Abstract
Mathematical models of transmission have become invaluable management tools in planning for the control of emerging infectious diseases. A key variable in such models is the reproductive number R. For new emerging infectious diseases, the value of the reproductive number can only be inferred indirectly from the observed exponential epidemic growth rate r. Such inference is ambiguous as several different equations exist that relate the reproductive number to the growth rate, and it is unclear which of these equations might apply to a new infection. Here, we show that these different equations differ only with respect to their assumed shape of the generation interval distribution. Therefore, the shape of the generation interval distribution determines which equation is appropriate for inferring the reproductive number from the observed growth rate. We show that by assuming all generation intervals to be equal to the mean, we obtain an upper bound to the range of possible values that the reproductive number may attain for a given growth rate. Furthermore, we show that by taking the generation interval distribution equal to the observed distribution, it is possible to obtain an empirical estimate of the reproductive number.
Keywords: basic reproduction ratio, epidemiology, influenza, Lotka–Euler equation, serial interval
1. Introduction
The past decade has seen a dramatic increase in the attention paid to infectious disease epidemics as a potential health threat. This is due in part to disease outbreaks in domestic livestock (Keeling et al. 2001), the fear of bioterrorist attacks with smallpox virus (Gani & Leach 2001), the emergence of severe acute respiratory syndrome (SARS) in 2003 (Lipsitch et al. 2003) and the risk of an influenza pandemic among human populations (Longini et al. 2004; Ferguson et al. 2005). Planning for the mitigation and control of such health threats relies increasingly on mathematical models of infection transmission.
One of the key parameters in mathematical transmission models is the reproductive number R0, defined as the number of secondary infections that arise from a typical primary case in a completely susceptible population. When infection is spreading through a population that may be partially immune, it is often more convenient to work with an effective reproductive number R, which is defined as the number of secondary infections that arise from a typical primary case. The magnitude of R is a useful indicator of both the risk of an epidemic and the effort required to control an infection (Anderson & May 1991; Roberts & Heesterbeek 2003; Heffernan et al. 2005). Accurate estimation of the value of the reproductive number is crucial to planning for the control of an infection.
For new emerging infections, such as SARS in 2003, the available information about the transmissibility of a new infectious disease epidemic is likely to be restricted to daily counts of new cases. It is well known that these counts increase exponentially in the initial phase of an epidemic. The rate of exponential growth, r, is defined as the per capita change in number of new cases per unit of time. The observed value of the growth rate r can be related to the value of reproductive number R through a linear equation: R=1+rTc (Anderson & May 1991; Pybus et al. 2001; Ferguson et al. 2005). Here, Tc is the mean generation interval, defined as the mean duration between time of infection of a secondary infectee and the time of infection of its primary infector (sometimes this is called the serial interval or generation time).
Demographers, ecologists and evolutionary biologists take a slightly different approach. They derive the growth rate from fecundity rates, survival rates and the reproductive number R according to the so-called Lotka–Euler equation (Dublin & Lotka 1925; Feller 1941; Metz & Diekmann 1986; Keyfitz & Caswell 2005). Ecological textbooks suggest simplifying this equation by ignoring variability in generation time (Begon et al. 1996). The result is, after rearranging, an exponential equation: R=exp(rTc). Here, Tc is the cohort generation time, a demographic analogue of the epidemiological mean generation interval.
Having two alternative equations for relating the desired value of reproductive number to the observed value for growth rate, we face the difficulty of choosing the most appropriate one. For example, the growth rate of the Hepatitis C epidemic is estimated to be r=0.96 per year, and the mean generation interval of this infection is of the order of Tc=20 years (Pybus et al. 2001). The value for the reproductive number by the linear equation is R=1+0.096×20=2.9, whereas the value that is obtained using the exponential equation is R=exp(0.096×20)=6.8. Such large discrepancies do matter in planning for public health interventions. There exist several other expressions that relate the reproductive number to the growth rate (Dublin & Lotka 1925; Lipsitch et al. 2003; Wearing et al. 2005) and expressions for estimating the reproductive number from time-series of case counts (Wallinga & Teunis 2004). How should we choose the most appropriate equation for inferring the reproductive number from observed growth rates for a particular infection?
We start by recapitulating the Lotka–Euler equation in terms of human demography, and we rephrase this equation into more convenient terms for infectious disease epidemiology, following Levin et al. (1996). We use the rephrased Lotka–Euler equation to examine the assumptions that underlie the alternative relationships between reproductive number and observed change in a number of cases. We will illustrate our findings by estimating the reproductive number R for influenza A infections. The key variables and their interpretation in ecological, demographical and epidemiological terms are presented in the electronic supplementary material.
2. Inferring R from r
(a) Deriving the Lotka–Euler equation
We introduce the Lotka–Euler equation using the human population as an example. For simplicity, we focus on female individuals, assuming that there is always a sufficient supply of males to ensure reproduction. We measure time and age in years, and we will refer to the present time as t=0, such that events in the past occurred when time t is negative and events in the future will occur when time t is positive. We assume that the population displays exponential growth at a fixed growth rate and that the age distribution of the population does not change over time.
The Lotka–Euler equation can be understood as the combination of two concepts that can be explained intuitively. First, if we add up the number of children born to mothers of all ages at a particular time, we get the total number of births at that time. The number of births to mothers of age a at time t is equal to the number of births at time t−a (the number of mothers, including those who have not survived) multiplied by the expected number of offspring per year for mothers of age a. Summing these births over all possible mothers' ages, we obtain the total number of births in year t,
| (2.1) |
where b(t) refers to birth rate of the population at time t and n(a) refers to the rate of production of female offspring by a mother at age a. This equation is a specific case of the renewal equation for the birth process (Feller 1941).
Second, because the population is growing exponentially with a stable age distribution, the number of births at any given time (say t) is equal to the number of births a time units ago, multiplied by the exponential growth of the population since then
| (2.2) |
Combining these two equations, we obtain an expression with b(t) on both sides
| (2.3) |
This equation has the intuitive interpretation that all the births in the past, multiplied by the number of offspring for individuals born at each time in the past, must add up to the current births.
The composite parameter n(a) is more familiarly known in demography as the product of the survivorship and fecundity functions, n(a)=l(a)m(a). Using this more familiar parameterization and removing b(t) from both sides of equation, we obtain the Lotka–Euler equation
| (2.4) |
(b) A moment generating function expression for the reproductive number R
While the Lotka–Euler equation is a basic part of demography, in which one may be interested in deriving population growth rates from life tables, a related problem in epidemiology is to estimate the reproductive number, R, from growth rates of a disease. We have previously defined n(a) as the rate of production of female offspring by a mother at age a. It is readily seen that if we integrate n(a) over the whole lifespan, we obtain the total number of female offspring produced by a mother over her lifespan, known as R
| (2.5) |
The rate n(a) can be normalized to a distribution g(a), which is the distribution of age at child bearing (cf. Metz & Diekmann 1986)
| (2.6) |
The generation interval distribution for an infectious disease is the probability distribution function for the time from infection of an individual to the infection of a secondary case by that individual. If we take the ‘age’ of an infection to be the time since infection, then in the notation above, the generation interval distribution is equivalent to g(a) above. Substituting this expression for the generation interval distribution g(a) into the Lotka–Euler equation (2.4), we obtain
| (2.7) |
The term that now appears in the right-hand side of this equation is familiar to mathematicians as the so-called Laplace transform of the function g(a). More specifically, it is known to statisticians as the moment generating function M(z) of the distribution g(a) (e.g. Mood et al. 1974)
| (2.8) |
We use this moment generating function to simplify the notation of the rephrased Lotka–Euler equation (2.7). Here, the argument z takes the value of minus the growth rate, −r:
| (2.9) |
provided that M(−r) exists.
A moment generating function, if it exists, uniquely characterizes the shape of the entire probability distribution: M(z) determines g(a) and, conversely, g(a) determines M(z). The biological corollary of this moment generating function expression is then that a relationship between growth rate r and reproductive number R uniquely characterizes the shape of the generation interval distribution and, conversely, the shape of the generation interval distribution determines the appropriate relationship between the reproductive number and the growth rate. The electronic supplementary material provides further mathematical details.
3. Specific generation interval distributions
(a) Epidemic models
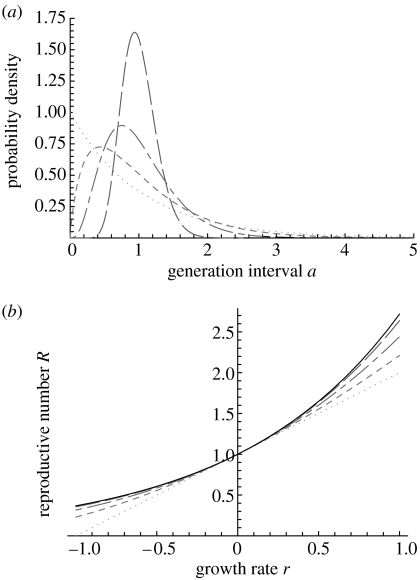
An epidemic model implicitly specifies a generation interval distribution. A simple epidemic model categorizes the population of hosts according to their infection status as either susceptible, infectious or recovered. This results in the so-called susceptible–infectious–recovered (SIR) class of epidemic models. The rate of leaving the infectious stage is denoted by b, and this rate is assumed constant. The rate of making contacts during this infectious stage is also assumed constant. The duration of a generation interval is thereby implicitly specified as an exponential distribution with mean Tc=1/b. Such an exponential distribution is illustrated in figure 1a, dotted line. We substitute the moment generating function for the exponential distribution M(z)=b/(b−z) with z=−r (see table 2 of the electronic supplementary material) into the moment generating function expression (2.9). Simplifying the result gives a linear relationship between growth rate r and reproductive number R,
| (3.1) |
provided that r>−b. This linear relationship between growth rate and reproductive number is shown in figure 1b, dotted line. The linear relationship is frequently used in infectious disease epidemiology, with the term 1/b interpreted as mean generation interval (Ferguson et al. 2005, 2006) or as duration of the infectious period (Anderson & May 1991; Pybus et al. 2001).
Figure 1.
Generation interval distributions uniquely characterize the relationship between the reproductive number R and the growth rate r: (a) generation interval distributions g(a) with identical mean Tc and increasing coefficient of variation σ/Tc from 0.25 (long dashes), 0.5 (long and short dashes), 0.75 (short dashes) to 1 (dots) and (b) the corresponding relationships between growth rate r and reproductive number R, the drawn curve indicates the upper bound obtained for the delta distribution.
To enhance realism of the epidemic models, we can add an exposed (infected but not yet infectious) stage. This results in the so-called susceptible–exposed–infectious–recovered (SEIR) class of epidemic models. The rate of leaving the exposed stage is b1, the rate of leaving the infectious stage is b2 and both rates are assumed constant. Thereby, the generation interval distribution is implicitly specified as a convolution of two exponential distributions with a mean Tc=1/b1+1/b2. Such a distribution has one mode with a long right tail (see figure 1a, short-dashed line). We substitute the moment generating function of this distribution (see electronic supplementary material) into the moment generating function expression (2.9). After rearranging, we obtain the relationship
| (3.2) |
with the proviso that r>min(−b1,−b2). This relationship is a quadratic increasing curve (see figure 1b, short-dashed line). The same equation has been derived by Lipsitch et al. (2003), using a different approach.
More complicated epidemic models have incorporated additional exposed and infectious stages. As an example, we consider the epidemic model proposed by Wearing et al. (2005) which has a number of x exposed stages, each with a rate b1, and a number of y infectious stages, each with a rate b2. We can compose the moment generating function of the generation interval from the generating functions of duration of each stage (see electronic supplementary material). Substitution of the resulting moment generating function into expression (2.9) gives
| (3.3) |
with the proviso that r>min(−b1,−b2). The same equation has been presented by Wearing et al. (2005).
(b) Normal distributions
For infections with a mean generation interval Tc and a standard deviation σ, the generation intervals may approximate a normal distribution (figure 1a, long-dashed line). Assuming a normally distributed generation interval yields the following relationship between growth rate r and reproductive number R (Dublin & Lotka 1925):
| (3.4) |
This relationship is a convex curve that approximates an exponential curve (see figure 1b, long-dashed line).
This equation also shows that a distribution which is more concentrated around the mean generation interval, with a lower value for σ, results in higher values for the reproductive number R (see figure 1b: the long-dashed line which corresponds to the more concentrated distribution is above the long and short-dashed line which corresponds to the more dispersed distribution).
(c) Delta distributions
For an infection where all secondary infections are exactly equal to the mean generation interval Tc, the distribution conforms to a so-called delta distribution. For this distribution, the moment generating function expression (2.9) reduces to a simple exponential relationship
| (3.5) |
This relationship is an exponentially increasing curve (figure 1b, drawn line).
The exponential equation (3.5) can be intuitively understood by realizing that the relative increase in the number of cases over an interval of Tc time units is per definition exp(rTc), and that, because in this specific case all generation intervals are of equal length, this relative increase is also exactly equal to R.
The delta distribution is the most concentrated distribution possible. We expect to find that for a given value of the growth rate and mean generation interval, the reproductive number attains the highest possible value. This can be illustrated by an example: if half of the cases will produce secondary infections a bit earlier than the average generation interval Tc and the other half will produce secondary infections a bit later than average, the additional number of secondary and tertiary cases which are due to the faster infection will more than compensate for the postponed cases that result from the slower infection. Therefore, epidemics with some variation in the duration of their generation intervals will increase at a higher growth rate r for a given reproductive number R than epidemics without any variation in generation interval. And likewise, epidemics without variation in generation interval will grow at higher reproductive number R for a given growth rate r than epidemics with variation in the duration of their generation intervals.
Our intuitive explanation of this upper bound is supported by a rigorous mathematical argument that invokes Jensen's inequality (see electronic supplementary material). Therefore, the exponential relationship in equation (3.5) provides an exact upper bound to the value that reproductive numbers can take. Hence, it is possible to indicate the range of values that the reproductive numbers R may attain for any shape of the generation interval distribution, using only the observed values for growth rate r and the mean generation interval Tc (table 1).
Table 1.
Bounds on the reproductive number.
| growth rate | reproductive number | |
|---|---|---|
| decline | r<0 | |
| stability | r=0 | R=1 |
| growth | r>0 |
We can even obtain a criterion for the relative overestimation of the reproductive number by equation (3.5). We take the ratio of equations (3.4) and (3.5), and rearrange. The relative difference between the upper bound and the actual value of the reproductive number remains below 5% whenever the standard deviation for generation intervals σ is smaller than 46% of the doubling time td=ln 2/r.
(d) Empirical distributions
We can observe the duration of generation intervals in a period of exponential epidemic growth, and approximate the generation interval distribution g(a) by a histogram of the observed durations. We denote the category bounds in such a histogram by a0, a1, …, an, and the observed relative frequencies of observed generation intervals within these bounds as y1, y2, …, yn. Substituting the observed distribution into the moment generating function expression (2.9) and calculating the integral gives
| (3.6) |
For an observed histogram of generation intervals (figure 2a), this relationship is a convex increasing curve (figure 2b).
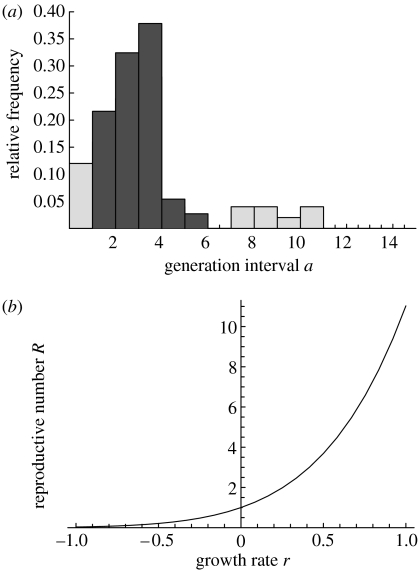
Figure 2.
Observed generation intervals for influenza A virus infections and the empirical relationship between reproductive number and growth rate: (a) histogram of observed generation intervals between two subsequent influenza cases in a household (after Hirotsu et al. 2004), the lighter shading indicates observations that could be due to co-primary or tertiary infections and (b) the corresponding empirical relationship between R and r, according to equation (3.6).
4. Tracking reproductive numbers
In a typical infectious disease management setting, such as the initial phase of the SARS outbreak of 2003, one of the important tasks of epidemiologists is to provide insight into the change in reproductive number R after control measures have been implemented. In such conditions, the assumption of a constant environment and exponential increase in new case counts is untenable.
Estimates for reproductive numbers can be obtained from the renewal equation for the birth process (equation (2.1)). We substitute observed birth rates for b(t), and a time-varying infection rate Rtg(a) for n(a). We assume that g(a) is independent of time t. After rearranging, we have an equation for the reproductive number Rt (C. Fraser 2006, personal communication)
| (3.6) |
This reproductive number Rt assigns its value to the time t at which the secondary cases are infected.
An alternative is to assign the value of the reproductive number to the time u at which the primary cases are infected. We find this value by integrating over all possible times t,
| (4.2) |
This equation has been used to estimate the time-varying reproductive number for SARS (Wallinga & Teunis 2004).
Both estimators are generalizations of the moment generating function expression (2.9): whereas the expressions for R invoke an integral transformation of the generation interval distribution by the exponential growth curve, the estimators for Rt and Ru invoke an integral transformation of the generation interval distribution by the observed growth curve. In all the cases, the value of the reproductive number depends on the shape of the generation interval distribution.
5. Application to influenza A
We use human infections with the influenza A virus to illustrate the impact that various assumptions about the shape of the generation interval distribution may have on the estimated value of the reproductive numbers for a given growth rate. Mills et al. (2004) analysed the observed growth rates for influenza A during the initial phase of the Spanish influenza pandemic in 1918 in 45 major cities in the USA. They found that the median of initial growth rates of this influenza epidemic was r=0.20 per day.
We focus on influenza within households and measure generation intervals as the duration from symptom onset of one household member back to the time of symptom onset of the first infected household member. We have to exclude observations where a household member developed symptoms simultaneously with the first infected household member (primary and co-primary cases), and observations where the household member was unlikely to be infected by the first infected household member (e.g. tertiary cases). Observed generation intervals for influenza A in a Japanese household study (Hirotsu et al. 2004), after exclusion of possible co-primary and tertiary cases, yield an estimated mean of Tc=2.85 days and an estimated standard deviation of σ=0.93 days (see figure 2a).
Without specific assumptions about the shape of the generation interval distribution (table 1), we find that the reproductive number of influenza A is larger than R=1, but smaller than or equal to R=1.77. Since the estimated standard deviation of σ=0.93 days is less than the criterion of 46% of the doubling time td=ln 2/0.20≈3.5 days, we know that the upper bound R=1.77 gives only a slight overestimation. We obtain a value of the reproductive number for influenza A of R=1.57 for the SIR epidemic model (equation (3.1)), a value of R=1.65 for the SEIR epidemic model (equation (3.2)) and a value of R=1.66 for the more complicated epidemic model with one latent stage and two infectious stages (equation (3.3), with x=1, y=2). Perhaps, the most accurate estimate is obtained with the empirical histogram (equation (3.6); figure 2a). This gives a value for the reproductive number of influenza A of R=1.73 secondary cases per primary case.
6. Discussion
New emerging diseases require inference of the reproductive number for an unknown infectious agent from the observed increase in case counts over time. We have shown that the existing relationships between observed epidemic growth rate and a reproductive number differ only with respect to their implicit presupposition about the precise shape of the generation interval distribution. Therefore, the agreement between the presupposed and the actual shape of the generation interval distribution determines the appropriateness of a relationship between observed epidemic growth rate and reproductive number.
Often, we have a poor knowledge of the precise shape of the generation interval distribution. Our results show that it is nonetheless possible to estimate the value of the reproductive number. Even in the absence of any information regarding the shape, we can indicate the upper bound to the possible values that a reproductive number can take (table 1). And with a few observed generation intervals, we can use the empirical relationship between observed growth rate and reproductive numbers (equation (3.6)).
The reproductive number has often been named as a key concept in epidemic theory (Anderson & May 1991; Roberts & Heesterbeek 2003; Heffernan et al. 2005). Since inference of the value of the reproductive number depends crucially on the generation interval distribution, it is surprising that very little is known about this distribution. There is potential for studying a variety of basic epidemiological and ecological questions. We address three of them here.
Firstly, the theoretical framework can be expanded to include discrete generation times, which might be more appropriate for diseases where the moment of infection is tied to discrete times. Such an expanded framework allows for estimation of reproductive numbers for a wide range of infections and organisms, and it could draw on the existing theory that has been developed for life-history analysis in ecology (Caswell 2001).
Secondly, it is possible to estimate the key transmission variables in epidemic models by fitting the modelled generation interval distribution to the observed generation interval distribution. We have followed this approach by fitting the epidemic models to the observed generation intervals for influenza.
Thirdly, there will be benefit in collecting data to assess distributions for generation intervals for various infections. It will be interesting to see whether generation intervals within households differ from generation intervals between households. Such a difference might explain the discrepancies between the various reported values for the mean generation interval of influenza A (Longini et al. 2004; Mills et al. 2004; Ferguson et al. 2005, 2006).
To conclude, the variety of equations that relate observed growth rate to reproductive number can be understood within the Lotka–Euler framework which embraces both a description of the infection cycle and a description of the change in number of new case counts. The observed generation intervals and the observed epidemic growth, when taken together, specify the appropriate value of the reproductive number, and therefore, the required control effort to contain the epidemic. This means that infectious disease surveillance systems which have an objective to inform health policy makers on the required control effort should monitor the symptom onset date of new cases as well as their generation interval for new emerging infections.
Acknowledgments
This work was supported by the National Institute of General Medical Sciences Models of Infectious Disease Agent Study (cooperative agreement 5U01GM076497 to ML), and the EC projects, INFTRANS (contract FP6-513715) and MODELREL (contract SANCO-790916); views and opinions expressed in this paper are those of the authors and do not necessarily reflect those of the funding agencies. We would like to thank Tommi Asikainen, Ted Cohen, Mirjam Kretzschmar, Christina Mills and Marcello Pagano for their useful comments on an earlier version of this manuscript.
Supplementary Material
Mathematical properties of moment generating functions and their biological implications (Section 1). Relation between reproduction number and growth rate for epidemic models with complex infection cycles (Section 2). Notation of key variables and their interpretation in ecology, demography and epidemiology (Table 1). Moment generating functions for several probability distributions (Table 2)
References
- Anderson R.M, May R.M. Oxford University Press; Oxford, UK: 1991. Infectious diseases of humans: dynamics and control. [Google Scholar]
- Begon M, Harper J.L, Townsend C.R. Blackwell; Oxford, UK: 1996. Ecology. [Google Scholar]
- Caswell H. 2nd edn. Sinauer Press; Sunderland, MA: 2001. Matrix population models: construction, analysis, and interpretation. [Google Scholar]
- Dublin L.I, Lotka A.J. On the true rate of natural increase, as exemplified by the population of the United States, 1920. J. Am. Stat. Assoc. 1925;150:305–339. doi:10.2307/2965517 [Google Scholar]
- Feller W. On the integral equation of renewal theory. Ann. Math. Stat. 1941;12:243–267. [Google Scholar]
- Ferguson N, et al. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature. 2005;437:209–214. doi: 10.1038/nature04017. doi:10.1038/nature04017 [DOI] [PubMed] [Google Scholar]
- Ferguson N, et al. Strategies for mitigating an influenza pandemic. Nature. 2006;442:448–452. doi: 10.1038/nature04795. doi:10.1038/nature04795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gani R, Leach S. Transmission potential of smallpox in contemporary populations. Nature. 2001;414:748–751. doi: 10.1038/414748a. doi:10.1038/414748a [DOI] [PubMed] [Google Scholar]
- Heffernan J.M, Smith R.J, Wahl L.M. Perspectives on the basic reproductive ratio. J. R. Soc. Interface. 2005;2:281–293. doi: 10.1098/rsif.2005.0042. doi:10.1098/rsif.2005.0042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu N, et al. Effects of antiviral drugs on viral detection in influenza patients and on the sequential infection to their family members—serial examination by rapid diagnosis (Capilia) and virus culture. Int. Congress Ser. 2004;1263:105–108. doi:10.1016/j.ics.2004.02.020 [Google Scholar]
- Keeling M.J, et al. Dynamics of the 2001 UK foot and mouth epidemic: stochastic dispersal in a heterogeneous landscape. Science. 2001;294:813–817. doi: 10.1126/science.1065973. doi:10.1126/science.1065973 [DOI] [PubMed] [Google Scholar]
- Keyfitz N, Caswell H. Springer; New York, NY: 2005. Applied mathematical demography. [Google Scholar]
- Levin B.R, Bull J.J, Stewart F.M. The intrinsic rate of increase of HIV/AIDS: epidemiological and evolutionary implications. Math. Biosci. 1996;132:69–96. doi: 10.1016/0025-5564(95)00053-4. doi:10.1016/0025-5564(95)00053-4 [DOI] [PubMed] [Google Scholar]
- Lipsitch M, et al. Transmission dynamics and control of severe acute respiratory syndrome. Science. 2003;300:1966–1970. doi: 10.1126/science.1086616. doi:10.1126/science.1086616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longini I.M, Halloran M.E, Nizam A, Yang Y. Containing pandemic influenza with antiviral agents. Am. J. Epidemiol. 2004;159:623–633. doi: 10.1093/aje/kwh092. doi:10.1093/aje/kwh092 [DOI] [PubMed] [Google Scholar]
- Metz J.A.J, Diekmann O. Springer; Berlin, Germany: 1986. The dynamics of physiologically structured populations. p. 153. [Google Scholar]
- Mills C, Robins J.M, Lipsitch M. Transmissibility of 1918 pandemic influenza. Nature. 2004;432:904–906. doi: 10.1038/nature03063. doi:10.1038/nature03063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mood A.M, Graybill F.A, Boes D.C. McGraw-Hill; Singapore: 1974. Introduction to the theory of statistics. [Google Scholar]
- Pybus O.G, et al. The epidemic behaviour of hepatitis C virus. Science. 2001;22:2323–2325. doi: 10.1126/science.1058321. doi:10.1126/science.1058321 [DOI] [PubMed] [Google Scholar]
- Roberts M.G, Heesterbeek J.A.P. A new method for estimating the effort required to control an infectious disease. Proc. R. Soc. B. 2003;270:1359–1364. doi: 10.1098/rspb.2003.2339. doi:10.1098/rspb.2003.2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallinga J, Teunis P. Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. Am. J. Epidemiol. 2004;160:509–516. doi: 10.1093/aje/kwh255. doi:10.1093/aje/kwh255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wearing H.J, Rohani P, Keeling M.J. Appropriate models for the management of infectious diseases. PLoS Med. 2005;2:e174. doi: 10.1371/journal.pmed.0020174. doi:10.1371/journal.pmed.0020174 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mathematical properties of moment generating functions and their biological implications (Section 1). Relation between reproduction number and growth rate for epidemic models with complex infection cycles (Section 2). Notation of key variables and their interpretation in ecology, demography and epidemiology (Table 1). Moment generating functions for several probability distributions (Table 2)