Abstract
Listeria monocytogenes is a human intracellular pathogen that is able to survive in the gastrointestinal environment and replicate in macrophages, thus bypassing the early innate immune defenses. Peptidoglycan (PG) is an essential component of the bacterial cell wall readily exposed to the host and, thus, an important target for the innate immune system. Characterization of the PG from L. monocytogenes demonstrated deacetylation of N-acetylglucosamine residues. We identified a PG N-deacetylase gene, pgdA, in L. monocytogenes genome sequence. Inactivation of pgdA revealed the key role of this PG modification in bacterial virulence because the mutant was extremely sensitive to the bacteriolytic activity of lysozyme, and growth was severely impaired after oral and i.v. inoculations. Within macrophage vacuoles, the mutant was rapidly destroyed and induced a massive IFN-β response in a TLR2 and Nod1-dependent manner. Together, these results reveal that PG N-deacetylation is a highly efficient mechanism used by Listeria to evade innate host defenses. The presence of deacetylase genes in other pathogenic bacteria indicates that PG N-deacetylation could be a general mechanism used by bacteria to evade the host innate immune system.
Keywords: cytokine, macrophage, pathogenesis, virulence, cell wall
The innate immune system is central for the early recognition and clearance of pathogens. Hence, a number of microbial pathogens have developed sophisticated strategies to evade or modulate the host response to their advantage. Listeria monocytogenes is an invasive pathogen that survives in the harsh gastrointestinal lumen and reaches the submucosa without inducing a vigorous inflammatory response. However, the mechanisms used by L. monocytogenes to survive during the very early steps of the infection are poorly understood. This bacterium then disseminates to various organs by spreading from cell to cell and by surviving in phagocytic cells such as macrophages. The ability of L. monocytogenes to internalize and escape into the cytosol requires key virulence factors such as the secreted toxin listeriolysin O (1), the surface protein ActA (2), and Internalin (3), which is anchored to its peptidoglycan (PG) (4), or InlB (5), which is attached to its lipoteichoic acids (ref. 6; for reviews see refs. 7 and 8). Hence, the cell wall of L. monocytogenes plays a central role in its virulence. Recently, the role of PG hydrolases in Listeria virulence has also been described and suggested to be important during infection (9, 10). PG is the pathogen-associated molecular pattern recognized by the innate immune system by some of the recently discovered intracellular pattern-recognition receptors such as the nucleotide-binding oligomerization domain (Nod) proteins (11). Recent work has reported a role for Nod1, Nod2 and Toll-like receptor (TLR)-2 in innate immune recognition of Listeria (12–15). However, little is known regarding the composition and structure of L. monocytogenes PG except that it is a mesodiaminopimelic acid containing PG and that N-acetylglucosamine residues are partially N-deacetylated into glucosamine (16). Whether these specific features play a role in infection has never been investigated.
Here, we report that N-deacetylation is a major modification of Listeria PG, conferring to this human pathogen the ability to evade the innate immune system. The inactivation of the single PG N-deacetylase of Listeria highlights the central role of this modification in Listeria virulence, through survival in the gastrointestinal track, in professional phagocytes, evasion to the action of host lysozyme, and modulation of inflammatory response.
Results and Discussion
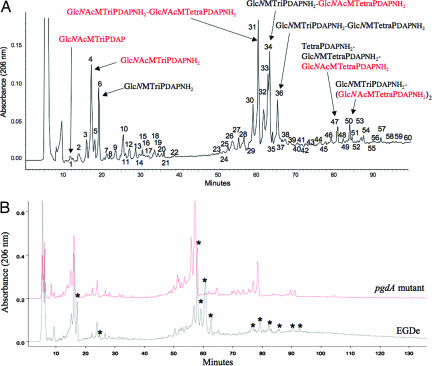
To address the role of Listeria PG in virulence, we first prepared highly purified PG from L. monocytogenes strain EGDe and analyzed its muropeptide composition by reverse-phase HPLC (Fig. 1A). Mass spectrometry analysis of each peak revealed that L. monocytogenes has several unusual modifications of its PG (Fig. 1A). Approximately 50% of its muropeptides featured a glucosamine residue instead of the canonical N-acetylglucosamine, definitively establishing that Listeria partially N-deacetylates its PG. Interestingly, PG N-deacetylation has been described as conferring resistance to host lysozyme (17), a major innate defense mechanism against bacterial infections.
Fig. 1.
Characterization of Listeria PG. (A) Each muropeptide peak highlighted by a number was purified by HPLC, desalted, and analyzed by MALDI-TOF. Structural assignment was done by muropeptide fragmentation using MALDI-postsource decay (PSD). The structure of major muropeptides is indicated by full arrows. Structures or substructures in red and black indicate fully acetylated and N-deacetylated moieties, respectively, of the different muropeptides. Peaks 1–22 correspond to monomeric muropeptides, peaks 23- 44 correspond to dimeric muropeptides, and 46 and over correspond to trimeric muropeptides. Approximately 50% of the muropeptides present a glucosamine residue instead of the canonical N-acetylglucosamine residue. (B) HPLC analysis of the muropeptide composition of Listeria WT EGDe strain and its pgdA isogenic mutant. Each muropeptide peak was purified by HPLC and analyzed by MALDI-PSD mass spectrometry. Muropeptide peaks indicated with an asterisk correspond to N-deacetylated muropeptides. N-deacetylated muropeptides characteristic of the parental strain EGDe were completely absent from the elution pattern of the pgdA mutant. GlcNAc, N-acetylglucosamine; GlcN, glucosamine; M, N-acetylmuramic acid, TriPDAP, l-alanyl-γ-d-glutamyl-mesodiaminopimelic acid; TriPDAPNH2, l-alanyl-γ-d-glutamyl-amidated mesodiaminopimelic acid; TetraPDAP, l-alanyl-γ-d-glutamyl-mesodiaminopimelyl-d-alanine; TetraPDAPNH2, l-alanyl-γ-d-glutamyl-amidated mesodiaminopimelyl-d-alanine.
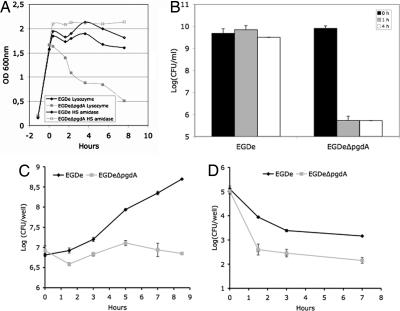
Analysis of the L. monocytogenes genome allowed identification of a gene encoding a putative PG deacetylase (lmo0415), the only putative PG N-deacetylase that could be identified. A deletion mutant of lmo0415 was generated. Analysis of its PG confirmed that lmo0415, renamed pgdA, encodes the unique PG N-deacetylase of L. monocytogenes because all peaks corresponding to N-deacetylated muropeptides disappeared from the HPLC pattern (Fig. 1B). As expected, the pgdA mutant showed sensitivity to lysozyme, resulting in a five-log decrease in viability compared with the parental strain EGDe in the presence of lysozyme (Fig. 2). Interestingly, the sensitivity of the pgdA mutant was specific for lysozyme, because the human serum amidase, another PG hydrolase produced by the host, had no bacteriolytic effect on the pgdA mutant (Fig. 2A). Also, the mutant was perfectly capable of growing in BHI broth and serum, indicating that pgdA inactivation had no effect on its growth [Fig. 2A and supporting information (SI) Fig. 6] or morphology (SI Fig. 7). Sensitivity to lysozyme was observed only in stationary phase and resulted in cell rounding (SI Fig. 7).
Fig. 2.
Effect of lysozyme on growth and impaired survival in macrophages of the pgdA mutant. Strain EGDe and its isogenic pgdA mutant were grown in BHI media and incubated with lysozyme (10 μg/ml) or the human serum amidase (1 μg/ml). (A) The pgdA mutant was selectively sensitive to the action of lysozyme upon entry into stationary phase. (B) Decrease of the optical density of the pgdA mutant correlated with a loss of viability, whereas the parental strain was insensitive to lysozyme. Lysozyme induced cell rounding of the pgdA mutant (see SI Fig. 7). (C and D) RAW264.7 macrophages (C) and PEM (D) were infected with WT EGDe and its pgdA mutant. Sensitivity of the pgdA mutant to lysozyme correlated with its impaired survival in macrophages.
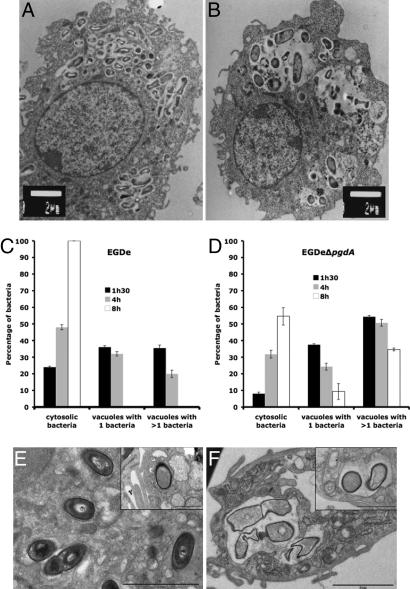
As shown above, the pgdA mutant is extremely sensitive to host lysozyme. Because lysozyme is particularly abundant in macrophages, to evaluate the possible impact of N-deacetylation on infection, we compared the ability of the pgdA mutant and that of strain EGDe to infect and survive in macrophages (Fig. 2 C and D). The pgdA mutant was severely impaired in its ability to survive and multiply in RAW264.7 macrophages (Fig. 2C), in peptone-elicited peritoneal macrophages (PEM) from C57/BL6J mice (Fig. 2D) and in bone marrow-derived macrophages from C57/BL6J mice (data not shown). For example, after 8 h of infection of RAW264.7 macrophages, the difference in viable bacteria between the pgdA mutant and strain EGDe was 1.5 log. Next, infected macrophages were observed by EM. Fig. 3 A and B illustrates EM images of macrophages infected with strain EGDe and the pgdA mutant, respectively, after 8 h of infection (see also Fig. 3 E and F). Whereas strain EGDe was present exclusively in the macrophage cytosol, the pgdA mutant accumulated in vacuoles, preventing its cytosolic multiplication. EM observations of infected macrophages at various time points after infection showed that the pgdA mutant was mainly detected in vacuoles compared with the parental strain EGDe (Fig. 3 C and D), suggesting an early intracellular killing. Strikingly, the pgdA mutant was found frequently in large vacuoles with multiple bacteria or bacterial debris (Fig. 3 A, B, E, and F). We also infected nonphagocytic Caco 2 epithelial cells with strain EGDe and its pgdA mutant. The pgdA mutant was not impaired in its ability to be internalized (data not shown), indicating that the pgdA mutant was not generally affected in bacterial stability. Moreover, the pgdA mutant produced normal amounts in listeriolysin O, showing that impaired escape from phagosomes in macrophages is not due to a defect of listeriolysin O production (data not shown).
Fig. 3.
Impaired survival of the pgdA mutant in macrophages. (A–D) RAW264.7 cells after 8 h of infection with the parental strain EGDe (A) and the pgdA mutant (B). Impaired survival was correlated with delay in escape of the pgdA mutant (C) from phagosomes compared with the WT strain EGDe (D). (E and F) PEM after 7 h of infection with the parental strain EGDe (E) and the pgdA mutant (F). [Scale bars: 2 μm and 1 μm (Insets).] Impaired survival correlated with delay in escape from phagosomes and bacterial lysis of the pgdA mutant compared with the WT strain EGDe.
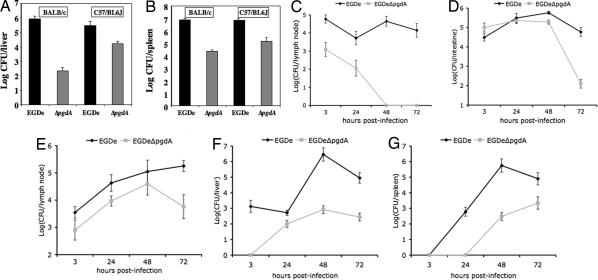
Virulence and dissemination of Listeria in its host strongly relies on survival in macrophages, as illustrated by the very strong attenuation of the listeriolysin O mutant (1). We thus performed in vivo challenge of BALB/c and C57/BL6J mice by the i.v. route with strain EGDe and the pgdA mutant. In both mouse backgrounds, the pgdA mutant was severely attenuated, with LD50s of 1.7 × 106 and 1.1 × 108 bacteria in C57/BL6J and BALB/c mice, respectively, compared with 2.3 × 104 and 1.7 × 104 bacteria, respectively, for the parental strain EGDe. Accordingly, bacterial counts for the pgdA mutant were lower than the WT in both liver and spleen (Fig. 4A and B). In addition, the pgdA mutant, although sensitive to lysozyme in vitro, was as resistant as the WT strain to the action of serum in the presence or absence of complement (SI Fig. 6). Hence, the impaired virulence of the pgdA mutant is not related to an impaired survival in the blood stream.
Fig. 4.
Impaired virulence of the pgdA mutant in vivo. (A and B) BALB/c and C57/BL6J mice were challenged by i.v. injection with the parental strain EGDe and its pgdA mutant with a sublethal dose (5 × 103 CFU per mouse). After 72 h, mice were killed, and bacterial counts in the liver (A) and the spleen (B) were determined. (C–G) Transgenic human E-cadherin mice were used as model for the oral route of infection, and colonization of several organs was followed after 3, 24, 48, and 72 h after challenge. Interestingly, the pgdA mutant was particularly vulnerable to persistence in the intestinal lumen as assayed by bacterial counts per grams of feces (C). Survival was more robust in the intestine (D) and the mesenteric lymph nodes (E) where colonization was particularly impaired after 72 h. The mutant was impaired in the survival of the liver (F) and spleen (G) at all time points as in the IV model shown in A and B.
The oral route is the natural route of Listeria infection. We thus also orally infected human E-cadherin (hEcad) transgenic mice, which are permissive to Listeria oral infection (18), and monitored bacterial counts in different organs at 3, 24, 48, and 72 h after infection (Fig. 4 C–G). The pgdA mutant was strongly impaired in surviving in the intestinal lumen, and, 48 h after infection, no bacteria could be detected in this compartment (Fig. 4C). Furthermore, bacterial counts of the pgdA mutant were compared with the WT. Those were lower in the intestine and in the mesenteric lymph nodes (Fig. 4 D and E, respectively) and, as observed after IV inoculation, in the liver and the spleen (Fig. 4 F and G, respectively). Hence, the pgdA mutant is attenuated in virulence at both very early stages of the infection and at later stages after dissemination from the intestine to the liver and the spleen. These results reveal that the pgdA mutant is highly sensitive to both the very first host innate immune responses and at later stages of the infection. This finding is in agreement with the fact that Paneth cells in the small intestine abundantly produce lysozyme and other antimicrobial agents.
We then investigated the impact that lack of PG N-deacetylation might have on the inflammatory response. Because the Nod proteins detect PG and PG fragments such as muropeptides (19, 20), we first analyzed the ability of highly purified PG from L. monocytogenes EGDe to activate the Nod pathways by measuring NF-κB activation with the classical luciferase reporter assay in HEK293T cells (21). The native PG of L. monocytogenes activated NF-κB poorly in both Nod1- and Nod2-dependent manners compared with Escherichia coli PG (SI Fig. 8) as reported (22). However, a complete predigestion with a muramidase increased induction of NF-κB in a Nod2-dependent (20-fold) and Nod1-dependent (6-fold) manner (SI Fig. 8A). The fully acetylated native PG from the pgdA mutant was better detected by the Nods (SI Fig. 8B), compared with the native PG of parental strain EGDe. Taken together, our results suggested that L. monocytogenes contains in its native PG Nod agonists but that these are not readily available to the host. Hence, our hypothesis that Listeria also N-deacetylates its PG as a strategy to avoid generation and presentation of cell-wall components to the pattern-recognition receptors such as the Nods or TLR2. As a proof of principle, we were able to show that an in vitro fully N-deacetylated PG from Helicobacter pylori completely lost its ability to be sensed by both Nod1 and Nod2 (SI Fig. 8C) compared with native H. pylori PG.
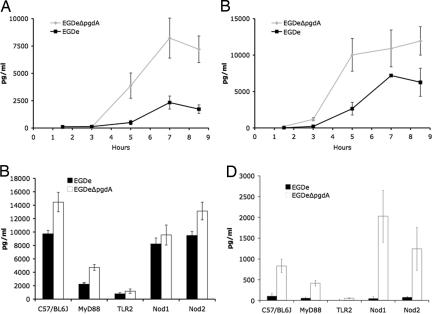
The cytokine response of macrophages to the pgdA mutant was then compared with WT. Despite the reduced number of viable mutant bacteria (see Fig. 3 A and B), the pgdA mutant induced a strikingly more vigorous cytokine response compared with WT (for example, higher amounts of IL-6 and IFN-β) than strain EGDe in RAW264.7 macrophages (Fig. 5A and B and SI Fig. 9). This enhanced cytokine response of the pgdA mutant compared with WT EGDe was also observed in PEM (Fig. 5 C and D, respectively). To our knowledge, the stimulation of IFN-β production by PEM at such a high level by the pgdA mutant is previously undescribed in Listeria.
Fig. 5.
Enhanced inflammatory response of the pgdA mutant. Cytokine production such as IL-6 (A) and IFN-β (B) by RAW264.7 macrophages was enhanced by the pgdA mutant compared with the parental strain EGDe (see also SI Fig. 9). PEM of WT C57/BL6J, Nod1−/−, Nod2−/− TLR2−/− and MyD88−/− mice were infected with strains EGDe and its isogenic pgdA mutant. After 7 h of infection, the inflammatory response was enhanced as measured by the amount of IL-6 (C) and, particularly, IFN-β (D) production. The cytokine response depended mainly on TLR2 and Myd88, although their contribution varied according to the cytokine measured. IFN-β production depended almost entirely on TLR2 in a MyD88-dependent and -independent manner (D). IL-6 production depended only partially on TLR2 and Myd88 (C) and also required Nod1. Surprisingly, Nod1 seemed to function as an inhibitor of IFN-β production, because the pgdA mutant induced 3-fold more IFN-β in Nod1−/− compared with C57/BL6J peritoneal macrophages (D). Note that the role of Nod1 is entirely restricted to the response to the pgdA mutant and does not participate in the response to the WT EGDe strain.
The role of different pattern-recognition receptors in the enhanced cytokine response to pgdA mutant infection was then addressed by using macrophages from Nod1−/−, Nod2−/−, MyD88−/−, and TLR2−/− mice. IL-6 and IFN-β production depended strongly on TLR2. The TLR2 response depended partially on MyD88, the master adaptor molecule myeloid differentiation factor 88. IL-6 production also partially depended on Nod1, which, surprisingly, negatively regulated IFN-β production by PEM (Fig. 5D). This negative regulatory impact on TLR2-mediated IFN-β production of the Nod1-pathway is reminiscent of the role of Nod2 as negative regulator of TLR2-mediated T helper type 1 responses (23). Together, our results suggested that the inflammatory response induced by the pgdA mutant is due to an enhanced accessibility of cell wall components to TLR2 and Nod1. These results also suggested, as shown in PEM, that WT Listeria exploit PG N-deacetylation to evade Nod1 sensing.
In conclusion, the pgdA mutant is a highly attenuated mutant of Listeria. Attenuation is in the same order of magnitude as that of listeriolysin O and ActA mutants, the most attenuated Listeria mutants (7, 8). Thus, N-deacetylation of Listeria's PG is a major virulence determinant. Listeria escapes recognition and killing by the host by preventing processing and optimal sensing of its PG by the host. Our observations in vitro and in vivo indicate that a major consequence of PG N-deacetylation is an increased survival at early and also later stages of the infectious process. We propose the following model (SI Fig. 10). Listeria PG N-deacetylation enhances the pathogen's ability to survive to first defenses of the host in the intestinal lumen by avoiding the bacteriolytic activity of lysozyme that is massively produced by Paneth cells in the intestine (24). Listeria PG N-deacetylation might also prevent induction of enhanced production of antimicrobial peptides synthesis by escaping Nod2 detection in the intestine as previously proposed, thus potentially coupling resistance to lysozyme to the Nod2 evasion (12). From the primary infection site, Listeria disseminates to target organs, owing to escape from lysozyme, survival in phagocytic cells, and down-regulation of the inflammatory response. The pgdA mutant sensitivity to lysozyme results in its enhanced destruction by phagocytes and release of its cell-wall components such as muropeptides and lipoteichoic acid (LTA). The pgdA mutant's LTA is readily available to membrane-bound TLR2, whereas muropeptides can be delivered to the cytosol by endogenous transporters such as hPepT1 (25) or after pore formation by listeriolysin O. Cytokine production would then ensue. PG N-deacetylation would thus function as a double-protection mechanism for Listeria against the innate immune defenses by escaping the action of lysozyme and providing a mechanism to evade TRL2 and the Nod proteins. The predominant TLR2-dependent phenotype is most probably due to the extreme sensitivity of the pgdA mutant to the phagosome environment before any major contribution of the cytosolic sensing mechanism by the Nods can occur. In perfect agreement with this report, N-deacetylation of pneumococcal PG had been associated with pneumococcal virulence, but the underlying mechanisms had not been studied in detail (26). PG N-deacetylation also occurs in other major Gram-positive pathogens such as Bacillus anthracis. Genome analysis of B. anthracis indicates the presence of 10 ORFs of putative deacetylases (27), suggesting that these might contribute to extreme virulence of this bioterrorism agent. Enterobacteria such as E. coli, Shigella flexneri, and Yersinia pestis have an unique lysozyme inhibitor, the small periplasmic polypeptide Ivy (28). Alternatively, Staphylococcus aureus, Neisseria meningitidis, and Mycobacterium tuberculosis have developed mechanisms such as PG O-acetylation and PG N-glycolylation of their muramic acid residues to counteract the activity of host lysozyme (29–31), suggesting that these major pathogens might also modulate the host innate immune response by modifications of their PGs. Our study reveals a mechanism by which pathogens modify their PG to escape from host defenses.
Materials and Methods
Bacterial and Cell Growth Conditions.
E. coli DH5α was used as host for the construction and preparation of plasmids. E. coli was cultivated in Luria Bertani solid or liquid media supplemented as appropriate with ampicillin (100 μg·ml−1). E. coli MC1061 (53338; American Type Culture Collection, Manassas, VA) (32) and H. pylori strain 26695 (700392; American Type Culture Collection) (33) were used to extracted PG. H. pylori was grown microaerobically at 37°C on blood agar plates or in liquid medium consisting of brain-heart infusion (BHI; Oxoid, Basingstock, Hampshire, U.K.) with 0.2% β-cyclodextrin (Sigma) supplemented with antibiotic–antifungal mix (34). L. monocytogenes EGDe (BUG1600; BAA-679; American Type Culture Collection) and its pgdA mutant (BUG 2288) were grown in BHI solid or liquid media. HEK293T (CRL-1573; American Type Culture Collection) cells and RAW264.7 (TIB-71; American Type Culture Collection) macrophages were cultured in DMEM (GIBCO, Paisley, Scotland, U.K.) containing 10% FCS (GIBCO). Before transfection, HEK293T cells were seeded into 24-well plates at a density of 105 cells per ml as described (35). RAW264.7 macrophages were seeded into either six-well plates or into 25-mm Petri dishes at 105 cells per ml and 106 cells per ml, respectively.
Construction of Listeria pgdA Mutant.
A DNA fragment containing 663 bp of the sequence upstream of lmo0415 was generated by PCR using oligonucleotides lmo0415–1 (5′-AACAGGATCCATAACTGGAGACACGGAGAC-3′) and lmo0415–2 (5′-AACAGAATTCATTATGCACCTCACCTCAG-3′). The fragment was cloned into BamHI-EcoRI digested pMAD (36). A DNA fragment containing 702 bp of the sequence downstream of lmo0415 was generated by PCR using oligonucleotides lmo0415–3 (5′-AACAGAATTCAAATCAGTAGCTAAGATGAGTTAA-3′) and lmo0415–4 (5′-AACAAGATCTGATTGTCAAACTTGAAATGG-3′). The fragment was cloned into EcoRI-BglII-digested pMAD containing the lmo0415 upstream fragment, constructing the pDC27. The construct was verified by sequencing. To achieve allelic exchange, pDC27 was electroporated into L. monocytogenes EGDe at 2,500 V, 200 Ω,and 25 μF. Transformants were selected at 30°C on BHI agar medium containing erythromycin (5 μg/ml) and X-Gal (50 μg/ml). One blue colony was grown in BHI-Ery broth at 43.5°C for 48 h, and the culture was plated onto BHI-Ery agar containing X-Gal at 43.5°C. One blue colony was selected and grown in BHI broth at 30°C. BHI broth was inoculated with 1 μl of a 1:10 dilution of the previous culture and incubated at 43.5°C. Tenfold serial dilutions of this culture were plated onto BHI-X-Gal agar and incubated at 43.5°C. White colonies were analyzed for erythromycin sensitivity, and PCR amplifications with oligonucleotides lmo0415–1 and lmo0415–4 and oligonucleotides lmo0415–5 (5′-GATGGACAGACTAATGAAAGACC-3′) and lmo0415–6 (5′-AAAGCACCTGTTTCTGCGTC-3′) were performed to confirm the gene deletion.
Lysozyme, PGRP-L, Serum Experiments, and Hemolytic Activity.
Listeria strains were grown in BHI media to which was added either hen egg lysozyme (10 μg/ml final concentration; Sigma-Aldrich), human serum amidase (or PGRP-L, kindly provided by Waldemar Vollmer, University of Tübigen, Tübigen, Germany; 1 μg/ml final concentration) or FCS (final concentration at 25%; GIBCO). When needed, decomplementation was done by heating the FCS at 65°C for 30 min. Growth was monitored by following the optical density at 600 nm and by determining the number of viable bacteria per milliliter. L. monocytogenes strains were grown on 5% horse blood agar plates. Plates were incubated at 37°C for 48 h and at 4°C for 24 h. The size of the clear zone around the colonies was measured, which indicated β-hemolytic activity.
PG Purification, HPLC Analysis, in Vitro N-Deacetylation and MALDI-postsource decay (PSD) Analysis.
E. coli, L. monocytogenes, and H. pylori PGs were prepared from exponentially growing bacteria and purified as described (22). From native PG, muropeptides were generated by using the muramidase mutanolysin M1 (Sigma-Aldrich), separated by HPLC, purified, and analyzed by MALDI-PSD as described (31). N-deacetylation of H. pylori PG was done as described by using the recombinant BC1960 deacetylase from Bacillus cereus (27).
Expression Plasmids, Transient Transfections, and NF-κB Activation Assays.
HEK293T cells were used for transfections as described (35). Synergistic activation of NF-κB by PGs, muramyl peptides, and related compounds in cells overexpressing Nod1 or Nod2 was studied as described by Inohara et al. (21). Data were standardized with positive controls: N-acetylmuramic acid-dipeptide for hNod2 and N-acetylmuramic acid-tripeptide for hNod1. hNod1 and hNod2 were activated with PG (0.3 μg/ml) as described (38). More detailed information is available in SI Text.
Infection of Murine Macrophages.
PEM and bone marrow-derived macrophages (BMDM) were isolated from BALB/c, C57/BL6J (Charles River Laboratories, L'Arbresle, France), TLR2−/−, MyD88−/−, Nod1−/−, Nod2−/− mice. BMDM preparation is detailed in SI Text. PEM were established as described (39). PEM or BMDM (106 cells per well) were infected with L. monocytogenes strains (MOI, 10), grown to an OD600 of 0.8, and incubated at 37°C for 1 h to allow bacterial phagocytosis. Extracellular bacteria were eliminated by three washes in RPMI medium 1640 supplemented with 10% FCS, and 10 μg/ml gentamicin was added for the time of infection. Macrophages were lysed with 0.2% Triton X-100 at various time points (1.5, 3, and 7 h) for 10 min, and the number of CFU was assessed by plating serial dilutions on BHI agar plates that were incubated at 37°C.
Epithelial Caco 2 Cells and RAW264.7 Macrophage Infections.
The Listeria strains were grown to an OD600 of 0.6–0.8, washed three times, and diluted in DMEM such that the MOI was ≈100 and 25 bacteria per Caco-2 cell and RAW264.7 macrophage, respectively (OD600 of 1 corresponds to ≈109 bacteria). Bacterial suspensions were added to mammalian cells for 1 h, and the cells were then washed and noninvasive bacteria were killed by adding 20 μg/ml gentamicin for 1.5 h. After washing three times, cells were lysed in 0.2% Triton X-100 at various desired time points (0, 1.5, 3, 5, 7, and 8.5 h), and the number of viable bacteria released from the cells was assessed after serial dilutions of the lysates on BHI agar plates. Supernatants were collected at the various time points to determine cytokine production. For EM observations, infected cells were recovered and fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer overnight at 4°C.
EM.
Fixed samples were rinsed three times with 0.1 M cacodylate buffer, postfixed with 2% osmium tetraoxide, 1% potassium ferricyanide in 0.1 M cacodylate buffer for 1 h at room temperature, and washed again with 0.1 M cacodylate buffer and water. The samples were dehydrated through a graded series of ethanol baths, one time with propylene oxide, and overnight in a mixture of Epon 812/propylene oxide at room temperature. After being embedded in Epon 812, samples were polymerized for 48 h at 60°C. Utrathin sections (70–80 nm) were cut with a diamond knife, stained with uranyl acetate and Reynolds lead citrate, and viewed in a JEM 1010 transmission electron microscope (Jeol, Tokyo, Japan) at 80 kV by using an Eloïse Mega View III camera and AnalySIS Pro Software version 3.1 (comEloïse SARL, Roissy, France).
Mouse Experiments.
Male mice 6 to 10 weeks old were used for this study. Detailed information on mice backgrounds is available in SI Text. LD50 were performed after i.v. challenge of mice with increasing doses of WT EGDe and its pgdA mutant. Survival was monitored during 3 weeks, and LD50 were calculated as described (40). Kinetics of infection was monitored i.v. in BALB/C or C57/BL6J mice at 72 h after challenge with a sublethal dose of strain EGDe and its pgdA mutant (5 × 103 bacteria per mouse). Oral infections were performed as described (18). Female iFABP-hEcad transgenic mice (6 to 8 weeks old) starved for 24 h were injected intragastrically with 5 × 109 bacteria mixed with PBS 150 mg/ml CaCO3. At 3, 24, 48, or 72 h after infection, the organs were dissected. The small intestine was rinsed and incubated for 2 h in 100 mg/liter gentamicin to kill extracellular bacteria from the intestinal lumen. Monitoring of Listeria in the intestinal lumen was done by determining the bacterial load of Listeria per gram of feces by using a Listeria-selective media (Oxford media; Oxoid).
Cytokine Production.
Supernatants of macrophages (RAW264.7 and PEM) infected by strain EGDe and isogenic pgdA mutant were used to determine the production of IL-6, IL-1β, TNF-α, and IFN-β using the Quantikine mouse IL-6, IL-1β, and TNF-α (R & D Systems, Minneapolis, MN) and mouse IFN β (PBL Biochemical Laboratories, Piscataway, NJ) ELISA kits, respectively, as recommended by the manufacturers.
Supplementary Material
Acknowledgments
This work was supported in part by Institut Pasteur (GPH No. 9, PTR No. 153), Institut National de la Recherche Agronomique, Institut National de la Santé et de la Recherche Médicale (INSERM), and Action Concertée Incitative Grants MIC 0312 and MIC 0321 from the Ministère Chargé de la Recherche (Program de Microbiologie Fondamentale et Appliquée, Maladies Infectieuses, Environnement et Bioterrorisme). D.C. and S.S. were supported by Grant POCI/SAU-MMO/60443/2004 from the Portuguese Science and Technology Foundation, S.E.G. was supported by the INSERM Avenir and INSERM U786 (Prof. Philippe Sansonetti), S.E.G. is a Canadian Research Chair holder, I.G.B. is an INSERM Research Scientist, and P.C. is an International Research Scholar of the Howard Hughes Medical Institute.
Abbreviations
- PEM
peritoneal macrophage
- PG
peptidoglycan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
See Commentary 691.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609672104/DC1.
References
- 1.Cossart P, Vicente MF, Mengaud J, Baquero F, Perez-Diaz JC, Berche P. Infect Immun. 1989;57:3629–3636. doi: 10.1128/iai.57.11.3629-3636.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. Cell. 1992;68:521–531. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- 3.Gaillard JL, Berche P, Frehel C, Gouin E, Cossart P. Cell. 1991;65:1127–1141. doi: 10.1016/0092-8674(91)90009-n. [DOI] [PubMed] [Google Scholar]
- 4.Lebrun M, Mengaud J, Ohayon H, Nato F, Cossart P. Mol Microbiol. 1996;21:579–592. doi: 10.1111/j.1365-2958.1996.tb02566.x. [DOI] [PubMed] [Google Scholar]
- 5.Dramsi S, Biswas I, Maguin E, Braun L, Mastroeni P, Cossart P. Mol Microbiol. 1995;16:251–261. doi: 10.1111/j.1365-2958.1995.tb02297.x. [DOI] [PubMed] [Google Scholar]
- 6.Jonquieres R, Bierne H, Fiedler F, Gounon P, Cossart P. Mol Microbiol. 1999;34:902–914. doi: 10.1046/j.1365-2958.1999.01652.x. [DOI] [PubMed] [Google Scholar]
- 7.Dussurget O, Pizarro-Cerda J, Cossart P. Annu Rev Microbiol. 2004;58:587–610. doi: 10.1146/annurev.micro.57.030502.090934. [DOI] [PubMed] [Google Scholar]
- 8.Hamon M, Bierne H, Cossart P. Nat Rev Microbiol. 2006;4:423–434. doi: 10.1038/nrmicro1413. [DOI] [PubMed] [Google Scholar]
- 9.Cabanes D, Dussurget O, Dehoux P, Cossart P. Mol Microbiol. 2004;51:1601–1614. doi: 10.1111/j.1365-2958.2003.03945.x. [DOI] [PubMed] [Google Scholar]
- 10.Lenz LL, Mohammadi S, Geissler A, Portnoy DA. Proc Natl Acad Sci USA. 2003;100:12432–12437. doi: 10.1073/pnas.2133653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meylan E, Tschopp J, Karin M. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, Flavell RA. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 13.Opitz B, Puschel A, Beermann W, Hocke AC, Forster S, Schmeck B, van Laak V, Chakraborty T, Suttorp N, Hippenstiel S. J Immunol. 2006;176:484–490. doi: 10.4049/jimmunol.176.1.484. [DOI] [PubMed] [Google Scholar]
- 14.Torres D, Barrier M, Bihl F, Quesniaux VJ, Maillet I, Akira S, Ryffel B, Erard F. Infect Immun. 2004;72:2131–2139. doi: 10.1128/IAI.72.4.2131-2139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi K, Inohara N, Hernandez LD, Galan JE, Nunez G, Janeway CA, Medzhitov R, Flavell RA. Nature. 2002;416:194–199. doi: 10.1038/416194a. [DOI] [PubMed] [Google Scholar]
- 16.Kamisango K, Saiki I, Tanio Y, Okumura H, Araki Y, Sekikawa I, Azuma I, Yamamura Y. J Biochem (Tokyo) 1982;92:23–33. doi: 10.1093/oxfordjournals.jbchem.a133918. [DOI] [PubMed] [Google Scholar]
- 17.Amano K, Hayashi H, Araki Y, Ito E. Eur J Biochem. 1977;76:299–307. doi: 10.1111/j.1432-1033.1977.tb11596.x. [DOI] [PubMed] [Google Scholar]
- 18.Lecuit M, Vandormael-Pournin S, Lefort J, Huerre M, Gounon P, Dupuy C, Babinet C, Cossart P. Science. 2001;292:1722–1725. doi: 10.1126/science.1059852. [DOI] [PubMed] [Google Scholar]
- 19.Girardin SE, Travassos LH, Herve M, Blanot D, Boneca IG, Philpott DJ, Sansonetti PJ, Mengin-Lecreulx D. J Biol Chem. 2003;278:41702–41708. doi: 10.1074/jbc.M307198200. [DOI] [PubMed] [Google Scholar]
- 20.Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, Fukase K, Inamura S, Kusumoto S, Hashimoto M, et al. J Biol Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 21.Inohara N, Ogura Y, Chen FF, Muto A, Nunez G. J Biol Chem. 2001;276:2551–2554. doi: 10.1074/jbc.M009728200. [DOI] [PubMed] [Google Scholar]
- 22.Travassos LH, Girardin SE, Philpott DJ, Blanot D, Nahori MA, Werts C, Boneca IG. EMBO Rep. 2004;5:1000–1006. doi: 10.1038/sj.embor.7400248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe T, Kitani A, Murray PJ, Strober W. Nat Immunol. 2004;5:800–808. doi: 10.1038/ni1092. [DOI] [PubMed] [Google Scholar]
- 24.Dommett R, Zilbauer M, George JT, Bajaj-Elliott M. Mol Immunol. 2005;42:903–912. doi: 10.1016/j.molimm.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 25.Vavricka SR, Musch MW, Chang JE, Nakagawa Y, Phanvijhitsiri K, Waypa TS, Merlin D, Schneewind O, Chang EB. Gastroenterology. 2004;127:1401–1409. doi: 10.1053/j.gastro.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 26.Vollmer W, Tomasz A. Infect Immun. 2002;70:7176–7178. doi: 10.1128/IAI.70.12.7176-7178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Psylinakis E, Boneca IG, Mavromatis K, Deli A, Hayhurst E, Foster SJ, Varum KM, Bouriotis V. J Biol Chem. 2005;280:30856–30863. doi: 10.1074/jbc.M407426200. [DOI] [PubMed] [Google Scholar]
- 28.Monchois V, Abergel C, Sturgis J, Jeudy S, Claverie JM. J Biol Chem. 2001;276:18437–18441. doi: 10.1074/jbc.M010297200. [DOI] [PubMed] [Google Scholar]
- 29.Bera A, Herbert S, Jakob A, Vollmer W, Gotz F. Mol Microbiol. 2005;55:778–787. doi: 10.1111/j.1365-2958.2004.04446.x. [DOI] [PubMed] [Google Scholar]
- 30.Raymond JB, Mahapatra S, Crick DC, Pavelka MS., Jr J Biol Chem. 2005;280:326–333. doi: 10.1074/jbc.M411006200. [DOI] [PubMed] [Google Scholar]
- 31.Antignac A, Rousselle JC, Namane A, Labigne A, Taha MK, Boneca IG. J Biol Chem. 2003;278:31521–31528. doi: 10.1074/jbc.M304749200. [DOI] [PubMed] [Google Scholar]
- 32.Casadaban MJ, Cohen SN. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 33.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, et al. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 34.Bury-Mone S, Thiberge JM, Contreras M, Maitournam A, Labigne A, De Reuse H. Mol Microbiol. 2004;53:623–638. doi: 10.1111/j.1365-2958.2004.04137.x. [DOI] [PubMed] [Google Scholar]
- 35.Girardin SE, Tournebize R, Mavris M, Page AL, Li X, Stark GR, Bertin J, DiStefano PS, Yaniv M, Sansonetti PJ, Philpott DJ. EMBO Rep. 2001;2:736–742. doi: 10.1093/embo-reports/kve155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnaud M, Chastanet A, Debarbouille M. Appl Environ Microbiol. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inohara N, Koseki T, del Peso L, Hu Y, Yee C, Chen S, Carrio R, Merino J, Liu D, Ni J, Nunez G. J Biol Chem. 1999;274:14560–14567. doi: 10.1074/jbc.274.21.14560. [DOI] [PubMed] [Google Scholar]
- 38.Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jehanno M, Viala J, Tedin K, Taha MK, Labigne A, Zahringer U, et al. Science. 2003;300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 39.Alford CE, King TE, Jr, Campbell PA. J Exp Med. 1991;174:459–466. doi: 10.1084/jem.174.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reed LJ, Muench H. Am J Hyg. 1938;27:493–497. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.