Intramembrane-cleaving proteases (I-CLiPs) are a class of hydrolytic enzymes that cleave hydrophobic substrates within the lipid bilayer (1). Unlike their well understood soluble or membrane-tethered counterparts, I-CLiPs are integral membrane proteins, with catalytic residues thought to lie within the confines of the membrane. Although the identities of these residues have suggested membrane-embedded active sites and mechanistic convergence with other proteases, I-CLiPs bear little or no sequence similarity to their soluble cousins, and their assignment into mechanistic categories has been tentative. Three new articles (2–4), including one by Ben-Shem et al. in this issue of PNAS (4), describe the first crystal structures of an I-CLiP, the Escherichia coli serine protease GlpG. Together, these reports provide complementary pictures that offer exciting insights into how an I-CLiP handles transmembrane substrates and carries out hydrolysis in a hydrophobic environment.
The known families of I-CLiPs include the site-2 protease (S2P) zinc metalloproteases, presenilin and presenilin-like aspartyl proteases, and the rhomboid family of serine proteases. These enzymes are found throughout the major kingdoms of life, are highly conserved, and play key roles in biology and disease. S2P-type proteases release membrane-tethered transcription factors controlling sterol and fatty acid biosynthesis in animals and sporulation in bacteria (5, 6). Presenilin is the catalytic engine of the four-component γ-secretase complex critical for cell-fate determinations and the pathogenesis of Alzheimer's disease (7). Signal peptide peptidase, the prototype presenilin-like protease, releases remnant signal peptides from the membrane but, unlike presenilin, does not require other protein cofactors (8).
E. coli GlpG is a homolog of Drosophila rhomboid-1, which was the first identified intramembrane serine protease (9). Rhomboid-1 releases the extracellular domain of the membrane-tethered epidermal growth factor-like protein Spitz as part of a key developmental signaling pathway in flies. The specific physiological roles of most rhomboids, including the seven homologs identified in the human genome and the many of bacterial and parasitic origin, are largely unknown, but research to date points to critical roles in mitochondrial membrane remodeling (10), apoptosis (11), quorum sensing (12), and cell invasion (13). Interestingly, even though GlpG has been shown to cleave Spitz (14), its normal function in E. coli remains elusive.
Careful biochemical and cell biological studies have contributed to our understanding of hydrolysis by intramembrane serine proteases since their discovery in 2001. Such studies have suggested a six- or seven-transmembrane architecture, a Ser–His catalytic dyad (14), and a critical Trp–Arg (WR) motif (9). Substrate cleavage is predicted to occur several residues into the transmembrane helix on the periplasmic/extracellular side and requires helix-destabilizing residues at or near the cleavage site (15). The crystal structure confirms and helps clarify several of these predicted features in molecular detail and also enables new structure-based hypotheses for catalysis by this family of enzymes.
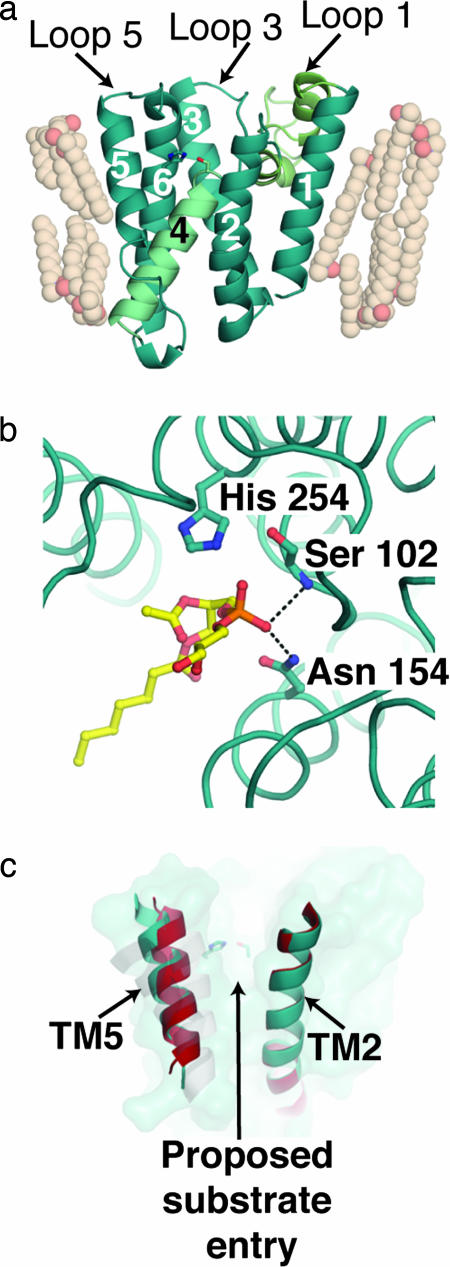
As predicted, the crystal structure of GlpG reveals a core molecular architecture composed of six transmembrane helices (TM) plus two helices, H1 and H2, that are part of the unusual loop 1 connecting TM1 and TM2 (Fig. 1a). Although membrane proteins are typically thought to form a bundle of uniform-length helices that cross the membrane and are parallel to each other, the TM helices in GlpG are of varying lengths and angles with respect to the membrane normal. The particular spatial arrangement creates a hydrophilic cavity within the protein and emphasizes the importance of the overall protein scaffold in forming the unique chemical environment amenable to hydrolysis. A major contribution of the GlpG structures is in establishing that residues implicated in catalysis by site-directed mutagenesis are indeed located within the boundaries of the membrane, ≈10 Å below the surface within a water-filled cavity on the periplasmic side of the membrane. These residues, Ser-201 and His-254, which form the proposed catalytic dyad, are hydrogen-bonded with a distance of ≈3.1 Å and are located at the start of TM4 and in the middle of TM6, respectively.
Fig. 1.
Views of GlpG. (a) Molecular architecture of E. coli GlpG [Protein Data Bank (PDB) ID code 2IRV]. TMs are numbered sequentially. TM4, which contains the catalytic residue Ser-201, is colored light cyan; and loop 1, which comprises two unusual helices H1 and H2, is colored light green. (b) Phospholipid binding in the proposed active site of GlpG. Dashed lines indicate hydrogen-bonding interactions to the phosphate group that likely parallel the oxyanion hole stabilization during catalysis. (c) GlpG surface (teal) and superimposed TM2 and TM5 helices from two molecules seen in structure solved by Ben-Shem et al. (red and teal). A grayscale overlay of TM5 for the other structures of GlpG (PDB ID codes 2NRF and 2IC8) is shown.
The active-site cavity is shaped by the angled and central TM4 (Fig. 1a), in which the helical structure begins only at Ser-201, well within the membrane. This substructure is caused by the invariant and helix-destabilizing glycine residues that flank Ser-201. In addition, surrounding TM helices contribute a number of polar residues and enable water molecules to be sequestered in this cavity. TM4 appears to act as a stopper to prevent the water molecules from penetrating beyond Ser-201. Below Ser-201, residues on TM4 interact with other hydrophobic residues from surrounding TM helices. Lipid entry to the active site is further prevented by interactions of surrounding loop residues. Although not conserved among all species, side chains from residues on loop 3 form hydrophobic interactions with similar residues in the connecting region between H1 and H2 of loop 1 and act as a further barrier to lipid entry into the hydrophilic cavity from that side. On the other side is loop 5, which may mediate substrate entry (see below) and is ordered in only one of the two independent views of GlpG in the Ben-Shem et al. (4) structure.
The ensemble of GlpG structures adds important catalytic insights. In support of site-directed mutagenesis studies (14), Asn-154, initially proposed to be the third residue of a catalytic triad (9), similar to the aspartate associated with many soluble serine proteases, is distant from the side chains of the catalytic residues. Instead, Asn-154 may provide the oxyanion hole that stabilizes the tetrahedral intermediate expected after nucleophilic attack of the peptide carbonyl by Ser-201. Seen only in the structure by Ben-Shem et al. (4), the phosphate head group of a lipid molecule found near Asn-154, His-254, and Ser-201 mimics the tetrahedral intermediate expected in the reaction and provides further support that this is the catalytic center of rhomboid-like proteases (Fig. 1b).
In addition, the presence of the ordered lipid molecule near the hydrophilic catalytic center and extending into the hydrophobic region of TM2 and TM5 suggests that this surface may be used for substrate presentation. TM5 is one of the least sequence-conserved regions among rhomboids and may play a role in tuning substrate specificity. Even among the available structures of GlpG, the greatest structural variation is in TM5 and the adjacent loop 5 (Fig. 1c). It is not known whether these differences are influenced by crystallization, but in the structure by Ben-Shem et al. (4), loop 5 is only ordered when the lipid is also present. How changes observed in TM5 and loop 5 might be triggered in vivo is unclear, but the variability of loop 5 and TM5 within the asymmetric unit demonstrates that such movement is possible.
The transmembrane helices in GlpG are of varying lengths and angles.
An alternative hypothesis is that the substrate is presented to the hydrophilic cavity after a substantial rearrangement of H1, H2, and the rest of loop 1 to reveal a large opening between TM1 and TM3 (2). The unusual short helices that protrude partway into the membrane contain the WR motif, are amphipathic, and thus may be metastable. However, this loop appears unaltered in all GlpG structures to date. Moreover, mutation of the WR motif, expected in this model to destabilize the unusual structure and therefore increase substrate access and activity, instead reduces or abolishes protease activity (9, 14). In addition, the catalytic residues do not appear poised for catalysis from this direction. Another possible purpose of this unusual region is in protein–protein interactions, which might transmit cell signals to control catalysis (4).
The first snapshots of GlpG have provided new clues about intramembrane proteolysis, and future efforts are likely to address details of the biochemical mechanism of this family of enzymes. To distinguish among various hypotheses for substrate entry, crystallographic trapping of a complex between GlpG and a substrate- or product-mimicking inhibitor, or a complex of a catalytically inactive GlpG and a substrate, would be invaluable. A cocrystal structure would provide evidence for any initial docking site for the substrate before presentation at the active site as well as residues involved in unwinding the helical substrate to help the nucleophile gain access to the cleavage site. Homology modeling and structures of other rhomboids are likely to expand our understanding of the functional landscape of this family of enzymes. Detailed knowledge of intramembrane proteolysis by rhomboids could eventually lead to the design of molecules to modulate pathological activities.
The sophisticated way in which rhomboid sequesters its active site, which requires hydrophilic residues and water, is likely to have parallels for other intramembrane proteases. For example, studies of presenilin and the γ-secretase complex point to a hydrophilic cavity in this intramembrane aspartyl protease (16–18). On the other hand, results using chemical probes of γ-secretase suggest that substrate access involves the interface between two distinct parts of presenilin (19), a mechanism that may be distinct from that proposed for rhomboid. Molecular structures of other I-CLiPs, such as presenilin/γ-secretase, signal peptide peptidase, and S2P, may reveal common features and general principles of protein hydrolysis in the membrane. Such future snapshots will eventually join those of rhomboid to form a family album of these fascinating enzymes.
Footnotes
The authors declare no conflict of interest.
See companion article on page 462.
References
- 1.Wolfe MS, Kopan R. Science. 2004;305:1119–1123. doi: 10.1126/science.1096187. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Zhang Y, Ha Y. Nature. 2006;444:179–180. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- 3.Wu Z, Yan N, Feng L, Oberstein A, Yan H, Baker RP, Gu L, Jeffrey PD, Urban S, Shi Y. Nat Struct Mol Biol. 2006;13:1084–1091. doi: 10.1038/nsmb1179. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Shem A, Fass D, Bibi E. Proc Natl Acad Sci USA. 2007;104:462–466. doi: 10.1073/pnas.0609773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rawson RB. Nat Rev Mol Cell Biol. 2003;4:631–640. doi: 10.1038/nrm1174. [DOI] [PubMed] [Google Scholar]
- 6.Rudner DZ, Fawcett P, Losick R. Proc Natl Acad Sci USA. 1999;96:14765–14770. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolfe MS. Biochemistry. 2006;45:7931–7939. doi: 10.1021/bi060799c. [DOI] [PubMed] [Google Scholar]
- 8.Martoglio B, Golde TE. Hum Mol Genet. 2003;12:R201–R206. doi: 10.1093/hmg/ddg303. [DOI] [PubMed] [Google Scholar]
- 9.Urban S, Lee JR, Freeman M. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 10.McQuibban GA, Saurya S, Freeman M. Nature. 2003;423:537–541. doi: 10.1038/nature01633. [DOI] [PubMed] [Google Scholar]
- 11.Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, Bartoli D, Polishuck RS, Danial NN, De Strooper B, et al. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 12.Gallio M, Sturgill G, Rather P, Kylsten P. Proc Natl Acad Sci USA. 2002;99:12208–12213. doi: 10.1073/pnas.192138799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brossier F, Jewett TJ, Sibley LD, Urban S. Proc Natl Acad Sci USA. 2005;102:4146–4151. doi: 10.1073/pnas.0407918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemberg MK, Menendez J, Misik A, Garcia M, Koth CM, Freeman M. EMBO J. 2005;24:464–472. doi: 10.1038/sj.emboj.7600537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urban S, Freeman M. Mol Cell. 2003;11:1425–1434. doi: 10.1016/s1097-2765(03)00181-3. [DOI] [PubMed] [Google Scholar]
- 16.Tolia A, Chavez-Gutierrez L, De Strooper B. J Biol Chem. 2006;281:27633–27642. doi: 10.1074/jbc.M604997200. [DOI] [PubMed] [Google Scholar]
- 17.Sato C, Morohashi Y, Tomita T, Iwatsubo T. J Neurosci. 2006;26:12081–12088. doi: 10.1523/JNEUROSCI.3614-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazarov VK, Fraering PC, Ye W, Wolfe MS, Selkoe DJ, Li H. Proc Natl Acad Sci USA. 2006;103:6889–6894. doi: 10.1073/pnas.0602321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kornilova AY, Bihel F, Das C, Wolfe MS. Proc Natl Acad Sci USA. 2005;102:3230–3235. doi: 10.1073/pnas.0407640102. [DOI] [PMC free article] [PubMed] [Google Scholar]