (Dobbs-McAuliffe et al., 2004; Kudoh et al., 2002; Sirbu et al., 2005), while both cyp26b1 and cyp26c1 mark, sequentially, the r4/5 and r6/7 boundaries. However it is important to note that cyp26a1 is also expressed in the anterior trunk mesoderm near the RA source, where it likely functions to reduce global RA levels (Emoto et al., 2005; Niederreither et al., 2002). We propose that the severe posteriorization of Cyp26-depleted embryos results from the combined effects of depleting segment-restricted Cyp26 activity within the hindbrain and increasing global RA levels due to loss of Cyp26a1 activity in the anterior trunk mesoderm (Fig. 7B).
Since hindbrain patterning is unaffected in cyp26b1; cyp26c1- depleted embryos when cyp26a1 is wild-type, we hypothesize that redundant mechanisms can control boundaries of RA-dependent gene expression in the hindbrain but that these mechanisms are overridden in cyp26a1 mutants in which global RA levels are elevated. One trivial possibility is that our MOs have not fully depleted Cyp26b1 and Cyp26c1 activity, however our validation experiments with these MOs indicate that they deplete over 95% of the wild-type gene products (supplementary material). A second possibility is that non-homogeneous expression of RARs or RXRs in the hindbrain may modulate RA responsiveness. During the stages when RA is patterning the zebrafish hindbrain, two receptors, RARaa and RARab are expressed throughout the hindbrain, but RARab mRNA levels are higher in presumptive r5 and r6 while RARaa mRNA levels are higher posterior to the presumptive r6/7 boundary (Hale et al., 2006). Furthermore, RXRγ, a RA co-receptor, is exclusively expressed posterior to the r6/7 boundary (Tallafuss et al., 2006). By increasing the RA response, these non-homogeneously distributed RARs and RXRs may help to establish the r4/5 and r6/7 boundaries.
A third possibility is that spatially restricted transcription factors repress RA responsive gene expression even when ligand and receptor are present. Iro7 is a TALE homeodomain protein expressed in the anterior hindbrain that represses vhnf1 expression anterior to the r4/5 boundary (Lecaudey et al., 2004). Other TALE homeodomain proteins have been shown to repress transcription from retinoid responsive elements by binding to RXR retinoid receptors and recruiting general co-repressors to the complex (Bartholin et al., 2006). Thus Iro7 may compensate for cyp26b1 and cyp26c1 by directly suppressing RA-responsive gene expression anterior to it the r4/5 boundary.
Finally, a diffusion gradient of RA from its source in the anterior trunk mesoderm may compensate for the absence of hindbrain Cyp26 expression. RA can act as a classical morphogen, specifying distinct rhombomere identities at different threshold concentrations (Dupe and Lumsden, 2001; Maves and Kimmel, 2005), and an RA gradient may initiate nested domains of RA-responsive gene expression in the hindbrain when cyp26b1 and cyp26c1 are depleted. We note that any or all of the mechanisms we have proposed above (a diffusion gradient; receptor expression; other transcription factors that modulate RAR activity) may contribute to hindbrain patterning under normal circumstances. However none of these mechanisms are sufficient to limit RA responsiveness in the hindbrain when all three Cyp26 enzymes are depleted.
Cyp26a1 is required to establish hindbrain pattern in the absence of a localized source of RA
We have shown that in the zebrafish, cyp26a1 is essential for the ability of exogenous RA to rescue embryos in which endogenous RA synthesis is inhibited. While 5 nM RA can fully rescue the hindbrain and anterior trunk patterning defects of wild-type embryos in which RA synthesis is inhibited with DEAB, in cyp26a1-/- embryos it causes a strong posteriorization similar to that normally caused by 40 times more RA. From this, we conclude that Cyp26a1 is responsible for the normal pattern generated in the presence of otherwise teratogenic amounts of RA.
How does Cyp26a1 protect the embryos from exogenous RA? We observe that in embryos treated with 5nM RA cyp26a1 expression expands throughout the epiblast. This expanded expression presumably eliminates the excess RA and allows the normal onset of expression of cyp26b1, cyp26c1, and other redundant factors described above which can modulate RA responsive gene expression in the hindbrain, and the hindbrain develops normally under these conditions. In cyp26a1 mutants treated with 5 nM RA, the excess RA is not eliminated, cyp26b1 and cyp26c1 expression is not initiated, and the entire brain is transformed to posterior hindbrain/anterior spinal cord identity. Unlike in untreated cyp26a1 mutants, cyp26b1 and cyp26c1 cannot compensate for the lack of cyp26a1 because they are not expressed.
The phenotype of cyp26a1-/- embryos treated with 5 nM RA is significantly more severe than that of embryos depleted of all three Cyp26 enzymes in the absence of exogenous RA. In the former, the entire brain is transformed to an r7/8 identity, while in the latter only the hindbrain territory is transformed. This difference may be because in untreated embryos, RA simply does not diffuse as far as the midbrain, so Cyp26 enzymes are not required to inactivate it there. However we have noted a surprisingly sharp anterior limit of RA-responsiveness in Cyp26-depleted embryos that corresponds with the posterior limit the presumptive cerebellum. It is possible that other mechanisms prevent RA signaling anterior to this boundary. The development of the mid- and forebrain has been shown to require active repression of gene expression by unliganded RARs (Koide et al., 2001), a mechanism that is expected to be easily destabilized by the presence of RA. It seems likely that multiple mechanisms exist that protect the mid- and forebrain from RA’s teratogenic effects.
Regulation of cyp26 expression
A major outstanding question is how cyp26 expression is normally regulated in the hindbrain. Kudoh et al. (2002) showed that the normal posterior limit of cyp26a1 in the hindbrain is established by signals (Fgfs and Wnts) from the margin, since in embryos treated with antagonists of these pathways the cyp26a1 boundary is shifted posteriorly. Although cyp26a1 is directly inducible by RA, its anterior neurectodermal expression arises independently of RA (Dobbs-McAuliffe et al., 2004; Sirbu et al., 2005)(this work). The factors that positively regulate cyp26a1 in the anterior neurectoderm remain to be identified.
We also do not know how cyp26b1 and cyp26c1 expression is initiated in r2-4, or what regulates their subsequent expansion into r5 and r6. In general, the mechanisms controlling gene expression in the anterior rhombomeres are poorly understood in any vertebrate (Moens and Prince, 2002). Like other anterior hindbrain genes, the initiation of cyp26b1 and cyp26c1 expression is independent of RA, since both genes are expressed in DEAB-treated embryos. Cyp26b1 and cyp26c1 expression is also independent of the prior establishment of hindbrain boundaries by Cyp26a1, since both genes are expressed normally in cyp26a1 mutants and in embryos in which cyp26a1 expression is globally up-regulated by sub-teratogenic concentrations of RA. The early expression domain of cyp26b1 and cyp26c1 in r3 and r4 is similar to that of iro7, suggesting that they may be downstream of, or co-regulated with, iro7 (Lecaudey et al., 2004). Modern genetic and genomic resources available for the zebrafish will allow the important question of cyp26 regulation to be addressed in the future.
Implications for the regulation of RA during hindbrain patterning
The model we propose for hindbrain patterning through localized RA inactivation by Cyp26 enzymes (Fig. 7B) accounts for a number of previously unexplained aspects of hindbrain patterning. First among these is the observation that embryos depleted of endogenous RA can be rescued by exogenous RA. This rescue can be achieved over a 20-fold range of RA concentrations, indicating that RA-dependent gene expression is also not strictly concentration dependent. By generating a stepwise pattern of RA degradation during hindbrain development, Cyp26 enzymes eliminate the need for a continuous RA gradient. Secondly, a major tenet of the RA morphogen model has been that more posterior RA-responsive genes such as hox-4 genes are less sensitive to RA than more anterior ones like hox-1 genes (Gould et al., 1998), however this has recently been challenged by the observation that in the context of the intact enhancer, the RARE of hoxd4 is no less sensitive to RA than the RARE of hoxa1 (Nolte et al., 2003). Furthermore, posterior RA-responsive genes do not require a longer exposure to RA than anterior ones as has been proposed (Dupe and Lumsden, 2001; Sirbu et al., 2005), since identical concentrations of RA applied shortly before the normal initiation of expression are sufficient to rescue this expression in RA-depleted embryos, irrespective of the anterior limit of the RA-responsive gene in question (Maves and Kimmel, 2005). According to our model, the anterior limit of hoxb1a, vhfn1 and hoxd4 are determined not by different RA concentrations or length of exposure of cells to RA, but simply by the posterior limit of Cyp26 activity at the time of their expression onset. In its most extreme version, each “step” in the model is essentially a binary decision, in which cells posterior to the Cyp26 domain experience RA and initiate RA-responsive gene expression appropriate for that developmental time while cells within the Cyp26 domain do not. What determines which RA-responsive genes are available to be expressed at a given time is the subject of ongoing studies. Recent work has shown that within a Hox cluster, the timing of hindbrain expression may be regulated by the progressive opening of chromatin rather than the local accumulation of active trans-acting factors (Chambeyron et al., 2005).
An RA-dependent patterning mechanism that does not require the formation of a stable gradient in either space or time is expected to be robust to environmental fluctuations. RA is a potent teratogen that is derived from dietary sources of vitamin A, so a robust mechanism for controlling its activity is particularly important. This control does not appear to be exerted at the level of RA biosynthesis, since we have observed that in the absence of Cyp26a1, even low amounts of precursor are highly teratogenic. Our step-wise model for hindbrain patterning by RA is robust in that it tolerates a broad range of environmental conditions.
While our model provides robustness to the hindbrain patterning process and explains how patterning can be established in the presence of uniform RA, our data does not rule out the possibility that other mechanisms act redundantly with RA degradation to pattern the hindbrain under normal circumstances. Indeed, our observation that hindbrain Cyp26b1 and Cyp26c1 activity is dispensable when global RA levels are kept in check by Cyp26a1 suggests that such mechanisms are at work. It is possible that an RA-responsive pre-pattern is established by a transient RA diffusion gradient, but that Cyp26 enzymes are required to “lock in” this pattern. As with other developmental processes, it is likely that RA-dependent nervous system patterning events are controlled by overlapping, redundant mechanisms that modulate RA signaling at multiple levels. Our work demonstrates that Cyp26-dependent RA degradation is a critical component of this complex regulation.
Acknowledgements
The cyp26a1 mutant strain was obtained from Dr. Yoshiyuki Imai under the support of the National Bioresource Project of Japan. The Cyp26 antagonist R115866 was provided by Janssen Pharmaceutica N.V. (Belgium). We thank Elizabeth Kwan for her helpful contribution to this work. We gratefully acknowledge Phil Soriano, Lisa Maves and the members of the Moens lab for their thoughtful comments during this work and on the manuscript. R.E.H. was supported by NIH Training Grants 2T32 HD07183 and NIGMS T32 07266 and a Cora May Poncin Scholarship and an ARCS-WRF Fellowship. This work was supported by NIH grant HD37909. C.B.M. is an Investigator with the Howard Hughes Medical Institute.
References
- Abu-Abed S, Dolle P, Metzger D, Beckett B, Chambon P, Petkovich M. The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes Dev. 2001;15:226–40. doi: 10.1101/gad.855001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Abed S, MacLean G, Fraulob V, Chambon P, Petkovich M, Dolle P. Differential expression of the retinoic acid-metabolizing enzymes CYP26A1 and CYP26B1 during murine organogenesis. Mech Dev. 2002;110:173–7. doi: 10.1016/s0925-4773(01)00572-x. [DOI] [PubMed] [Google Scholar]
- Bartholin L, Powers SE, Melhuish TA, Lasse S, Weinstein M, Wotton D. TGIF inhibits retinoid signaling. Mol Cell Biol. 2006;26:990–1001. doi: 10.1128/MCB.26.3.990-1001.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien J, Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Begemann G, Schilling TF, Rauch GJ, Geisler R, Ingham PW. The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development. 2001;128:3081–94. doi: 10.1242/dev.128.16.3081. [DOI] [PubMed] [Google Scholar]
- Begemann G, Marx M, Mebus K, Meyer A, Bastmeyer M. Beyond the neckless phenotype: influence of reduced retinoic acid signaling on motor neuron development in the zebrafish hindbrain. Dev Biol. 2004;271:119–29. doi: 10.1016/j.ydbio.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Chambeyron S, Da Silva NR, Lawson KA, Bickmore WA. Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development. 2005;132:2215–23. doi: 10.1242/dev.01813. [DOI] [PubMed] [Google Scholar]
- Costaridis P, Horton C, Zeitlinger J, Holder N, Maden M. Endogenous retinoids in the zebrafish embryo and adult. Dev Dyn. 1996;205:41–51. doi: 10.1002/(SICI)1097-0177(199601)205:1<41::AID-AJA4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Dobbs-McAuliffe B, Zhao Q, Linney E. Feedback mechanisms regulate retinoic acid production and degradation in the zebrafish embryo. Mech Dev. 2004;121:339–50. doi: 10.1016/j.mod.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Dupe V, Lumsden A. Hindbrain patterning involves graded responses to retinoic acid signalling. Development. 2001;128:2199–208. doi: 10.1242/dev.128.12.2199. [DOI] [PubMed] [Google Scholar]
- Emoto Y, Wada H, Okamoto H, Kudo A, Imai Y. Retinoic acid-metabolizing enzyme Cyp26a1 is essential for determining territories of hindbrain and spinal cord in zebrafish. Dev Biol. 2005;278:415–27. doi: 10.1016/j.ydbio.2004.11.023. [DOI] [PubMed] [Google Scholar]
- Fujii H, Sato T, Kaneko S, Gotoh O, Fujii-Kuriyama Y, Osawa K, Kato S, Hamada H. Metabolic inactivation of retinoic acid by a novel P450 differentially expressed in developing mouse embryos. Embo J. 1997;16:4163–73. doi: 10.1093/emboj/16.14.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale E, Zile M, Maden M. Hindbrain respecification in the retinoid-deficient quail. Mech Dev. 1999;89:43–54. doi: 10.1016/s0925-4773(99)00202-6. [DOI] [PubMed] [Google Scholar]
- Gavalas A. ArRAnging the hindbrain. Trends Neurosci. 2002;25:61–4. doi: 10.1016/s0166-2236(02)02067-2. [DOI] [PubMed] [Google Scholar]
- Gould A, Itasaki N, Krumlauf R. Initiation of rhombomeric Hoxb4 expression requires induction by somites and a retinoid pathway. Neuron. 1998;21:39–51. doi: 10.1016/s0896-6273(00)80513-9. [DOI] [PubMed] [Google Scholar]
- Grandel H, Lun K, Rauch GJ, Rhinn M, Piotrowski T, Houart C, Sordino P, Kuchler AM, Schulte-Merker S, Geisler R, et al. Retinoic acid signalling in the zebrafish embryo is necessary during pre-segmentation stages to pattern the anterior-posterior axis of the CNS and to induce a pectoral fin bud. Development. 2002;129:2851–65. doi: 10.1242/dev.129.12.2851. [DOI] [PubMed] [Google Scholar]
- Gu X, Xu F, Wang X, Gao X, Zhao Q. Molecular cloning and expression of a novel CYP26 gene (cyp26d1) during zebrafish early development. Gene Expr Patterns. 2005;5:733–9. doi: 10.1016/j.modgep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Hale LA, Tallafuss A, Yan YL, Dudley L, Eisen JS, Postlethwait JH. Characterization of the retinoic acid receptor genes raraa, rarab and rarg during zebrafish development. Gene Expr Patterns. 2006;6:546–55. doi: 10.1016/j.modgep.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Hernandez RE, Rikhof HA, Bachmann R, Moens CB. vhnf1 integrates global RA patterning and local FGF signals to direct posterior hindbrain development in zebrafish. Development. 2004;131:4511–20. doi: 10.1242/dev.01297. [DOI] [PubMed] [Google Scholar]
- Idres N, Marill J, Flexor MA, Chabot GG. Activation of retinoic acid receptor-dependent transcription by all-trans-retinoic acid metabolites and isomers. J Biol Chem. 2002;277:31491–8. doi: 10.1074/jbc.M205016200. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Raya A, Raya RM, Rodriguez-Esteban C, Belmonte JC. Retinoic acid signalling links left-right asymmetric patterning and bilaterally symmetric somitogenesis in the zebrafish embryo. Nature. 2005;435:165–71. doi: 10.1038/nature03512. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Koide T, Downes M, Chandraratna RA, Blumberg B, Umesono K. Active repression of RAR signaling is required for head formation. Genes Dev. 2001;15:2111–21. doi: 10.1101/gad.908801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudoh T, Wilson SW, Dawid IB. Distinct roles for Fgf, Wnt and retinoic acid in posteriorizing the neural ectoderm. Development. 2002;129:4335–46. doi: 10.1242/dev.129.18.4335. [DOI] [PubMed] [Google Scholar]
- Lampert JM, Holzschuh J, Hessel S, Driever W, Vogt K, von Lintig J. Provitamin A conversion to retinal via the beta,beta-carotene-15,15′-oxygenase (bcox) is essential for pattern formation and differentiation during zebrafish embryogenesis. Development. 2003;130:2173–86. doi: 10.1242/dev.00437. [DOI] [PubMed] [Google Scholar]
- Lecaudey V, Anselme I, Rosa F, Schneider-Maunoury S. The zebrafish Iroquois gene iro7 positions the r4/r5 boundary and controls neurogenesis in the rostral hindbrain. Development. 2004;131:3121–31. doi: 10.1242/dev.01190. [DOI] [PubMed] [Google Scholar]
- Loudig O, Babichuk C, White J, Abu-Abed S, Mueller C, Petkovich M. Cytochrome P450RAI(CYP26) promoter: a distinct composite retinoic acid response element underlies the complex regulation of retinoic acid metabolism. Mol Endocrinol. 2000;14:1483–97. doi: 10.1210/mend.14.9.0518. [DOI] [PubMed] [Google Scholar]
- Lumsden A. The cellular basis of segmentation in the developing hindbrain. Trends Neurosci. 1990;13:329–35. doi: 10.1016/0166-2236(90)90144-y. [DOI] [PubMed] [Google Scholar]
- MacLean G, Abu-Abed S, Dolle P, Tahayato A, Chambon P, Petkovich M. Cloning of a novel retinoic-acid metabolizing cytochrome P450, Cyp26B1, and comparative expression analysis with Cyp26A1 during early murine development. Mech Dev. 2001;107:195–201. doi: 10.1016/s0925-4773(01)00463-4. [DOI] [PubMed] [Google Scholar]
- Maden M, Gale E, Kostetskii I, Zile M. Vitamin A-deficient quail embryos have half a hindbrain and other neural defects. Curr Biol. 1996;6:417–26. doi: 10.1016/s0960-9822(02)00509-2. [DOI] [PubMed] [Google Scholar]
- Marshall H, Studer M, Popperl H, Aparicio S, Kuroiwa A, Brenner S, Krumlauf R. A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature. 1994;370:567–71. doi: 10.1038/370567a0. [DOI] [PubMed] [Google Scholar]
- Maves L, Kimmel CB. Dynamic and sequential patterning of the zebrafish posterior hindbrain by retinoic acid. Dev Biol. 2005;285:593–605. doi: 10.1016/j.ydbio.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Mic FA, Haselbeck RJ, Cuenca AE, Duester G. Novel retinoic acid generating activities in the neural tube and heart identified by conditional rescue of Raldh2 null mutant mice. Development. 2002;129:2271–82. doi: 10.1242/dev.129.9.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens CB, Cordes SP, Giorgianni MW, Barsh GS, Kimmel CB. Equivalence in the genetic control of hindbrain segmentation in fish and mouse. Development. 1998;125:381–91. doi: 10.1242/dev.125.3.381. [DOI] [PubMed] [Google Scholar]
- Moens CB, Prince VE. Constructing the hindbrain: insights from the zebrafish. Dev Dyn. 2002;224:1–17. doi: 10.1002/dvdy.10086. [DOI] [PubMed] [Google Scholar]
- Nelson DR. A second CYP26 P450 in humans and zebrafish: CYP26B1. Arch Biochem Biophys. 1999;371:345–7. doi: 10.1006/abbi.1999.1438. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Subbarayan V, Dolle P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat Genet. 1999;21:444–8. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Vermot J, Schuhbaur B, Chambon P, Dolle P. Retinoic acid synthesis and hindbrain patterning in the mouse embryo. Development. 2000;127:75–85. doi: 10.1242/dev.127.1.75. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Abu-Abed S, Schuhbaur B, Petkovich M, Chambon P, Dolle P. Genetic evidence that oxidative derivatives of retinoic acid are not involved in retinoid signaling during mouse development. Nat Genet. 2002;31:84–8. doi: 10.1038/ng876. [DOI] [PubMed] [Google Scholar]
- Njar VC. Cytochrome p450 retinoic acid 4-hydroxylase inhibitors: potential agents for cancer therapy. Mini Rev Med Chem. 2002;2:261–9. doi: 10.2174/1389557023406223. [DOI] [PubMed] [Google Scholar]
- Njar VC, Gediya L, Purushottamachar P, Chopra P, Vasaitis TS, Khandelwal A, Mehta J, Huynh C, Belosay A, Patel J. Retinoic acid metabolism blocking agents (RAMBAs) for treatment of cancer and dermatological diseases. Bioorg Med Chem. 2006 doi: 10.1016/j.bmc.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Nolte C, Amores A, Nagy Kovacs E, Postlethwait J, Featherstone M. The role of a retinoic acid response element in establishing the anterior neural expression border of Hoxd4 transgenes. Mech Dev. 2003;120:325–35. doi: 10.1016/s0925-4773(02)00442-2. [DOI] [PubMed] [Google Scholar]
- Pijnappel WW, Hendriks HF, Folkers GE, van den Brink CE, Dekker EJ, Edelenbosch C, van der Saag PT, Durston AJ. The retinoid ligand 4-oxo-retinoic acid is a highly active modulator of positional specification. Nature. 1993;366:340–4. doi: 10.1038/366340a0. [DOI] [PubMed] [Google Scholar]
- Prince VE, Moens CB, Kimmel CB, Ho RK. Zebrafish hox genes: expression in the hindbrain region of wild-type and mutants of the segmentation gene, valentino. Development. 1998;125:393–406. doi: 10.1242/dev.125.3.393. [DOI] [PubMed] [Google Scholar]
- Reijntjes S, Gale E, Maden M. Generating gradients of retinoic acid in the chick embryo: Cyp26C1 expression and a comparative analysis of the Cyp26 enzymes. Dev Dyn. 2004;230:509–17. doi: 10.1002/dvdy.20025. [DOI] [PubMed] [Google Scholar]
- Reijntjes S, Blentic A, Gale E, Maden M. The control of morphogen signalling: regulation of the synthesis and catabolism of retinoic acid in the developing embryo. Dev Biol. 2005;285:224–37. doi: 10.1016/j.ydbio.2005.06.019. [DOI] [PubMed] [Google Scholar]
- Rossant J, Zirngibl R, Cado D, Shago M, Giguere V. Expression of a retinoic acid response element-hsplacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 1991;5:1333–44. doi: 10.1101/gad.5.8.1333. [DOI] [PubMed] [Google Scholar]
- Russo JE, Hauguitz D, Hilton J. Inhibition of mouse cytosolic aldehyde dehydrogenase by 4-(diethylamino)benzaldehyde. Biochem Pharmacol. 1988;37:1639–42. doi: 10.1016/0006-2952(88)90030-5. [DOI] [PubMed] [Google Scholar]
- Sakai Y, Meno C, Fujii H, Nishino J, Shiratori H, Saijoh Y, Rossant J, Hamada H. The retinoic acid-inactivating enzyme CYP26 is essential for establishing an uneven distribution of retinoic acid along the anterio-posterior axis within the mouse embryo. Genes Dev. 2001;15:213–25. doi: 10.1101/gad.851501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Arcioni L, Andrews PW, Boncinelli E, Mavilio F. Sequential activation of HOX2 homeobox genes by retinoic acid in human embryonal carcinoma cells. Nature. 1990;346:763–6. doi: 10.1038/346763a0. [DOI] [PubMed] [Google Scholar]
- Sirbu IO, Gresh L, Barra J, Duester G. Shifting boundaries of retinoic acid activity control hindbrain segmental gene expression. Development. 2005;132:2611–22. doi: 10.1242/dev.01845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppie P, Borgers M, Borghgraef P, Dillen L, Goossens J, Sanz G, Szel H, Van Hove C, Van Nyen G, Nobels G, et al. R115866 inhibits all-trans-retinoic acid metabolism and exerts retinoidal effects in rodents. J Pharmacol Exp Ther. 2000;293:304–12. [PubMed] [Google Scholar]
- Studer M, Popperl H, Marshall H, Kuroiwa A, Krumlauf R. Role of a conserved retinoic acid response element in rhombomere restriction of Hoxb-1. Science. 1994;265:1728–32. doi: 10.1126/science.7916164. [DOI] [PubMed] [Google Scholar]
- Swindell EC, Thaller C, Sockanathan S, Petkovich M, Jessell TM, Eichele G. Complementary domains of retinoic acid production and degradation in the early chick embryo. Dev Biol. 1999;216:282–96. doi: 10.1006/dbio.1999.9487. [DOI] [PubMed] [Google Scholar]
- Tahayato A, Dolle P, Petkovich M. Cyp26C1 encodes a novel retinoic acid-metabolizing enzyme expressed in the hindbrain, inner ear, first branchial arch and tooth buds during murine development. Gene Expr Patterns. 2003;3:449–54. doi: 10.1016/s1567-133x(03)00066-8. [DOI] [PubMed] [Google Scholar]
- Taimi M, Helvig C, Wisniewski J, Ramshaw H, White J, Amad M, Korczak B, Petkovich M. A novel human cytochrome P450, CYP26C1, involved in metabolism of 9-cis and all-trans isomers of retinoic acid. J Biol Chem. 2004;279:77–85. doi: 10.1074/jbc.M308337200. [DOI] [PubMed] [Google Scholar]
- Tallafuss A, Hale LA, Yan YL, Dudley L, Eisen JS, Postlethwait JH. Characterization of retinoid-X receptor genes rxra, rxrba, rxrbb and rxrg during zebrafish development. Gene Expr Patterns. 2006;6:556–65. doi: 10.1016/j.modgep.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Waskiewicz AJ, Rikhof HA, Hernandez RE, Moens CB. Zebrafish Meis functions to stabilize Pbx proteins and regulate hindbrain patterning. Development. 2001;128:4139–51. doi: 10.1242/dev.128.21.4139. [DOI] [PubMed] [Google Scholar]
- Weidinger G, Stebler J, Slanchev K, Dumstrei K, Wise C, Lovell-Badge R, Thisse C, Thisse B, Raz E. dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr Biol. 2003;13:1429–34. doi: 10.1016/s0960-9822(03)00537-2. [DOI] [PubMed] [Google Scholar]
- White JA, Guo YD, Baetz K, Beckett-Jones B, Bonasoro J, Hsu KE, Dilworth FJ, Jones G, Petkovich M. Identification of the retinoic acid-inducible all-trans-retinoic acid 4-hydroxylase. J Biol Chem. 1996;271:29922–7. doi: 10.1074/jbc.271.47.29922. [DOI] [PubMed] [Google Scholar]
- Woods IG, Wilson C, Friedlander B, Chang P, Reyes DK, Nix R, Kelly PD, Chu F, Postlethwait JH, Talbot WS. The zebrafish gene map defines ancestral vertebrate chromosomes. Genome Res. 2005;15:1307–14. doi: 10.1101/gr.4134305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiro K, Zhao X, Uehara M, Yamashita K, Nishijima M, Nishino J, Saijoh Y, Sakai Y, Hamada H. Regulation of retinoic acid distribution is required for proximodistal patterning and outgrowth of the developing mouse limb. Dev Cell. 2004;6:411–22. doi: 10.1016/s1534-5807(04)00062-0. [DOI] [PubMed] [Google Scholar]
- Zhang F, Nagy Kovacs E, Featherstone MS. Murine hoxd4 expression in the CNS requires multiple elements including a retinoic acid response element. Mech Dev. 2000;96:79–89. doi: 10.1016/s0925-4773(00)00377-4. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Dobbs-McAuliffe B, Linney E. Expression of cyp26b1 during zebrafish early development. Gene Expr Patterns. 2005;5:363–9. doi: 10.1016/j.modgep.2004.09.011. [DOI] [PubMed] [Google Scholar]