Abstract
We describe self-assembly of ssDNA brushes that exploits the intrinsic affinity of adenine nucleotides (dA) for gold surfaces. The grafting density and conformation of these brushes is deterministically controlled by the length of the anchoring dA sequences, even in the presence of thymine nucleotides (dT). We produce and characterize brushes of model block-oligonucleotides, d(Tm-An), with systematically varied lengths m and n of the thymine and adenine blocks [denoted d(Tm) and d(An), respectively]. The hairpin conformation, dominant for self-complementary d(Tm-An) oligos in solution, is disrupted by the high preferential affinity of dA for gold surfaces. As a result, the d(Tm-An) oligos adsorb as a brush of d(T) strands immobilized via the d(A) blocks. Quantitative analysis by FTIR spectroscopy and x-ray photoelectron spectroscopy (XPS) reveals a unique feature of DNA immobilization via d(A) blocks: The surface density of dA nucleotides is close to saturation and is nearly independent of d(A) block length. Accordingly, the lateral spacing (grafting density) of the d(T) blocks is determined by the length of the d(A) blocks. The d(T) blocks extend away from the surface in a brush-like conformation at a lateral spacing 2–3 times larger (a grafting density 5–10 times lower) than in analogous films immobilized via standard thiol linkers. This combination of brush-like conformation and low saturation grafting density is expected to increase the efficiency of DNA hybridization at surfaces. Therefore, immobilization via d(A) blocks offers a method of producing DNA brushes with controlled properties for applications in biotechnology and nanotechnology.
Keywords: FTIR, gold, immobilization, oligonucleotides, x-ray photoelectron spectroscopy
The properties of surfaces functionalized with ssDNA exhibit a remarkable richness that underlies the versatility of these surfaces in a wide range of applications. The ssDNA brushes described in this work offer unique properties for two types of applications: control of nanoscale self-assembly (1, 2) and design and operation of biosensors (3–8). In general, a critical attribute of a ssDNA-modified surface is efficient and reproducible hybridization with target DNA. Model studies using thiolated DNA probes have shown that efficient hybridization occurs when the spacing between immobilized DNA probes is large and the orientation of the probes is upright (4–11). Unfortunately, reproducible preparation of DNA films possessing both of these qualities remains challenging (12–14). For example, it is generally observed that when the grafting density is low (<1013 cm−2), i.e., the spacing between probes is comparable to their length, DNA immobilizes in a flat conformation through nonspecific adsorption (9, 15, 16). This observation can be largely explained by conventional polyelectrolyte brush theory, which predicts that negatively charged DNA strands should only assume upright conformations in densely packed films, where repulsive interactions force the strands to extend away from the surface (7).
Surface passivation is a common strategy used to decouple the DNA conformation from grafting density. Passivation prevents nonspecific interactions between the surface and DNA or other biomolecules, which enables widely spaced DNA probes to maintain an upright orientation. In a common implementation of this strategy, films of thiol-anchored DNA can be exposed to a solution containing a competing molecule, such as mercaptohexanol (MCH), which displaces most of the DNA from the surface and forces the remaining DNA strands into an upright conformation (10, 17–19). Alternatively, grafting densities can be adjusted by coupling functionalized DNA to a bifunctional self-assembled monolayer (6).
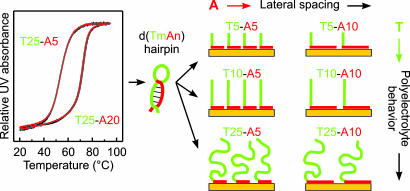
Our objective in this study is to control independently and deterministically both the conformation and grafting density of DNA. We realize this objective by introducing anchoring sequences of adenine nucleotides (dA), which in our previous work have been shown to have a high intrinsic affinity for gold surfaces (20). Here, the function of the adenine blocks [(dA)] extends, however, beyond simple anchoring: They preferentially bind to gold surfaces and block nearly all of the adsorption sites, preventing nonspecific binding of other sequences to these surfaces (an effect similar to that of using the MCH posttreatment or a bifunctional self-assembled monolayer). To demonstrate these characteristics, we use d(Tm-An) block-oligonucleotides [thymine d(T) and adenine d(A) blocks consisting of m and n nucleotides, respectively]. The d(Tm-An) oligos adopt the L-shaped conformation shown in Fig. 1: d(A) blocks (red) adsorb on the gold, and d(T) blocks (green) extend away from the surface. Quantitatively, the model in Fig. 1 predicts that the grafting density of oligos should be determined by the length of the d(A) blocks: The density decreases as the length of the d(A) blocks increases.
Fig. 1.
Idealized schematic of d(Tm-An) oligos in solution (hairpins) and after adsorption on gold (L-shape model). (Left) The UV absorbance at 260 nm for d(T25-An) hairpins was measured in 1 M CaCl2-TE buffer at 1°C per minute of heating (red lines) or cooling (black lines). (Right) In the L-shape model, the d(A) blocks (red) adsorb flat on the gold surface and the d(T) blocks (green) extend away from the surface. The surface density of dA nucleotides on gold is approximately independent of the d(A) length. Increasing the length of the d(A) block therefore decreases the grafting density of the oligos. Increasing the length of the d(T) block increases the polyelectrolyte effects in the brush layer.
The choice of using d(Tm-An) block-oligonucleotides as the model system may appear surprising in some respects. Foremost, the self-complementary d(Tm-An) sequences can form a number of hybrids, including hairpins (Fig. 1 Left), which may interfere with immobilization. Additionally, nonspecific nucleotide adsorption can be expected to dominate in the absence of linker groups for specific surface attachment (16–18, 20–22), and the immobilization efficiency should be suppressed for oligos longer than 30 nt (15). Despite these caveats, two factors actually make d(Tm-An) a convenient model system. First, adenine exhibits the highest affinity for gold compared with that of other nucleotides (20–22). Second, the unique spectroscopic signatures of dA and dT allow for unambiguous determination of the conformation and grafting density of d(Tm-An) oligos using FTIR spectroscopy and x-ray photoelectron spectroscopy (XPS) (16, 20, 23, 24).
Results
FTIR Results.
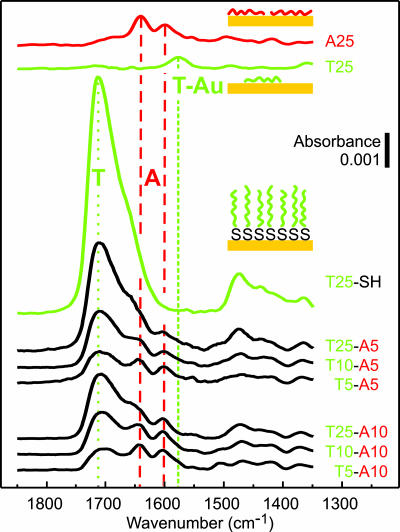
The features used to interpret the reflectance FTIR data from the d(Tm-An) oligo films on gold are identified in the reference spectra obtained from (dT)25 and (dA)25 homo-oligos and thiol-modified (dT)25-SH (top three spectra in Fig. 2). The features at ≈1,600 and ≈1,650 cm−1 in the (dA)25 spectrum (Fig. 2, red dashed lines) are characteristic of dA adsorbed on gold (16). Carbonyl features in the (dT)25 spectrum, located between 1,550 and 1,600 cm−1 (Fig. 2, short dash green line), are attributed to thymine bases chemisorbed on the gold substrate (16). The dominant carbonyl feature at ≈1,700 cm−1 in the (dT)25-SH spectrum is characteristic of thymine bases that do not directly interact with the gold surface (hereafter referred to as the “nonchemisorbed” dT feature) (Fig. 2, dotted green line) (16). The absence of the chemisorbed feature in the (dT)25-SH spectrum indicates that (dT)25-SH oligos anchor on gold via the thiol group and that few dT nucleotides directly adsorb on the gold.
Fig. 2.
FTIR spectra obtained from ssDNA adsorbed on gold. The adsorption conditions for the d(Tm-A5), d(Tm-A10), and (dT)25-SH oligos were 3 μM ssDNA in 1 M CaCl2-TE, pH 7, at 35°C for 2,400 min. Reference spectra are presented for (dA)25 and (dT)25 oligo films. The feature at ≈1,700 cm−1 (green dotted line) corresponds to C O groups in nonchemisorbed dT nucleotides. Features between 1,550 and 1,600 cm−1 (green short dash line) are attributed to chemisorbed dT nucleotides. Features at ≈1,600 and ≈1,650 cm−1 (red dashed lines) are associated primarily with dA nucleotides.
O groups in nonchemisorbed dT nucleotides. Features between 1,550 and 1,600 cm−1 (green short dash line) are attributed to chemisorbed dT nucleotides. Features at ≈1,600 and ≈1,650 cm−1 (red dashed lines) are associated primarily with dA nucleotides.
We observe four trends in the FTIR spectra obtained from the d(Tm-An) samples. First, absorbances of the dA features (Fig. 2, red dashed lines) for all six samples are very similar to those measured for the reference (dA)25 homo-oligo. Second, the chemisorbed dT features are small in each d(Tm-An) spectrum. Third, for a series of oligos with fixed d(A) block length [e.g., d(Tm-A5)], the intensity of the nonchemisorbed dT feature increases as the length of the d(T) block increases. Finally, fixing the length of the d(T) block and increasing the length of the d(A) block [e.g., from d(T10-A5) to d(T10-A10)] leads to a decrease in the absorbance of the nonchemisorbed dT feature.
XPS Results.
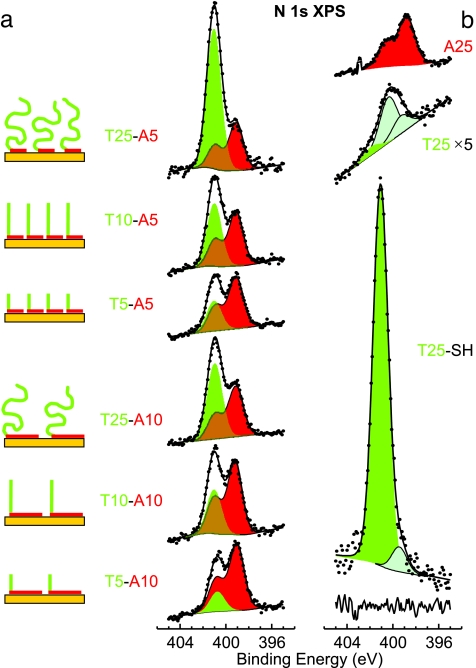
The features used to interpret the N 1s spectra from the d(Tm-An) oligos are identified in the reference spectra of (dT)25 and (dA)25 homo-oligos and thiol-modified (dT)25-SH (Fig. 3b). The two main features in the d(Tm-An) spectra in Fig. 3a are emphasized by shading. First, a single peak that has binding energy (BE) of ≈401 eV (Fig. 3, green shading), which corresponds to nonchemisorbed dT, using the terminology from the previous section [compare with the (dT)25-SH spectrum in Fig. 3b]. Second, a pair of peaks (BE ≈ 399 eV) (Fig. 3, red shading) that corresponds to a typical N 1s spectral envelope for dA [compare with the (dA)25 spectrum in Fig. 3b]. This N 1s spectral signature of dA is invariant with respect to the adsorption conditions and to the film structure; therefore, deconvolution of N 1s spectra can be simplified by fixing the dA spectral envelope. The reference spectra for (dT)25 and (dT)25-SH oligos also contain lower BE components associated with dT chemisorbed on gold (Fig. 3b, light-green shading with black outline).
Fig. 3.
XPS spectra of the N 1s region for ssDNA films adsorbed on gold. (a) d(Tm-A5) and d(Tm-A10) oligo series. (b) Reference spectra for (dA)25, (dT)25, and (dT)25-SH oligos. The adsorption conditions are analogous to those in Fig. 2. All spectra have been normalized to the Au 4f substrate peak intensities. Symbols, lines, and solid shading indicate data, fits, and fit components, respectively. The line at the bottom of b indicates a (dT)25-SH fit residual. Fit components are color-coded: The single green peak is ≈401 eV BE for dT, and the pair of red peaks are ≈399 eV BE for dA.
The spectra are shown normalized by the respective Au 4f substrate peak intensities, such that N 1s peak areas are approximately proportional to surface densities of nucleotides. dA contains five N atoms, whereas dT contains only two; therefore, the ratio between the areas of the dA (Fig. 3, red shading) and dT (Fig. 3, green shading) components is ≈2.5 for blocks of equal length [e.g., see the spectra for d(T5-A5) or d(T10-A10)].
Discussion
Conformation of Adsorbed d(Tm-An).
The FTIR and XPS results provide complementary evidence for the formation of brush layers where the d(A) blocks adsorb on the gold surface and the d(T) blocks extend away from the surface, as illustrated by the schematic diagrams in Figs. 1 and 3a. In each d(Tm-An) spectrum, the dominance of the nonchemisorbed dT feature (1,700 cm−1 in FTIR and 401 eV in XPS) and the absence of the chemisorbed dT features (1,550–1,600 cm−1 in FTIR and 398–399 eV in XPS) indicates that few thymine bases are attached to the gold surface; the d(T) blocks thus extend away from the substrate, leaving the d(A) blocks as the only means of surface attachment.
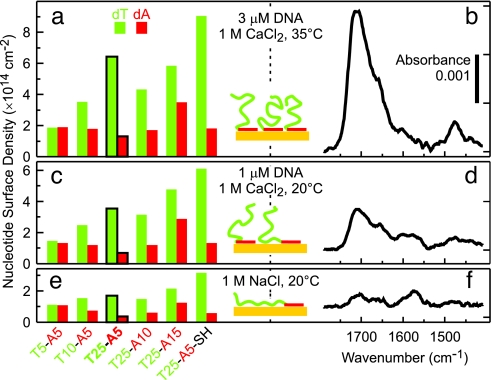
The observation that d(Tm-An) oligos adsorb in this fashion may seem surprising, especially because their self-complementary sequences allow for a variety of hybridized structures. In fact, the UV melting curves shown in Fig. 1 indicate that many of the d(Tm-An) oligos in solution have a folded hairpin conformation, which can be expected to impede immobilization via d(A) blocks. However, as we have previously shown, the dA–Au interactions are sufficiently strong to induce the denaturing of (dA)·(dT) hybrids (20). In control experiments summarized in Fig. 4, we clearly observe spectral features associated with chemisorbed dT for low d(Tm-An) grafting densities. The d(Tm-An) oligos therefore initially adsorb denatured and flat against the surface (Fig. 4 e and f) but ultimately adopt the L-shaped conformation (Figs. 3a and 4 a and b) as additional d(A) blocks compete for and adsorb onto the surface.
Fig. 4.
Saturation nucleotide coverages calculated from XPS data for d(Tm-An) films and corresponding FTIR spectra for d(T25-A5) after immobilization under different conditions. The following immobilization conditions were used in the three series of experiments: (a and b) 3 μM DNA in 1 M CaCl2-TE at 35°C for 2,400 min; (c and d) 1 μM DNA in 1M CaCl2-TE at 20°C for 1,200 min; and (e and f) 1 μM DNA in 1 M NaCl-TE at 20°C for 1,200 min.
Grafting Density of Short d(Tm-An).
The short d(Tm-An) oligos follow the model in Fig. 1, and the primary factor controlling their grafting density is the length of the d(A) block. The key assumption of this model is that the surface density of dA nucleotides on gold is largely independent of the d(A) block length. XPS measurements for (dA) homo-oligo films support this assumption: a 400% length increase between (dA)5 and (dA)25 results in an increase of surface density of dA nucleotides by only 20% (1.8 × 1013 vs. 2.2 × 1013 cm−2). Furthermore, the grafting density of d(T5-A5) and d(T10-A5) is nearly identical to that of (dA)5 (Fig. 5a), exactly as the model predicts. In the FTIR spectra (Fig. 2), for a fixed d(A) length, increasing the d(T) length from 5 to 10 nt results in a doubling of the dT feature and no measurable change in the dA feature, in agreement with the model prediction that the grafting density is constant when the length of d(A) is fixed. For a fixed d(T) length, increasing the d(A) length from 5 to 10 nt leads to a decrease in the absorbance of the dT feature in the FTIR spectra in Fig. 2, consistent with the grafting density decrease of ≈35% as measured by XPS (Fig. 5a).
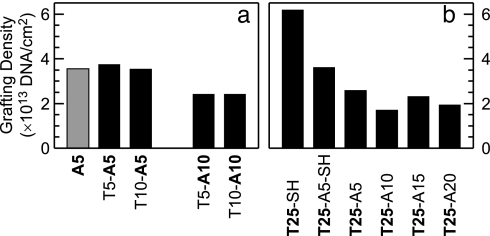
Fig. 5.
DNA grafting densities calculated from XPS data for as-deposited films of d(Tm-An) oligos. (a) Comparison of grafting densities for short oligos d(T5-A5), d(T10-A5), d(T5-A10), and d(T10-A10). The reference value for a (dA)5 film is shown shaded in gray. (b) Comparison of d(T25) grafting densities obtained by using different attachment strategies. Our model predicts that the grafting density should decrease as the number of nucleotides in the d(A) block increases. Note that higher than predicted grafting densities measured for the as-deposited d(T25-A15) and d(T25-A20) samples are attributed to (dA)·(dT) hybrid formation (see Fig. 6 and text for discussion).
The Grafting Density of d(T25)-X.
Fig. 5b shows the effect of increasing the length of the anchoring group, X [X = −SH or d(An)], on the grafting density of brushes of the longest d(T) blocks in our study [d(T25)]. Thiol linkers have slightly higher affinity for gold than does dA, so the two thiolated oligos [(dT)25-SH and d(T25-A5)-SH] form the highest density brushes. An analogous trend is observed between (dA)n-SH and (dA)n surface densities [supporting information (SI) Fig. 10].
For a d(Tm-A5) or d(Tm-A10) series, the quantitative prediction of the L-shape model is that grafting density should be independent of d(Tm) length. A comparison between Fig. 5 b and a shows that the grafting density is actually ≈30% lower for d(T25-A5) oligos than for the shorter d(T5-A5) and d(T10-A5). This trend is also observed in the d(Tm-A10) series. We attribute the effect to the larger electrostatic and entropic forces that must be overcome to uncoil and pack the longer d(T25) brush strands (Fig. 1), similar to the trend observed for films of thiolated DNA (15, 19, 24). As the d(T) length increases, the saturation grafting density is determined not solely by the length of the d(A) blocks, but also by the polyelectrolyte effects in the d(T) brush.
As expected for polyelectrolyte brushes, the saturation grafting densities of d(Tm-An) oligos strongly depend on the immobilization conditions (Fig. 4). The main factor limiting surface densities appears to be the barrier for DNA to diffuse from solution through the negatively charged DNA brush to the gold surface (4, 5, 7). Immobilization of oligos with the longest d(T) blocks is therefore very sensitive to the ionic strength of the solution. For example, when d(T25-A5) is immobilized in 1 M CaCl2-TE buffer, the saturation surface density is approximately double that achieved in 1 M NaCl-TE. We attribute this difference to the more effective electrostatic screening in buffers with higher ionic strength (25), which lowers the repulsive barrier for the molecules diffusing toward the surface. In NaCl-TE buffer, the repulsion created by the initial adsorption of both d(A) and d(T) blocks on the surface (note the chemisorbed dT feature at ≈1,550 cm−1) is sufficient to inhibit further immobilization, whereas, in CaCl2-TE buffer, a quantity of oligos sufficient for creating a brush can diffuse and attach to the surface. In other words, repulsion between long d(T) “tails” and immobilized DNA inhibits the ability of d(A) blocks to diffuse to the surface in buffers with low ionic strength. In addition to providing the enhanced screening, the high concentration of divalent Ca2+ counterions in 1 M CaCl2-TE buffer may help to stabilize the extended brush configuration indicated in Fig. 4 a–d (25).
Increasing the temperature to 35°C enhances diffusion over the electrostatic barrier both thermodynamically and by exciting conformational fluctuations of the d(T) blocks (thereby lowering the average barrier). Agitating the solution during immobilization does not change the resulting surface density, indicating that the increase measured at 35°C is not due to convective mixing. The UV data in Fig. 1 clearly show that 35°C is still below the melting transition for the d(T25-A5) hairpins, indicating that their stability is not directly affected by such a small increase of temperature. Independently increasing the DNA concentration also increases the surface density, but the effect is smaller than that of temperature (data not shown).
For the d(T25-An) series, the quantitative prediction of our model is that the oligos with the longest d(A) blocks, d(T25-A15) and d(T25-A20), should have the lowest grafting densities. The grafting densities in Fig. 5b for the as-deposited films of these two sequences, however, are actually as high as those for the d(T25-A5) and d(T25-A10) films, respectively. We believe that these apparently high d(T25-A15) and d(T25-A20) grafting densities are due to the presence of hybrids in as-deposited films (Fig. 6b). Our ability to observe such hybrids by ex situ spectroscopy depends on their stability in the postdeposition stringency rinse, which in turn is determined by the lengths of the d(A) and d(T) blocks. Hybrids of d(Tm-An) oligos with dT and dA blocks of ≤10 nt are likely to be disrupted by the deionized water rinse because they have melting points below room temperature at low salt concentration. In contrast, the d(T25-A15) and d(T25-A20) hybrids with overlaps up to 15 and 20 nt, respectively, may be stable enough to survive the rinse.
Fig. 6.
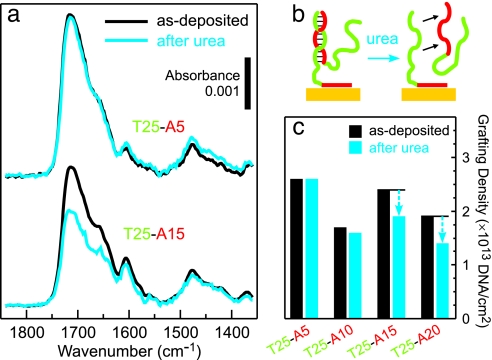
Detecting coadsorbed d(T25-An) hybrids. (a and c) FTIR spectra (a) and XPS grafting densities (c) obtained for d(T25-An) brushes before (black) and after (cyan) 30 min of exposure to an 8 M urea denaturing solution. (b) Schematic of a possible conformation of a d(T25-A15) partial hybrid. The adsorption conditions were 3 μM ssDNA in 1 M CaCl2-TE at 35°C for 2,400 min.
To test for the presence of d(Tm-An) hybrids, a set of d(T25-An) samples was soaked in an 8 M urea denaturing solution for 30 min. After the urea treatment, the d(T25-A15) and d(T25-A20) films showed nucleotide losses of ≈20% and ≈25%, respectively (Fig. 6c). In contrast, d(T25-A5) and d(T25-A10) films were unaffected by the denaturing solution, indicating that hybrids had not been present in these two samples after the deionized water rinse (Fig. 6 a and c). Control experiments confirmed that (dA)15 oligos could be hybridized to an immobilized d(T25-An) film (Fig. 7b and c) and subsequently removed by soaking the film in urea (Fig. 7d). The results of the experiments shown in Figs. 6 and 7 strongly support the interpretation that as-deposited d(T25-A15) and d(T25-A20) films in Fig. 5b consist of anchored d(Tm-An) oligos and coadsorbed hybrids, which leads to an apparent grafting density higher than that of the shorter oligos.
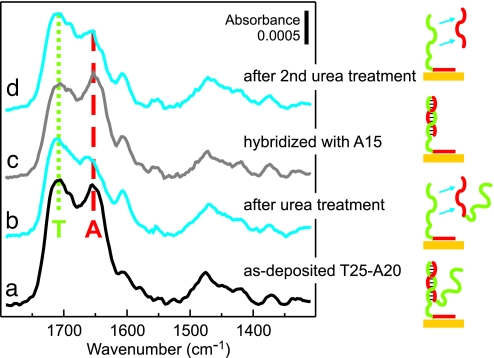
Fig. 7.
Hybridization properties of d(T25-A20) films. FTIR spectra for d(T25-A20) films as deposited (a), after a 30-min soak in 8 M urea denaturing solution (cyan) (b), after a subsequent overnight hybridization with 1 μM (dA)15 in 1 M CaCl2-TE buffer (gray) (c), and after a second 30-min soak in 8 M urea (cyan) (d). Spectral features primarily attributed to dT and dA nucleotides are indicated by green dotted and red dashed lines, respectively.
After removal of the coadsorbed hybrids by urea, the d(T25-A20) film has the lowest grafting density, as predicted by the model. Importantly, we observe that the spectrum of d(T25-A20) contains no evidence of chemisorbed dT. In contrast, at comparably low grafting densities of thiol-modified (dT)25-SH oligos, many d(T) strands are nonspecifically adsorbed on the substrate (16). Thus, the minimal grafting density at which d(T) brush strands can maintain an upright orientation is lower for oligos anchored via d(A) blocks than for oligos immobilized via thiol linkers. Presumably, the near-saturation surface density of dA nucleotides blocks most surface adsorption sites, thereby preventing nonspecific interactions between dT nucleotides and gold. Additionally, the dA-covered surface may retain a negative charge, which would further promote the extending of d(T) blocks into solution.
Practical Considerations.
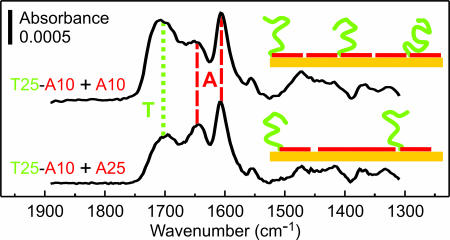
The lowest “saturation” grafting density we measured was ≈1013 cm−2 [for d(T25-A20)]. Achieving lower grafting densities apparently requires the use of oligos having d(A) blocks significantly longer than 25 nt, which may be impractical. An alternative method, which we have explored, is coadsorption from a mixture of (dA)n and d(Tm-An) oligos, whereby (dA)n homo-oligos compete for adsorption sites and ultimately act as lateral spacers, as shown in the inset diagrams of Fig. 8. The FTIR data presented in Fig. 8 support the feasibility of this strategy. Whereas the dA features are nearly identical for the two films in Fig. 8, the dT feature is smaller for the film codeposited with the longer (dA)25 oligo, consistent with the (dA)n oligos acting as lateral spacers. Additionally, the absorbance of the dT feature (green dotted line) for both coadsorbed films in Fig. 8 is smaller than for d(T25-A20) in Fig. 7, indicating that we produced brushes having grafting density <1013 cm−2, considered optimal for hybridization assays (3–5, 9).
Fig. 8.
Using (dA)n as lateral spacers in d(T25-A10) films. Oligos were coadsorbed on gold from solution mixtures [1 μM d(T25-A10) plus 1 μM (dA)n in 1 M CaCl2-TE at 35°C for 2,400 min], and the resulting films were soaked in 8 M urea denaturing solution before obtaining FTIR spectra. Green dotted and red dashed lines denote positions of spectral features attributed primarily to dT and dA nucleotides, respectively.
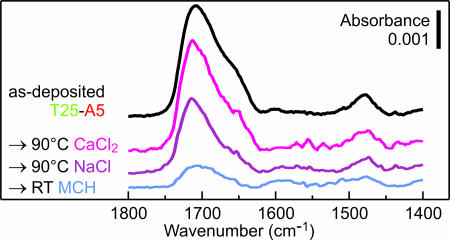
To further demonstrate the practicality of the d(A) attachment, we have examined the stability of d(T25-A5) brushes at elevated temperature and against displacement by MCH (Fig. 9). Attachment via a short d(A) block produces a grafting density similar to that observed in typical brushes of thiolated ssDNA; therefore, the stability of the two attachment mechanisms can be directly compared by using the d(T25-A5) as a model. The losses of DNA from the surface in buffers at 90°C in the case of the d(A) attachment were comparable or smaller than those reported for the thiol attachment (1, 13, 14). We attribute this stability to the effect of multipoint attachment to the gold, similar to the enhanced stability of attachment via multiple vs. single thiol groups (13, 14). The lower thermal loss in CaCl2-TE compared with NaCl-TE likely results from enhanced electrostatic screening typically observed in buffers with high ionic strength (25). Exposure to MCH is clearly the most effective method of disrupting the d(A) attachment, consistent with the higher affinity to gold of MCH compared with that of dA. But even against the rather aggressive displacement by small thiolated molecules, the d(A) attachment is at least as stable as attachment via thiol linkers (8, 13, 17).
Fig. 9.
Stability of the d(A) attachment in aggressive environments. FTIR spectra of d(T25-A5) films before (black) and after (colors) exposure to 90°C for 30 min in CaCl2-TE buffer (magenta) or NaCl-TE buffer (purple), or in 1 mM MCH solution at room temperature for 1 min (blue).
Conclusions
For all of the d(Tm-An) compositions studied, the FTIR and XPS results support a model in which the d(A) blocks preferentially adsorb n the gold surface and the d(T) blocks extend away from the substrate. For short d(Tm-An) oligos, the immobilization behavior is almost exclusively controlled by the d(A) block; the surface density of dA nucleotides remains approximately constant and is close to that observed for (dA)n homo-oligos on gold. For d(Tm-An) oligos with long d(T) blocks, the control of grafting density switches to the polyelectrolyte behavior limit. The polyelectrolyte behavior is most pronounced for the d(T25-A5) oligos, in which the d(A) block is considerably shorter than the d(T) block. Remarkably, even when stable hybrids can form between block-oligos, e.g., for d(T25-A20), attachment still occurs primarily through the dA nucleotides.
Attachment of ssDNA to gold surfaces via d(A) blocks has a number of potential advantages over using thiol linkers. Omitting the thiol-modification step in the DNA synthesis lowers the cost and avoids practical problems, e.g., disulfide formation and sample contamination (26). The dA component effectively blocks most DNA–Au binding sites; thus, the functionalized surfaces are inherently resistant to the nonspecific adsorption of DNA. In addition, the probe portions of the oligos attached via d(A) blocks are forced away from the substrate and therefore maintain upright brush-like conformations at grafting densities significantly lower than do thiolated ssDNA probes. Tuning the grafting density by simply changing the length of the d(A) attachment block introduces the possibility of controlling the effects of repulsion between anionic sugar–phosphate backbones (4, 5).
Direct and indirect effects of the strong interactions between oligo(dA) and gold have been observed on different types of gold substrates and by using several different techniques (1, 20–22, 27–29). A full explanation of these surprising observations will require an understanding of both the structure of oligo(dA) and d(A)–Au interactions in ionic solutions; note that the behavior of adenine bases on gold in vacuum (30–32) does not directly correlate with solution-phase observations. In addition, investigating the attachment of DNA strands of varying composition and length will be required to determine the generality of the d(A) block anchoring method.
Finally, we note that the differences in adsorption properties of the DNA bases could be exploited to form other interesting DNA structures on gold, for example, by using d(AxTyAz) oligos to create surface loops or d(TxAyTz) oligos to generate brushes with pairs of strands at a fixed distance. This type of immobilization strategy expands the potential for using DNA as a molecular-scale building material to generate unique, ordered structures on surfaces.
Materials and Methods
Materials.
Commercial custom oligonucleotides were synthesized and HPLC-purified by the vendor and used as-received without further purification. The d(Tm-An) oligos [Tm-An in the figures] are written in the 3′-to-5′ direction, where m and n are the number of nucleotides in the d(T) and the d(A) blocks, respectively. The 5′ thiol-modified oligonucleotides were used without removing the protective S-(CH2)6OH group from the 5′ end. For brevity, these oligos are referred to as (dT)25-SH, (dA)5-SH, (dA)25-SH, and d(T25-A5)-SH. Buffer solutions denoted as NaCl-TE and CaCl2-TE contained 1 M NaCl or 1 M CaCl2, respectively (1× TE denotes a mixture of 10 mM Tris·HCl and 1 mM EDTA adjusted to pH 7 with HCl).
Preparation of DNA Films on Gold Substrates.
Polycrystalline gold substrates on Si(100) wafers were prepared as described previously (16). Substrates were cleaned with piranha solution (70% H2SO4/30% H2O2 in H2O) and rinsed thoroughly with deionized water (18.3 MΩ) immediately before adsorption of DNA. Note that piranha solution is extremely oxidizing, reacts violently with organics, and should be stored in loosely covered containers to avoid pressure buildup. For the d(Tm-An) deposition, 1-cm2 gold substrates were immersed for 2,400 min in 2 ml of 3 μM DNA in CaCl2-TE buffer at 35°C, which were the conditions we found to produce the highest density films. Reference (dA)25 and (dT)25 films were deposited from 1 μM ssDNA solutions in CaCl2-TE buffer at 20°C for 1,200 min. After deposition, each sample was rinsed with deionized water to remove salts and loosely bound DNA and then blown dry under flowing nitrogen.
XPS Analysis.
Quantitative analysis of DNA films on gold by XPS is described in detail in refs. 16, 23, and 33. XPS measurements were performed by using a commercial spectrometer equipped with a monochromatic Al Kα source, hemispherical electron energy analyzer (58° angle between monochromator and analyzer), and magnetic electron lens (23). Normal emission angle-integrated scans with 15- to 20-eV windows and 20-eV pass energy (0.36-eV nominal analyzer energy resolution) were acquired in the Au 4f and 4d, N 1s, P 2p, C 1s, and O 1s regions.
N 1s Peak Parameters.
The main dT peak BE value was 401.1 eV, and the full width at half-maximum was 1.6 eV. For a pair of dA peaks, the BE values were 398.8 and 400.6 eV with an intensity ratio of 2:1 and a full width at half-maximum of 1.6 eV. The XPS data from d(Tm-An) films were fit by using a single peak, with BE ≈ 401 eV for the dT component, and a pair of peaks, with BE ≈ 399 eV (fixed 1.8 eV BE difference and 2:1 intensity ratio) for the dA component. The intensity ratio of the dT and dA components was fixed in each fit based on the dT:dA (m:n) ratio for the corresponding d(Tm-An) oligo.
DNA surface densities were determined by using the method from ref. 23 modified for a two-component system (SI Fig. 11). The measured peak intensities for DNA elements and the gold substrate (bare and with DNA film) were analyzed for each sample by using a uniform overlayer model, which included attenuation of photoelectrons (23). DNA grafting density was calculated from the N surface density (amount of substance) and known stoichiometry of d(Tm-An) oligos.
FTIR Measurements.
Infrared reflection absorption spectra were obtained by using an FTIR spectrometer equipped with a wire grid infrared polarizer (p-polarized) and a variable angle specular reflectance accessory (75° grazing incidence angle) as described previously (16). FTIR measurements were performed in a nitrogen-purged environment using freshly prepared samples and a gold substrate cleaned with piranha solution as a reference.
Supplementary Material
Acknowledgments
We thank Dr. T. D. Clark for discussions of interactions in DNA films, Ms. C. L. Cole for advice on DNA hybridization, and Dr. P. Vallone for assistance with melting experiments. The research done at the Naval Research Laboratory was supported by the Office of Naval Research and the Air Force Office of Scientific Research. A.O. was a National Research Council Postdoctoral Associate at the National Institute of Standards and Technology.
Abbreviations
- dT
thymine nucleotide
- dA
adenine nucleotide
- MCH
mercaptohexanol
- XPS
x-ray photoelectron spectroscopy
- BE
binding energy.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0608568103/DC1.
References
- 1.Dillenback LM, Goodrich GP, Keating CD. Nano Lett. 2006;6:16–23. doi: 10.1021/nl0508873. [DOI] [PubMed] [Google Scholar]
- 2.Pinto YY, Le JD, Seeman NC, Musier-Forsyth K, Taton TA, Kiehl RA. Nano Lett. 2005;5:2399–2402. doi: 10.1021/nl0515495. [DOI] [PubMed] [Google Scholar]
- 3.Nelson BP, Grimsrud TE, Liles MR, Goodman RM, Corn RM. Anal Chem. 2001;73:1–7. doi: 10.1021/ac0010431. [DOI] [PubMed] [Google Scholar]
- 4.Vainrub A, Pettitt BM. J Am Chem Soc. 2003;125:7798–7799. doi: 10.1021/ja035020q. [DOI] [PubMed] [Google Scholar]
- 5.Hagan MF, Chakraborty AK. J Chem Phys. 2004;120:4958–4968. doi: 10.1063/1.1645786. [DOI] [PubMed] [Google Scholar]
- 6.Ladd J, Boozer C, Yu QM, Chen SF, Homola J, Jiang S. Langmuir. 2004;20:8090–8095. doi: 10.1021/la049867r. [DOI] [PubMed] [Google Scholar]
- 7.Halperin A, Buhot A, Zhulina EB. Biophys J. 2005;89:796–811. doi: 10.1529/biophysj.105.063479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thaxton CS, Hill HD, Georganopoulou DG, Stoeva SI, Mirkin CA. Anal Chem. 2005;77:8174–8178. doi: 10.1021/ac0514265. [DOI] [PubMed] [Google Scholar]
- 9.Peterson AW, Heaton RJ, Georgiadis RM. Nucleic Acids Res. 2001;29:5163–5168. doi: 10.1093/nar/29.24.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong ELS, Chow E, Gooding JJ. Langmuir. 2005;21:6957–6965. doi: 10.1021/la050725m. [DOI] [PubMed] [Google Scholar]
- 11.Gong P, Lee CY, Gamble LJ, Castner DG, Grainger DW. Anal Chem. 2006;78:3326–3334. doi: 10.1021/ac052137j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterson AW, Wolf LK, Georgiadis RM. J Am Chem Soc. 2002;124:14601–14607. doi: 10.1021/ja0279996. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Jin RC, Mirkin CA, Letsinger RL. Nucleic Acids Res. 2002;30:1558–1562. doi: 10.1093/nar/30.7.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson PA, Levicky R. Langmuir. 2003;19:10288–10294. [Google Scholar]
- 15.Steel AB, Levicky RL, Herne TM, Tarlov MJ. Biophys J. 2000;79:975–981. doi: 10.1016/S0006-3495(00)76351-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrovykh DY, Kimura-Suda H, Whitman LJ, Tarlov MJ. J Am Chem Soc. 2003;125:5219–5226. doi: 10.1021/ja029450c. [DOI] [PubMed] [Google Scholar]
- 17.Herne TM, Tarlov MJ. J Am Chem Soc. 1997;119:8916–8920. [Google Scholar]
- 18.Levicky R, Herne TM, Tarlov MJ, Satija SK. J Am Chem Soc. 1998;120:9787–9792. [Google Scholar]
- 19.Rant U, Arinaga K, Fujita S, Yokoyama N, Abstreiter G, Tornow M. Langmuir. 2004;20:10086–10092. doi: 10.1021/la0492963. [DOI] [PubMed] [Google Scholar]
- 20.Kimura-Suda H, Petrovykh DY, Tarlov MJ, Whitman LJ. J Am Chem Soc. 2003;125:9014–9015. doi: 10.1021/ja035756n. [DOI] [PubMed] [Google Scholar]
- 21.Wolf LK, Gao Y, Georgiadis RM. Langmuir. 2004;20:3357–3361. doi: 10.1021/la036125+. [DOI] [PubMed] [Google Scholar]
- 22.Storhoff JJ, Elghanian R, Mirkin CA, Letsinger RL. Langmuir. 2002;18:6666–6670. [Google Scholar]
- 23.Petrovykh DY, Kimura-Suda H, Tarlov MJ, Whitman LJ. Langmuir. 2004;20:429–440. doi: 10.1021/la034944o. [DOI] [PubMed] [Google Scholar]
- 24.Petrovykh DY, Perez-Dieste V, Opdahl A, Kimura-Suda H, Sullivan JM, Tarlov MJ, Himpsel FJ, Whitman LJ. J Am Chem Soc. 2006;128:2–3. doi: 10.1021/ja052443e. [DOI] [PubMed] [Google Scholar]
- 25.Rant U, Arinaga K, Fujiwara T, Fujita S, Tornow M, Yokoyama N, Abstreiter G. Biophys J. 2003;85:3858–3864. doi: 10.1016/S0006-3495(03)74800-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee CY, Canavan HE, Gamble LJ, Castner DG. Langmuir. 2005;21:5134–5141. doi: 10.1021/la0472302. [DOI] [PubMed] [Google Scholar]
- 27.Demers LM, Mirkin CA, Mucic RC, Reynolds RA, Letsinger RL, Elghanian R, Viswanadham G. Anal Chem. 2000;72:5535–5541. doi: 10.1021/ac0006627. [DOI] [PubMed] [Google Scholar]
- 28.Anne A, Bouchardon A, Moiroux J. J Am Chem Soc. 2003;125:1112–1113. doi: 10.1021/ja028640k. [DOI] [PubMed] [Google Scholar]
- 29.Ray SG, Cohen H, Naaman R, Rabin Y. J Am Chem Soc. 2005;127:17138–17139. doi: 10.1021/ja055201n. [DOI] [PubMed] [Google Scholar]
- 30.Demers LM, Östblom M, Zhang H, Jang NH., Liedberg B, Mirkin CA. J Am Chem Soc. 2002;124:11248–11249. doi: 10.1021/ja0265355. [DOI] [PubMed] [Google Scholar]
- 31.Östblom M, Liedberg B, Demers LM, Mirkin CA. J Phys Chem B. 2005;109:15150–15160. doi: 10.1021/jp051617b. [DOI] [PubMed] [Google Scholar]
- 32.Rapino S, Zerbetto F. Langmuir. 2005;21:2512–2518. doi: 10.1021/la047091o. [DOI] [PubMed] [Google Scholar]
- 33.Petrovykh DY, Kimura-Suda H, Opdahl A, Richter LJ, Tarlov MJ, Whitman LJ. Langmuir. 2006;22:2578–2587. doi: 10.1021/la050928a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.