Abstract
Archaeological maize specimens from Andean sites of southern South America, dating from 400 to 1400 years before present, were tested for the presence of ancient DNA and three microsatellite loci were typed in the specimens that gave positive results. Genotypes were also obtained for 146 individuals corresponding to modern landraces currently cultivated in the same areas and for 21 plants from Argentinian lowland races. Sequence analysis of cloned ancient DNA products revealed a high incidence of substitutions appearing in only one clone, with transitions prevalent. In the archaeological specimens, there was no evidence of polymorphism at any one of the three microsatellite loci: each exhibited a single allelic variant, identical to the most frequent allele found in contemporary populations belonging to races Amarillo Chico, Amarillo Grande, Blanco and Altiplano. Affiliation between ancient specimens and a set of races from the Andean complex was further supported by assignment tests. The striking genetic uniformity displayed by the ancient specimens and their close relationship with the Andean complex suggest that the latter gene pool has predominated in the western regions of southern South America for at least the past 1400 years. The results support hypotheses suggesting that maize cultivation initially spread into South America via a highland route, rather than through the lowlands.
Keywords: ancient DNA, landraces, maize, microsatellites, South America
1. Introduction
Maize (Zea mays ssp. mays) is the principal domesticated crop of the Americas. Although its Mesoamerican origin has been clearly established (e.g. Doebley 1990; Benz 2001; Matsuoka et al. 2002b), its time of arrival and trajectory of spread through South America is still uncertain. According to Piperno & Pearsall (1998), maize was already present in southern Central and northern South America between 7700 and 6000 years ago, but this scenario, primarily based upon plant microfossil evidence, has been questioned by Staller & Thompson (2002) who proposed a more recent introduction on archaeological and palaeoethnobotanical grounds (1200–2200 BC). The pattern of spread of maize cultivation into South America has been inferred by cytogenetic and molecular studies of landraces and other varieties, but again with contradictory results. McClintock et al. (1981), following an extensive examination of heterochromatic bands (chromosome knobs), concluded that maize was initially introduced into the central Andes and then spread extensively throughout the highland and lowland regions of the continent, not being significantly supplemented by other types of maize until new genotypes spread southwards along the east coast of Brazil in relatively recent times. In contrast, analysis of microsatellite variation suggests that maize was first introduced into the lowlands of South America, reaching the Andes at a later stage (Matsuoka et al. 2002b).
The large number of landraces currently cultivated throughout South America (more than 300), many of which are directly descended from the crops grown by natives and are maintained by traditional farmers with little or no input from commercial inbred lines, constitute a valuable source of material from which to reconstruct the origins and spread of maize. Additional insights might also be obtained by examination of ancient DNA (aDNA) preserved in archaeological specimens: such studies enabling genetic variation to be assessed over time (Jaenicke-Després et al. 2003) and avoiding the complicating factors caused by the movement of genotypes during the post-Columbian period. By analysis of a short segment of the Adh2 locus in archaeological specimens from eastern Brazil, Peru and northern Chile, Freitas et al. (2003) found evidence for two separate expansions of maize cultivation into South America, one from Central America into the Andean regions and a second along the lowlands of the northeast coast of the continent. This examination of aDNA therefore enabled a rapport to be reached between the differing outcomes of the cytogenetic and molecular studies of modern material.
Ancient DNA therefore has considerable potential in developing an understanding of the history of maize cultivation in South America, particularly so given that the morphological attributes needed for racial identification are not always present or sufficiently well preserved in charred cobs and kernels, the types of material generally recovered from archaeological sites. The aim of the present study was to use this approach to assess the genetic affiliation of archaeological specimens and extant landraces in northwestern Argentina. This region comprises the southernmost area of maize distribution and possesses a diverse variety of landraces, some of which are thought to be related to primitive maize forms. Indeed, Argentine popcorn, a collection introduced into the United States from Argentina 50 years ago, is thought to more closely resemble the earliest maize remains from the Tehuacan Valley, Mexico, than any extant race thus far examined (Benz & Iltis 1990). Apart from its present importance as a reservoir of morphological and genetic variation, northwestern Argentina is also noted for providing extraordinarily good preservation conditions for human, animal and plant remains, with the Llullaillaco mummies being one of the most remarkable examples (Previgliano et al. 2003). Desiccated and charred maize cobs and kernels have been recovered from several archaeological sites ranging in age from 300 to 2000 years before present (BP) and have been related to different human cultures with contrasting degrees of social and technological complexity, from the early agro-pastoral societies to the Inca and Spanish occupations (e.g. Sempé 1977; Tarragó 1980; Korstanje & Wurschmidt 1999).
The extensive microsatellite survey conducted by Matsuoka et al. (2002a,b), which encompasses the entire pre-Columbian range of maize distribution, enables comparison of microsatellite alleles throughout the Americas and provides a framework for the analysis of genetic variation in both extant and archaeological specimens. The short lengths of the polymerase chain reaction (PCR) amplicons needed to type most microsatellite markers make them suitable for aDNA analysis, aDNA typically being recovered as short fragments less than 200 bp (Allaby et al. 1997), and identification of alleles by amplicon size circumvents problems caused by diagenetic changes when single nucleotide polymorphisms are typed in aDNA (Pääbo 1989; Poinar 2003). The aim of this project was therefore to extend the microsatellite data obtained by Matsuoka et al. (2002a,b) to key archaeological maize specimens from northwestern Argentina, in order to establish the genetic affiliations between these ancient plants and the crops currently grown in this region.
2. Material and methods
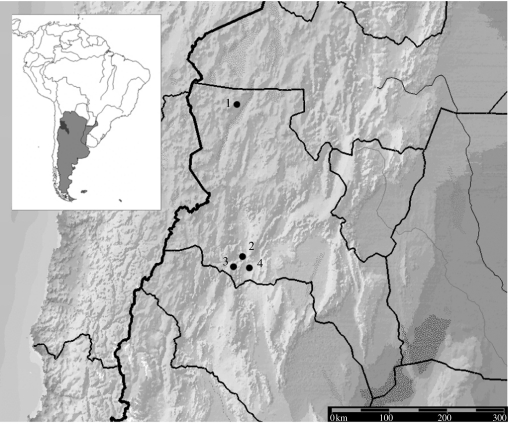
Collection sites, race, voucher information, age, type of remains and number of individuals examined in archaeological and modern maize landraces are described in table 1. Location of archaeological sites is presented in figure 1. Archaeological samples were dated according to the contextual evidence available for each site. Accelerator mass spectroscopy was used to confirm the antiquity of the specimens belonging to sites Lorohuasi, Tebenquiche and Punta Colorada (Beta Analytic).
Table 1.
Maize specimens used in this study.
| collection site | voucher/IDa | raceb | altitude (m.a.s.l.) | age (years BP) | type of remains | contextc | number of samples | |
|---|---|---|---|---|---|---|---|---|
| archaeological specimens | ||||||||
| Tebenquiche | Quebrada de Tebenquiche Chico, Dpto. Antofagasta de la Sierra, Catamarca, Argentina | TC | Blanco y Ocho Rayas–Ocho Rayas | 3650 | 360±40 | cobs | D | 12 |
| Lorohuasi | National Route 60, Paraje Lorohuasi, Dpto. Tinogasta, Catamarca, Argentina | LH | Pisingallo–Capia Morocho–Chaucha Rosita Colorado | 2150 | 440±50 | kernels | F | 25 |
| Punta Colorada | 12 km east of Las Angosturas, left margin of the river Guanchin-Chaschuil, Dpto. Tinogasta, Catamarca, Argentina | PC | Capia Pisingallo | 2255 | 1320±40 | cobs | F | 8 |
| Batungasta | Quebrada de la Troya, Dpto. Tinogasta, Catamarca | BT | Capia Pisingallo | 1500 | ca 400 | cobs | D | 6 |
| modern specimens | ||||||||
| pop. 6473 | Susques, Dpto. Susques, Jujuy, Argentina | VAV 6473 | Altiplano | 3620 | — | — | — | 20 |
| pop. 6167 | El Puesto, Dpto. Santa Victoria, Salta, Argentina | VAV 6167 | Altiplano | 3000 | — | — | — | 14 |
| pop. 6484 | Tumbaya, Dpto. Tumbaya, Jujuy, Argentina | VAV 6484 | Amarillo Chico | 2010 | — | — | — | 20 |
| pop. 6476 | Termas de Reyes, Dpto. Capital, Jujuy, Argentina | VAV 6476 | Amarillo Chico | 1690 | — | — | — | 14 |
| pop. 6480 | La Ciénaga de Pumamarca, Dpto. Tumbaya, Jujuy, Argentina | VAV 6480 | Amarillo Grande | 2420 | — | — | — | 25 |
| pop. 6485 | Colonia San José, Dpto. Tilcara, Jujuy, Argentina | VAV 6485 | Blanco | 2670 | — | — | — | 13 |
| pop. 6313 | Los Toldillos, Piedras Blancas, Dpto. Ambato, Catamarca, Argentina | VAV 6313 | Pisingallo | 1600 | — | — | — | 16 |
| pop. 6482 | La Candelaria, Dpto. Candelaria, Salta, Argentina | VAV 6482 | Orgullo Cuarentón | 910 | — | — | — | 24 |
| pop. 6607 | Aldea Alecrin, Dpto. Eldorado, Misiones, Argentina | VAV 6607 | Pipoca Colorado | — | — | — | — | 10 |
| pop. 6614 | Aldea Guavirá Poty, Dpto. San Pedro, Misiones, Argentina | VAV 6614 | Pororó Grande | — | — | — | — | 11 |
TC, Tebenquiche Chico; LH, Lorohuasi; PC, Punta Colorada; BT, Batungasta. Voucher specimens of modern landraces are deposited at the Laboratorio de Recursos Genéticos Vegetales N.I. Vavilov (VAV), Faculty of Agronomy, University of Buenos Aires, Argentina.
Taxonomic identification based on morphological criteria.
Context from which archaeological specimens were recovered: D, domestic; F, funerary.
Figure 1.
Locations of the archaeological sites from which ancient maize specimens were collected: 1, Tebenquiche; 2, Lorohuasi; 3, Punta Colorada; and 4, Batungasta.
DNA from desiccated cobs and kernels (100 mg) was isolated by the silica-based spin column method described by Yang et al. (1998) with minor modifications. Three microsatellite loci were examined, phi127, phi029 and phi059 (nomenclature of Matsuoka et al. (2002a,b); the loci are on linkage groups 2, 3 and 10, respectively), using primer sequences described in the Maize Genetics and Genomics Database (http://www.maizegdb.org/locus.php). Reaction mixtures contained 2–10 μl DNA extract, 90 ng of each primer, 125 μM of each dNTP, 2 mM MgCl2, 150 μg ml−1 bovine serum albumen (BSA), 1.25 units Taq DNA polymerase (Fermentas), 1× PCR buffer with (NH4)2SO4 and sterile double distilled water to a final volume of 50 μl. Cycling conditions were: 4 min at 94°C; 35 cycles of 1 min at 94°C; 1 min at the annealing temperature; 1 min at 72°C; and a final elongation step of 6 min at 72°C. The annealing temperatures were 47, 50 and 55°C for phi127, phi029 and phi059, respectively. Reactions containing fragments of the expected size were further purified (QIAquick PCR Purification Kit, Qiagen), reamplified under the same conditions but with no addition of BSA and cloned using the TOPO TA Cloning System (Invitrogen). At least five clones were sequenced per specimen (247 clones in total) with M13 universal primers using the ABI Prism BigDye Terminator Cycle Sequencing Kit and an Applied Biosystems 377 DNA sequencer. Specimens that gave the same sequence in every amplification product were considered to be homozygous, although heterozygosity cannot be excluded. Precautions were taken to minimize the risk of contaminating ancient material with modern DNA: ancient extracts were prepared in an isolated room not used for handling modern DNA or PCR products; PCR mixes were set up in a third laboratory in a laminar flow cabinet (HEPA filter, Class 100, conforming to BS 5295 and 5726) used for no other purpose; standard precautions were taken regarding pipette types; solutions and work surfaces were sterilized by autoclaving and ultraviolet irradiation, respectively; and all aDNA amplifications were accompanied by an extraction and water blank. Reproducibility of positive results was confirmed by independent rounds of extraction and amplification.
For modern plants, DNA was extracted from 2- to 3-day-old seedlings according to Dellaporta et al. (1983). Maize landraces were genotyped at loci phi127, phi029 and phi059 in order to calculate contemporary allelic frequencies. Reaction mixtures contained 1 μl DNA extract, 30 ng of each primer, 125 μM of each dNTP, 1.5 mM MgCl2, 0.5 units Taq DNA polymerase (Promega), 1× PCR buffer and sterile double distilled water to a final volume of 25 μl. A touchdown cycling profile (annealing temperature 65–55°C) was used and PCR products were separated on a 6% denaturing polyacrylamide gel (8 M urea) following standard procedures. Gels were silver stained with Silver Sequence DNA Staining Reagents (Promega). Alleles were identified by comparison with products of known size using GelPro Analyzer v. 4.0 (Media Cybernetics) and further confirmed by direct sequencing of homozygous specimens representative of each allelic class. Allelic variants, 112 (phi127) and 154 (phi029), corresponding to teosintes Z. mays ssp. parviglumis and Z. mays ssp. mexicana were also sequenced for comparison with archaeological specimens. All Zea sequences obtained in this study have been deposited in GenBank under accession numbers AY965913–AY965994.
Sequences were aligned using ClustalW (Higgins et al. 1994) followed by minor manual modifications. Allelic frequencies of modern maize populations were calculated by the direct count method. Genic differentiation among populations and genotypic linkage equilibria were evaluated using Fisher's exact test as implemented in genepop (Raymond & Rousset 1995). Departures from Hardy–Weinberg proportions were assessed using the score test (U-test) provided by the same software package. Amplification products not corresponding to the microsatellite loci under study were analysed by comparison with the public sequence databases as both nucleotide and amino acid translations using blastn, blastx and tblastx. A sequence was classified as a known element when retrieved with an E-value of less than 10−5 following the criteria of Meyers et al. (2001). The identity and E-values of the highest score identifiable matches are provided as electronic supplementary material. Assignment of archaeological specimens to modern landraces was performed according to the assignment test developed by Paetkau et al. (1995). The method assigns an individual to the population in which its genotype is most likely to occur. As summarized in Cornuet et al. (1999), given J independent loci typed in I reference populations (and in the individuals to be assigned), the frequency of allele k at locus j in population i is pijk. Assuming Hardy–Weinberg equilibrium, the likelihood of a genotype AkAk′ occurring in the ith population at the jth locus is proportional to (pijk)2 if k=k′ and to 2 pijkpijk′ otherwise. Since the J loci are assumed to be independent, the likelihood of a multilocus genotype occurring in a given population is the product of likelihoods for each locus. The probability of drawing random samples of individuals of a given genotype from different populations was calculated with Microsoft Excel assuming a binomial distribution (see §3).
3. Results
(a) Ancient DNA sequences
DNA extracted from archaeological specimens was consistently of low molecular weight (100–500 bp) as expected for aDNA (Pääbo 1989; Poinar 2003). However, high molecular weight fragments (approx. 20 kb) were also retrieved from Tebenquiche samples. Amplicons of the expected size and sequence were obtained for 9 out of 51 archaeological samples examined (table 2). The remaining samples failed to give positive results despite repeated rounds of extraction and amplification. No positive amplifications were obtained from the Batungasta samples. All negative controls including mock extractions were always devoid of PCR product.
Table 2.
Microsatellite alleles found in archaeological maize specimens from northwestern Argentina.
| sample | site | type of remains | racea | allele | no. of clonesb | |
|---|---|---|---|---|---|---|
| Phi127 | ||||||
| TC7C | Tebenquiche | cob | n.a. | 112 | 5/10 | |
| TC54 | Tebenquiche | cob | n.a. | 112 | 8/20 | |
| LH20 | Lorohuasi | kernel | Pisingallo | 112 | 5/39 | |
| LH22 | Lorohuasi | kernel | Capia | 112 | 2/53 | |
| LH38 | Lorohuasi | kernel | Morocho | 112 | DS | |
| LH39 | Lorohuasi | kernel | Chaucha | 112 | 5/18 | |
| LH40 | Lorohuasi | kernel | Chaucha | 112 | 1/5 | |
| PC51 | Punta Colorada | cob | undetermined | 112 | 5/5 | |
| PC52 | Punta Colorada | cob | undetermined | 112 | 9/11 | |
| Phi029 | ||||||
| LH38 | Lorohuasi | kernel | Morocho | 154 | 8/11 | |
| LH40 | Lorohuasi | kernel | Chaucha | 154 | 1/2 | |
| Phi059 | ||||||
| TC54 | Tebenquiche | cob | n.a. | 157 | DS |
Taxonomic identification based on morphological criteria; n.a., could not be assigned to any extant race.
No. of clones, number of clones carrying an amplicon of the targeted microsatellite locus/total number of clones examined; DS, genotype obtained via direct sequencing.
At locus phi127, all the nine archaeological specimens that gave results were homozygous for allelic variant 112 (table 2). This allele was also present in modern populations, along with four others ranging in size from 114 to 126 bp (table 3). Analysis of 37 clones from archaeological sequences revealed sequence variations at a total of 23 nucleotide positions (see electronic supplementary material). The majority of variations (21/25) were singletons (substitutions appearing only in one clone), with 43% corresponding to putative C/G→T/A transitions. Alterations that occur in only a few sequences of a cloned PCR product can result either from misincorporation due to aDNA degradation or from erroneous Taq polymerase activity (Hansen et al. 2001). In our results, the strong bias towards C/G→T/A transitions indicates amplification of degraded templates with a high incidence of cytosine deamination (Hofreiter et al. 2001). Consensus sequences reconstructed from those individuals represented by multiple clones were identical to the allelic variant 112 present in contemporary samples, with one exception. This was specimen PC51, which had substitutions at positions 88 and 96 in the majority of clones. These could be authentic polymorphisms, but the observed pattern is also consistent with a small number of degraded template molecules initiating the PCR (Hofreiter et al. 2001). To evaluate the possible evolutionary significance of these substitutions, allele variant 112 was sequenced from Z. mays ssp. parviglumis and Z. mays ssp. mexicana. No evidence of polymorphism was detected in these teosintes (see electronic supplementary material). However, the same substitution was observed at position 96 of allele 126 from contemporary samples.
Table 3.
Allele frequencies observed at microsatellite loci phi127, phi029 and phi059 for the eight modern maize populations from northwestern Argentina.
| alleles | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pop. | locus phi127 | locus phi029 | locus phi059 | |||||||||||||||
| 112 | 114 | 120 | 124 | 126 | 150 | 151 | 154 | 156 | 158 | 159 | 160 | 161 | 148 | 152 | 154 | 157 | 161 | |
| 6473 | 0.971 | 0 | 0 | 0.029 | 0 | 0.028 | 0 | 0.972 | 0 | 0 | 0 | 0 | 0 | 0.583 | 0 | 0 | 0.417 | 0 |
| 6167 | 0.923 | 0 | 0 | 0.077 | 0 | 0.286 | 0 | 0.678 | 0 | 0.036 | 0 | 0 | 0 | 0.423 | 0.038 | 0 | 0.462 | 0.077 |
| 6484 | 0.895 | 0 | 0 | 0 | 0.105 | 0.325 | 0 | 0.650 | 0 | 0 | 0 | 0.025 | 0 | 0.350 | 0 | 0 | 0.625 | 0.025 |
| 6476 | 0.542 | 0.042 | 0.042 | 0.374 | 0 | 0.167 | 0.083 | 0.708 | 0.042 | 0 | 0 | 0 | 0 | 0.409 | 0 | 0.136 | 0.364 | 0.091 |
| 6480 | 0.780 | 0 | 0 | 0.220 | 0 | 0.333 | 0 | 0.625 | 0.042 | 0 | 0 | 0 | 0 | 0.460 | 0 | 0 | 0.460 | 0.080 |
| 6485 | 1 | 0 | 0 | 0 | 0 | 0.423 | 0 | 0.539 | 0 | 0 | 0.038 | 0 | 0 | 0.417 | 0 | 0 | 0.500 | 0.083 |
| 6313 | 0.438 | 0 | 0 | 0.406 | 0.156 | 0.250 | 0 | 0.214 | 0 | 0.072 | 0 | 0 | 0.464 | 0.594 | 0 | 0 | 0.219 | 0.187 |
| 6482 | 0.478 | 0 | 0 | 0.500 | 0.022 | 0.417 | 0.146 | 0.417 | 0 | 0.020 | 0 | 0 | 0 | 0.761 | 0.043 | 0.023 | 0.130 | 0.043 |
The two ancient specimens typed at locus phi029 were homozygous for allelic variant 154 (table 2), which was also present in all the modern populations examined. In total, eight alleles were detected in contemporary specimens, with allele 154 being found at high frequency in several populations (table 3). The consensus sequence obtained from specimen Lorohuasi 38 was identical to that retrieved from modern plants. Nineteen base differences were identified in single clones at random positions and, as previously described for locus phi127, C/G→T/A transitions were the prevalent nucleotide substitutions (14/19; see electronic supplementary material). The allelic variant reconstructed from specimen Lorohuasi 40 showed 3% uncorrected sequence divergence with respect to its contemporary counterpart. Five nucleotide substitutions were detected, four of which were present in both clones examined. Modern specimens display sequence variation within the part of the amplicon containing these substitutions, but the specific variations seen in Lorohuasi 40 were not found in any modern maize or teosinte (see electronic supplementary material).
Alleles from locus phi059 were refractory to cloning and had to be analysed by direct sequencing. In the archaeological specimens, a single allele was detected corresponding to variant 157 of contemporary landraces (tables 2 and 3). The archaeological sequence was identical to that observed for modern populations 6473 and 6476, but it showed 0.5% sequence divergence from the alleles from populations 6480, 6484 and 6482.
Clone analysis revealed several amplification products not corresponding to the expected microsatellite loci. After BLAST searches, most of these were found to be of microbial or unknown origin (see electronic supplementary material), but highly significant matches (E-values≤e−26) were also obtained for sequences identified as members of the Huck (SanMiguel et al. 1996) and Zeon (Hu et al. 1995) maize retrotransposon families, which are among the most abundant sequences of the maize genome (Meyers et al. 2001). It is noteworthy that all four putative retrotransposon sequences were retrieved from kernels (Lorohuasi 20, Lorohuasi 38 and Lorohuasi 40), while the majority of contaminating micro-organisms were associated with cobs from the Tebenquiche site. The presence of high molecular weight DNA in the Tebenquiche extracts is concordant with these observations, as ancient extracts with DNA fragments up to 10–20 kb are frequently indicative of microbial contamination (Rollo et al. 1994). The cobs from this site were the only ones recovered from domestic areas, whereas Lorohuasi and Punta Colorada specimens were retrieved from funerary, more protected, settings.
(b) Assignment of archaeological specimens to modern landraces
Genotyping of 146 individuals belonging to eight extant maize landraces currently cultivated in northwestern Argentina was conducted to provide a reference framework for determination of the genetic affiliations between modern and archaeological specimens. Following from the assumptions of Paetkau's assignment method, both panmixia and linkage equilibria were evaluated in the eight modern populations. No deviations from genotypic linkage equilibrium between pairs of loci were detected, as determined by the Fisher's exact test. Hardy–Weinberg proportions were confirmed for all population–locus combinations, except for the homozygote excess observed in population 6482 at locus phi059 (score test p=0.0001). Allelic frequencies for contemporary populations are given in table 3.
Despite being cultivated in the same region (northwestern Argentina), the eight landraces can be placed in three groups according to morphological and cytogenetic evidence (Poggio et al. 1998): (i) Andean complex (Altiplano populations 6473, 6167; Amarillo Chico populations 6476, 6484; Amarillo Grande population 6480; Blanco population 6485), (ii) South American popcorns (Pisingallo population 6313), and (iii) incipient races derived from the introduction of commercial germplasm into local varieties approximately 40 years ago (Orgullo Cuarentón population 6482). Application of the assignment test to locus phi127 places each archaeological specimen in race Blanco (table 4). However, the allelic frequencies obtained for this population are not significantly different from those corresponding to races Altiplano, Amarillo Grande and population 6484 of race Amarillo Chico. Similarly, when phi059 is considered, the highest probability of occurrence of the archaeological genotype is shown by population 6484 of race Amarillo Chico, but the populations belonging to races Amarillo Grande, Altiplano and Blanco, as well as population 6476 of race Amarillo Chico, show no significant differences in their allelic distributions. In contrast, phi029 assigns the highest probability to population 6473 of race Altiplano, with no other population displaying such a pattern of allelic distribution. As individuals Lorohuasi 38, Lorohuasi 40 and TC54 could be simultaneously genotyped for two loci, the probabilities of the combined genotypes (phi127112/112–phi029154/154; phi127112/112–phi059157/157) were also calculated (table 4). Once again, the populations from the Andean complex exhibit the highest probabilities of occurrence, while those corresponding to races Pisingallo and Orgullo Cuarentón are greatly diminished. To examine possible affiliations with non-Andean landraces, plants representative of racial complexes from the lowlands (populations VAV6607 and VAV6614; table 1) were typed at loci phi127 and phi029. Three alleles were found at phi127: 112 (f, 0.545); 124 (f, 0.137); and 126 (f, 0.318). Two alleles were found at phi029: 150 (f, 0.650); and 154 (f, 0.350). Therefore, for locus phi127, the probability of a genotype 112/112 being sampled from the lowland region was 0.297. For locus phi029, the probability of occurrence of genotype 154/154 in this area was 0.123, with the combined genotype phi127112/112–phi029154/154 having a likelihood of 0.037.
Table 4.
Probability of occurrence of archaeological genotypes in modern maize landraces from northwestern Argentina according to the method of Paetkau et al. (1995). (SA, South American; Am. Ch., Amarillo Chico; Am. Gde., Amarillo Grande; Org. C., Orgullo Cuarentón.)
| archaeological genotype | modern landraces | |||||||
|---|---|---|---|---|---|---|---|---|
| Andean races | SA popcorns | incipient races | ||||||
| Altiplano 6473 | Am. Ch. 6476 | Blanco 6485 | Am. Gde. 6480 | Am. Ch. 6484 | Altiplano 6167 | Pisingallo 6313 | Org. C. 6482 | |
| Phi127112/112 | 0.941a | 0.292b | 1a | 0.608a | 0.801a | 0.852a | 0.191b | 0.228b |
| Phi029154/154 | 0.945 | 0.502a | 0.290b | 0.391a | 0.422a | 0.460a | 0.046 | 0.174b |
| Phi059157/157 | 0.174a | 0.132a | 0.250a | 0.212a | 0.391a | 0.187a | 0.048 | 0.017 |
| Phi127112/112–Phi029154/154 | 0.889 | 0.146 | 0.404 | 0.238 | 0.335 | 0.390 | 0.008 | 0.035 |
| Phi127112/112–Phi059157/157 | 0.163 | 0.035 | 0.213 | 0.129 | 0.309 | 0.155 | 0.008 | 0.003 |
Populations designated by the same letter show no significant differences in the distribution of their allelic frequencies at the corresponding locus.
Generally, the results of the assignment test indicate that the archaeological specimens are more closely affiliated to the races of the Andean complex than to South American popcorns or incipient races, even though these are all currently cultivated in the same area, or to populations from the lowland regions of South America. Given the small sample sizes, it is conceivable that germplasm from lineages other than the Andean complex were present historically, but not represented in the archaeological samples that were studied. To evaluate this possibility, data from extant populations were combined into overall allele frequencies and the probability of drawing allele samples equal to those observed in the archaeological specimens was calculated. Taking advantage of the fact that a single allele size was sampled from archaeological specimens at all the three loci examined, population frequencies were recalculated considering only two allele categories (e.g. 112 and non-112 for locus phi127) and probabilities were computed according to the binomial distribution. For locus phi127, the probability of drawing eighteen 112 alleles was: 0.0047 for the combined gene pool comprising the populations from the Andean complex, Pisingallo and Orgullo Cuarenton; 0.0298 for the Andean complex–lowland region assembly; 1.8×10−5 for the lowland region alone; and 0.0578 for the Andean complex alone. For locus phi029, these probabilities were 0.1302, 0.1975, 0.015 and 0.2374, respectively.
4. Discussion
(a) Ancient DNA
Plant aDNA studies have lagged behind animal and human investigations, but several nuclear loci have successfully been amplified from a variety of domesticated crops (e.g. O'Donoghue et al. 1996; Allaby et al. 1997; Brown et al. 1998; Manen et al. 2003), including maize (Rollo et al. 1991; Goloubinoff et al. 1993; Jaenicke-Després et al. 2003; Freitas et al. 2003). The microsatellite sequences reported here provide another example of single-copy regions being amplified from archaeological plant remains. Several observations support the authenticity of these ancient sequences: (i) those DNA extracts that were not heavily contaminated with microbial DNA contained only low molecular weight material, consistent with the expected molecular properties of aDNA, (ii) sequences of cloned PCR products contained singleton substitutions typical in nature and distribution to damage artefacts present in ancient DNA, (iii) the results were corroborated by independent rounds of DNA extraction and amplification, and (iv) the results are phylogenetically meaningful. However, the lack of genetic variation seen among the archaeological specimens could be regarded as unnatural and hence due to uniform contamination of these specimens or extracts (Hadly et al. 2003). As previously stated, no evidence of contamination was detected at any stage of the experimental procedures and although cross-contamination between PCRs is always a possibility, there is no reason to believe that it should exclusively affect archaeological samples and not extraction or amplification controls. Therefore, it is reasonable to conclude that the genotypes obtained for the archaeological specimens are authentic.
A thorough understanding of the chemical lesions present in aDNA is of fundamental importance for the identification of authentic nucleotide substitutions in sequences from archaeological and other preserved specimens. Two main types of damage are likely to affect DNA in archaeological deposits. First, hydrolytic damage will result in depurination and depyrimidination and in deamination of bases. Second, oxidative damage will result in a variety of base modifications (Höss et al. 1996). Deoxyuridine residues appear to be the most frequent miscoding lesion in aDNA extracted from both animal and human specimens (Hansen et al. 2001; Hofreiter et al. 2001; Orlando et al. 2003), but no such bias has been reported for plant remains. The prevalence of C/G→T/A transitions in the archaeological sequences studied here (see electronic supplementary material) support the hypothesis that cytosine deamination is indeed the predominant miscoding modification in many if not most aDNA samples.
O'Donoghue et al. (1996) suggested that the microenvironment within desiccated seeds is conducive to enhanced preservation of lipids and other biomolecules. Interestingly, five of the nine archaeological specimens genotyped at locus phi127 were kernels, and so were the two individuals typed at locus phi029.
(b) Genotypes of archaeological specimens and relationships with modern landraces
All the three microsatellite loci examined in the archaeological specimens exhibited a single allelic variant, identical in size to the most frequent allele found in contemporary populations belonging to races Amarillo Chico (6476, 6484), Amarillo Grande (6480), Blanco (6485) and Altiplano (6167). This genetic homogeneity is remarkable when considering the diversity of the archaeological sites included in this study (table 1). These not only encompass a time period of nearly 1000 years, but they also cover different socio-historical periods each characterized by a distinctive pattern of agricultural production and interregional exchange. Furthermore, the specimens from Punta Colorada and Lorohuasi were found in association with funerary artefacts, whereas the Tebenquiche specimens were retrieved from households. Contrasting climatic conditions could also be regarded as a factor promoting genetic differentiation among the races cultivated at each site. The mountain slopes of the Abaucán Valley (Lorohuasi, Punta Colorada) provide a fertile environment for the development of cultivars with little or no need for artificial irrigation. Most of the sites from this area are located along extant riversides or at the verge of ancient riverbeds. In contrast, the agricultural activities in a high-altitude desert such as the Atacama Plateau (Tebenquiche, 3650 m.a.s.l.) are strongly conditioned by water availability.
Although the genetic homogeneity displayed by these archaeological samples is remarkable, the results are not unprecedented. Vásquez et al. (in press) report homogeneity at five microsatellite loci in 400–600-year-old maize remains from two sites, 220 km apart, associated with the Chimú culture of the north Peruvian coast. They suggest that homogeneity at a particular site could arise owing to an annual founder effect, the inhabitants saving just a few cobs every year for sowing, rather than flailing the crop and choosing the best grains as the seeds for the following year. Similar farming practices are observed in northwestern Argentina, suggesting that founder effects could also explain the homogeneity of the archaeological specimens from this area. A founder effect is most likely to fix the most common allele variants in a particular population, so the homogeneity observed in samples from Lorohuasi, Tebenquiche and Punta Colorada could indicate that the original maize populations at each of these three sites had similar genetic structures. If this hypothesis is correct, then the assignment tests suggest that these ancestral populations were similar to the modern Andean complex.
An alternative explanation of the homogeneity is that this is an artefact of the small sample size which is an inevitable constraint of aDNA studies. If only a few individuals are sampled from a population within which there is a prevalence of certain allelic variants, then these sampled individuals will most probably exhibit the most frequent alleles. If the prevalent alleles are the same in each of the several source populations, then individuals sampled from these different populations might appear identical. If this is the explanation of the homogeneity of the archaeological samples then, of the modern races studied, those of the Andean complex are the ones most likely to resemble the archaeological populations, the probability of drawing gene samples equal to those observed in the archaeological specimens from extant populations being higher for the Andean complex than for the other population combinations examined. For instance, it is 10 times more likely to draw eighteen 112 alleles (locus phi127) from the Andean complex alone than from the gene pool comprising the Andean complex, Pisingallo and Orgullo Cuarenton, and almost two times more likely when the Andean complex–lowland assembly is considered. This explanation of the observed homogeneity implies that the ancient populations had similar genetic structures despite the temporal, socio-historical and geographical differences between the sites. This could also arise if significant gene flow occurred in the past to preclude genetic differentiation and maintain a similar population structure throughout time and space. With kernels being the principal means of dispersal, food exchange among neighbouring human populations could easily produce such an effect. In fact, high levels of intra- and inter-regional exchange have been extensively documented for northwestern Argentina since very early in the archaeological record (10 000 BP; Castro & Tarragó 1992; Albeck 2000).
Whichever of the above explanations is correct, it appears that, regardless of their site of origin, each archaeological specimen is more closely related to the races of the Andean complex than to the South American popcorns, the incipient races or the lowland races included in this study. The actual assignment of the archaeological specimens to the Andean complex depends on two assumptions, both of which we can demonstrate to be correct. The first assumption is that a certain amount of phylogenetic signal has been retained through time and that allelic frequencies have not been significantly altered by deterministic forces. Genetic differentiation and cluster analyses of a total of 18 microsatellite loci in the modern populations included in this study strongly suggest that this has been the case (Lia 2004). Moreover, Bayesian analysis of a combined data matrix including multilocus genotypes (10 loci) of individuals from these populations and those examined by Matsuoka et al. (2002b) showed that individuals corresponding to populations 6473, 6167, 6476, 6480, 6484 and 6485 and a set of accessions defined as ‘core Andean’ share a similar genetic constitution, whereas populations 6313 and 6482 are significantly different (Lia 2004). The second assumption concerns allele size convergence. To be meaningful, affiliations should be deduced from a set of alleles that are not only identical in state but also, and more importantly, identical by descent. The mutation rate per generation for maize dinucleotide microsatellite loci has been estimated as 7.7×10−4, with microsatellites having repeats of more than 2 bp showing an upper limit of 5.1×10−5 (Vigouroux et al. 2002). Therefore, considering that maize is an annual cultigen and accepting that its domestication took place between 6500 and 9000 years ago (Piperno & Flannery 2001; Matsuoka et al. 2002b), the amount of time elapsed is not sufficient for allele size convergence to have distorted the genetic assignments presented here.
(c) Origins of the Andean complex
Although maize racial classification is far from clear, and the term Andean complex may seem rather vague, the validity of this complex as a significant evolutionary unit has been repeatedly stressed by morphological, cytogenetic and genetic evidence (Goodman & Bird 1977; McClintock et al. 1981; Goodman & Brown 1988; Matsuoka et al. 2002b). The conclusion of the assignment tests, as described earlier, is that each of the nine archaeological specimens for which aDNA sequences were obtained is a member of the Andean complex and that this gene pool has therefore predominated in the western regions of southern South America for at least the last 1300 years. Although interpretation of genetic data, especially the necessarily limited data obtainable from aDNA analysis, in the context of crop dispersal is not straightforward, our results are pertinent to the competing hypotheses regarding the spread of maize cultivation into and through South America. Taking into account the location of northwestern Argentina at the extreme southern range of maize distribution, meaning that it was presumably one of the last areas of South America reached by cultivation and considering that the genetic similarities displayed by the archaeological specimens must predate the oldest age of these specimens (1400 years) by at least several centuries, it appears likely that the genetic ancestors of the Andean complex became established in northwestern Argentina soon after the first arrival of maize cultivation to South America. The prevailing view that the Andean complex is a highland rather than lowland population therefore supports a highland origin for maize cultivation in this region, consistent with the views of McClintock et al. (1981) and Freitas et al. (2003). According to McClintock et al. (1981), the extensive spread of the chromosome constitution characteristic of the Andean complex across such a vast territory (i.e. from Colombia to Northern Chile and the highlands of Argentina) would be expected if this was the type initially introduced into the region and if the introduction of later varieties was delayed, the race Pisingallo (Pisinkalla) from Bolivia and Argentina being an example of the latter. The slightly different proposal of Freitas et al. (2003), that there were separate expansions of maize cultivation into South America, one from Central America into the Andean regions and a second along the lowlands of the northeast coast of the continent, is still consistent with the cytogenetic evidence presented by McClintock and also allows that maize from the Andean region was the first to be introduced in South America.
The antiquity of the Andean complex is not however compatible with the interpretation of Matsuoka et al. (2002b) that maize cultivation reached the Andes at a presumably late stage, only after its initial introduction to the lowlands of South America. If the races from the lowlands of South America were ancestral to those of the Andean complex, then at least some indication of their presence might be expected at archaeological sites from northwestern Argentina, but no evidence for the presence of germplasm from sources other than the Andean complex were found within the samples that we analysed. Both Paetkau's assignment test and the random sampling probabilities calculated according to the binomial distribution show that it is very unlikely for the archaeological specimens to have been derived from a lowland gene pool. Further studies of archaeological specimens, in particular from lowland regions of southern South America, will clarify these issues.
Acknowledgments
We thank Dra. Carlota Sempé and Dr Alejandro Haber for kindly providing the archaeological specimens from the Punta Colorada and Tebenquiche sites, respectively, and we thank Dr Victor Vásquez for allowing us to cite his work on Peruvian maize. We are also in debt to Keri Brown for her assistance in the radiocarbon dating of the samples. Financial support for this work was provided by the Consejo Nacional de Investigaciones Científicas y Técnicas (Argentina), the Agencia Nacional de Promoción Científica y Tecnológica (Argentina), the Universidad de Buenos Aires and the Natural Environment Research Council (UK).
Supplementary Material
BLAST analysis of non-microsatellite nucleotide sequences retrieved from archaeological maize specimens
Allele sequences of locus phi127 from archaeological and extant maize specimens. Clones (c) and direct sequencing (ds) products corresponding to archaeological specimens are highlighted in gray. Z.m.p.: Zea mays ssp parviglumis; Z.m.m.: Zea mays ssp mexicana.
Allele sequences of locus phi029 from archaeological and extant maize specimens. Clones (c) and direct sequencing (ds) products corresponding to archaeological specimens are highlighted in gray. Z.m.p.: Zea mays ssp parviglumis; Z.m.m.: Zea mays ssp mexicana.
References
- Albeck M.E. La vida agraria de los Andes del Sur. In: Tarragó M.N, editor. Los pueblos originarios y la conquista. Editorial Sudamericana; Buenos Aires, Argentina: 2000. pp. 187–227. [Google Scholar]
- Allaby R.G, O'Donoghue K, Sallares R, Jones M.K, Brown T.A. Evidence for the survival of ancient DNA in charred wheat seeds from European archaeological sites. Ancient Biomol. 1997;1:119–129. [Google Scholar]
- Benz B.F. Archaeological evidence of teosinte domestication from Guilá Naquitz, Oaxaca. Proc. Natl Acad. Sci. USA. 2001;98:2104–2106. doi: 10.1073/pnas.98.4.2104. doi:10.1073/pnas.98.4.2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz B.F, Iltis H.H. Studies in archaeological maize I: the ‘wild’ maize from San Marcos cave reexamined. Am. Antiq. 1990;55:500–511. doi:10.2307/281280 [Google Scholar]
- Brown T.A, Allaby R.G, Sallares R, Jones G. Ancient DNA in charred wheats: taxonomic identification of mixed and single grains. Ancient Biomol. 1998;2:185–193. [Google Scholar]
- Castro R.V, Tarragó M.N. Los inicios dela producción de alimentos en el Cono Sur de América. Revista de Arqueología Americana. 1992;6:91–124. [Google Scholar]
- Cornuet J.-M, Piry S, Luikart G, Estoup A, Solignac M. New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics. 1999;153:1989–2000. doi: 10.1093/genetics/153.4.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta I.K, Wood J, Hicks J.B. A plant DNA minipreparation: version II. Plant Mol. Biol. Rep. 1983;1:19–21. [Google Scholar]
- Doebley J.F. Molecular systematics of Zea (Gramineae) Maydica. 1990;35:143–150. [Google Scholar]
- Freitas F.O, Bendel G, Allaby R.G, Brown T.A. DNA from primitive maize landraces and archaeological remains: implications for the domestication of maize and its expansion into South America. J. Archaeol. Sci. 2003;30:901–908. doi:10.1016/S0305-4403(02)00266-2 [Google Scholar]
- Goloubinoff P, Pääbo S, Wilson A. Evolution of maize inferred from sequence diversity of an Adh2 gene segment from archaeological specimens. Proc. Natl Acad. Sci. USA. 1993;90:1997–2001. doi: 10.1073/pnas.90.5.1997. doi:10.1073/pnas.90.5.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M.M, Bird R.M. The races of maize. IV: tentative grouping of 219 Latin American races. Econ. Bot. 1977;31:204–221. [Google Scholar]
- Goodman M.M, Brown W.L. Races of corn. In: Sprague G.F, Dudley J.W, editors. Corn and corn improvement. American Society for Agronomy; Madison, WI: 1988. pp. 33–79. [Google Scholar]
- Hadly E, van Tunen M, Chan Y, Heiman K. Ancient DNA evidence of prolonged population persistence with negligible genetic diversity in an endemic tuco–tuco (Ctenomys sociabilis) J. Mammal. 2003;84:403–417. doi:10.1644/1545-1542(2003)084<0403:ADEOPP>2.0.CO;2 [Google Scholar]
- Hansen A.J, Willerslev E, Wiuf C, Mourier T, Arctander P. Statistical evidence for miscoding lesions in ancient DNA templates. Mol. Biol. Evol. 2001;18:262–265. doi: 10.1093/oxfordjournals.molbev.a003800. [DOI] [PubMed] [Google Scholar]
- Higgins D.G, Thompson J.D, Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreiter M, Jaenicke V, Serre D, von Haeseler A, Pääbo S. DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res. 2001;29:4793–4799. doi: 10.1093/nar/29.23.4793. doi:10.1093/nar/29.23.4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höss M, Jaruga P, Zastawny T, Dizdaroglu M, Pääbo S. DNA damage and DNA sequence retrieval from ancient tissues. Nucleic Acids Res. 1996;24:1304–1307. doi: 10.1093/nar/24.7.1304. doi:10.1093/nar/24.7.1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Das O.P, Messing J. Zeon-1, a member of a new maize retrotransposon family. Mol. Gen. Genet. 1995;248:471–480. doi: 10.1007/BF02191647. doi:10.1007/BF02191647 [DOI] [PubMed] [Google Scholar]
- Jaenicke-Després V, Buckler E.S, Smith B, Gilbert M.T.P, Cooper A, Doebley J, Pääbo S. Early allelic selection in maize as revealed by ancient DNA. Science. 2003;302:1206–1208. doi: 10.1126/science.1089056. doi:10.1126/science.1089056 [DOI] [PubMed] [Google Scholar]
- Korstanje M.A, Wurschmidt A. Producir y recolectar en los valles altos del NOA: los Viscos como caso de estudio. In: Aschero C.A, Korstanje M, Vuoto P, editors. En los tres reinos: prácticas en el Cono Sur de América. Ediciones Magna Publicaciones; Buenos Aires, Argentina: 1999. pp. 151–160. [Google Scholar]
- Lia, V. V. 2004 Diversidad genética y estructura poblacional en razas nativas de maíz (Zea mays ssp. mays) del Noroeste Argentino: presente y pasado del germoplasma autóctono. Ph.D. thesis, Facultad de Ciencias Exactas y Naturales, Universidad de Buenos Aires, Argentina.
- Manen J.F, Bouby L, Dalnoki O, Marinval P, Turgay M, Schlumbaum A. Microsatellites from archaeological Vitis vinifera seeds allow a tentative assignment of the geographical origin of ancient cultivars. J. Archaeol. Sci. 2003;30:721–729. doi:10.1016/S0305-4403(02)00244-3 [Google Scholar]
- Matsuoka Y, Mitchell S.E, Kresovich S, Goodman M.M, Doebley J.F. Microsatellites in Zea—variability, patterns of mutations, and use for evolutionary studies. Theor. Appl. Genet. 2002a;104:436–450. doi: 10.1007/s001220100694. doi:10.1007/s001220100694 [DOI] [PubMed] [Google Scholar]
- Matsuoka Y, Vigouroux Y, Goodman M.M, Sanchez J, Buckler G.E, Doebley J. A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl Acad. Sci. USA. 2002b;99:6080–6084. doi: 10.1073/pnas.052125199. doi:10.1073/pnas.052125199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B, Kato T.A, Blumenschein A. Colegio de Postgraduados; Chapingo, Mexico: 1981. Chromosome constitution of the races of maize. Its significance in the interpretation of relationships between races and varieties in the Americas. [Google Scholar]
- Meyers B.C, Tingey S.V, Morgante M. Abundance, distribution and transcriptional activity of repetitive elements in the maize genome. Genome Res. 2001;11:1660–1676. doi: 10.1101/gr.188201. doi:10.1101/gr.188201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donoghue K, Clapham A, Evershed R, Brown T.A. Remarkable preservation of biomolecules in ancient radish seeds. Proc. R. Soc. B. 1996;263:541–547. doi: 10.1098/rspb.1996.0082. [DOI] [PubMed] [Google Scholar]
- Orlando L, Leonard J.A, Thenot A, Laudet V, Guerin C, Hänni C. Ancient DNA analysis reveals woolly rhino evolutionary relationships. Mol. Phylogenet. Evol. 2003;28:485–499. doi: 10.1016/s1055-7903(03)00023-x. doi:10.1016/S1055-7903(03)00023-X [DOI] [PubMed] [Google Scholar]
- Pääbo S. Ancient DNA: extraction, characterization, molecular cloning, and enzymatic amplificaton. Proc. Natl Acad. Sci. USA. 1989;86:1939–1943. doi: 10.1073/pnas.86.6.1939. doi:10.1073/pnas.86.6.1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetkau D, Calvert W, Stirling I, Strobeck C. Microsatellite analysis of population structure in Canadian polar bears. Mol. Ecol. 1995;4:347–354. doi: 10.1111/j.1365-294x.1995.tb00227.x. [DOI] [PubMed] [Google Scholar]
- Piperno D.R, Flannery K.V. The earliest archaeological maize (Zea mays L.) from highland Mexico: new accelerator mass spectrometry dates and their implications. Proc. Natl Acad. Sci. USA. 2001;98:2101–2103. doi: 10.1073/pnas.98.4.2101. doi:10.1073/pnas.98.4.2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno D.R, Pearsall D.M. Academic Press; San Diego, CA: 1998. The origins of agriculture in the lowland Neotropics. [Google Scholar]
- Poggio L, Rosato M, Chiavarino A.M, Naranjo C.A. Genome size and environmental correlations in maize (Zea mays ssp. mays, Poaceae) Ann. Bot. 1998;82:107–115. doi:10.1006/anbo.1998.0757 [Google Scholar]
- Poinar H.N. International congress series. Vol. 1239. 2003. The top 10 list: criteria of authenticity for DNA from ancient and forensic samples; pp. 575–579. [Google Scholar]
- Previgliano C.H, Ceruti C, Reinhard J, Araoz F.A, Diez J.G. Radiologic evaluation of the Llullaillaco mummies. Am. J. Roentgenol. 2003;181:1473–1479. doi: 10.2214/ajr.181.6.1811473. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J. Hered. 1995;86:248–249. [Google Scholar]
- Rollo F, Venanzi F.M, Amici A. Nucleic acids in mummified plant seeds: biochemistry and molecular genetics of pre-Columbian maize. Genet. Res. 1991;58:193–201. doi: 10.1017/s0016672300029943. [DOI] [PubMed] [Google Scholar]
- Rollo F, Venanzi F.M, Amici A. DNA and RNA from ancient plant seeds. In: Herrmann B, Hummel S, editors. Recovery and analysis of genetic material from paleontological, archaeological, museum, medical, and forensic specimens. Springer; New York, NY: 1994. pp. 218–236. [Google Scholar]
- SanMiguel P, et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. doi:10.1126/science.274.5288.765 [DOI] [PubMed] [Google Scholar]
- Sempé C. Caracterización de la cultura Saujil. Obra del Centenario del Museo de La Plata. Antropología. 1977;II:211–235. [Google Scholar]
- Staller J.E, Thompson R.G. A multidisciplinary approach to understanding the initial introduction of maize into coastal Ecuador. J. Archaeol. Sci. 2002;29:33–50. doi:10.1006/jasc.2001.0750 [Google Scholar]
- Tarragó M.N. Actas del V Congreso Nacional de Arqueología Argentina. Vol. 1. 1980. El proceso de agriculturización en el Noroeste Argentino, Zona Valliserrana; pp. 181–218. [Google Scholar]
- Vásquez, V., Arceo, J. M., Rosales, T., Koschmieder, K., Caballero, J. L. & Dorado, G. In press. Archaeological and contemporary native breeds of corn (Zea mays) from North Peru: phylogeny by microsatellite (STR) fingerprinting. In Molecular markers, PCR, bioinformatics and ancient DNA–technology and applications (ed. G. Dorado). New York, NY: Science Publishers.
- Vigouroux Y, Jaqueth J.S, Matsuoka Y, Smith O.S, Beavis W.D, Smith J.S.C, Doebley J. Rate and pattern of mutation at microsatellite loci in maize. Mol. Biol. Evol. 2002;19:1251–1260. doi: 10.1093/oxfordjournals.molbev.a004186. [DOI] [PubMed] [Google Scholar]
- Yang D, Eng B, Waye J, Dudar J.C, Saunders S.R. Improved DNA extraction from ancient bones using silica-based spin columns. Am. J. Phys. Anthropol. 1998;105:539–543. doi: 10.1002/(SICI)1096-8644(199804)105:4<539::AID-AJPA10>3.0.CO;2-1. doi:10.1002/(SICI)1096-8644(199804)105:4<539::AID-AJPA10>3.0.CO;2-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BLAST analysis of non-microsatellite nucleotide sequences retrieved from archaeological maize specimens
Allele sequences of locus phi127 from archaeological and extant maize specimens. Clones (c) and direct sequencing (ds) products corresponding to archaeological specimens are highlighted in gray. Z.m.p.: Zea mays ssp parviglumis; Z.m.m.: Zea mays ssp mexicana.
Allele sequences of locus phi029 from archaeological and extant maize specimens. Clones (c) and direct sequencing (ds) products corresponding to archaeological specimens are highlighted in gray. Z.m.p.: Zea mays ssp parviglumis; Z.m.m.: Zea mays ssp mexicana.