Abstract
Bordetella pertussis, the causative agent of whooping cough, has many well-studied virulence factors and a characteristic clinical presentation. Despite this information, it is not clear how B. pertussis interaction with host cells leads to disease. In this study, we examined the interaction of B. pertussis with a human bronchial epithelial cell line (BEAS-2B) and measured host transcriptional profiles by using high-density DNA microarrays. The early transcriptional response to this pathogen is dominated by altered expression of cytokines, DNA-binding proteins, and NFκB-regulated genes. This previously unrecognized response to B. pertussis was modified in similar but nonidentical fashions by the antiinflammatory agents dexamethasone and sodium salicylate. Cytokine protein expression was confirmed, as was neutrophil chemoattraction. We show that B. pertussis induces mucin gene transcription by BEAS-2B cells then counters this defense by using mucin as a binding substrate. A set of genes is described for which the catalytic activity of pertussis toxin is both necessary and sufficient to regulate transcription. Host genomic transcriptional profiling, in combination with functional assays to evaluate subsequent biological events, provides insight into the complex interaction of host and pathogen.
Bordetella pertussis is a Gram-negative coccobacillus and the causative agent of whooping cough in humans. It is a well-studied pathogen with a number of potent virulence factors. However, little is known about the responses elicited by this organism in human cells, especially at the level of gene transcription.
Autopsy studies of pertussis victims performed in the early 20th century revealed a diffuse bronchopneumonia with increased secretion of mucus, associated with airway plugging and atelectasis (1). Bacteria were seen tightly packed between the cilia of epithelial cells, which desquamated during infection. Rodent models of Bordetella pertussis infection have revealed infiltrates of monocytes, neutrophils, and lymphocytes in the lung (2).
B. pertussis expresses several virulence factors directed at the host epithelium. Filamentous hemagglutinin is the major adhesin of B. pertussis for bronchial epithelial cells (3). B. pertussis also produces several toxins, including pertussis toxin (PT). PT is associated with a panoply of biological effects, many of which are linked to its ADP–ribosyltransferase activity. ADP ribosylation of the Gα family of host proteins prevents their usual regulatory response to G-protein-linked receptor engagement (4). B. pertussis also secretes an adenylate cyclase toxin that enters host cells and raises intracellular cAMP concentrations (5). Tracheal cytotoxin (TCT), a muramyl tetrapeptide bacterial cell wall fragment, in combination with lipopolysaccharide, paralyzes respiratory epithelial cilia and ultimately causes cell death and extrusion through pathways involving IL-1 and nitric oxide synthase (iNOS) (6). The molecular mechanisms through which B. pertussis exerts these effects and by which host cells attempt to counter this pathogenic strategy remain largely undescribed.
The goal of this study was to understand the pathology caused by B. pertussis and to explore whether early transcriptional responses reveal pathogenic mechanisms and pathology at a molecular level. Our approach involved genome-wide expression analysis of a human bronchial epithelial cell line exposed to B. pertussis, coupled with functional analyses.
Materials and Methods
Bacterial Strains, Toxins, and Cell Lines.
Wild-type B. pertussis BP536 (3) and an isogenic derivative expressing a catalytically inactive PT, BP-9K/129G (7), were grown at 37°C on Bordet–Gengou agar (Difco), supplemented with sheep blood (13% v/v) and 100 μg/ml streptomycin. Bordetellae were suspended in Eagle's minimal essential medium (EMEM) without FCS overnight at 37°C in 5% CO2. They were then added to 2 × 107 epithelial cells at a ratio of 50 bacteria per cell. Purified wild-type and catalytically inactive PT (7) were added to 2 × 107 epithelial cells at a concentration of 1 μg/ml. The SV40-transformed human bronchial epithelial cell-line BEAS-2B (8) was grown in EMEM with 10% FCS at 37°C in 5% CO2. A human colon carcinoma cell line, HM3, was stably transfected with a construct of the MUC2 or MUC5AC promoter fused to a luciferase reporter gene. Transfected cells were maintained in DMEM with high glucose and 10% FCS.
Infection Scheme.
BEAS-2B cells (1–1.5 × 107) were incubated for 8 h, then some were treated for 1 hr with 10 mM sodium salicylate or 1 μM dexamethasone. The cells were then infected with B. pertussis, exposed to PT, or mock infected, and incubated at 37°C for 3 h. Cells were washed with Hanks' balanced salt solution (HBSS), released from the flask with 5 mM EDTA in HBSS, pelleted at 200 × g for 5 min and frozen at −80°C.
Microarray Expression Analysis.
Total RNA was extracted from cell pellets by using Trizol (Life Technologies, Rockville, MD) and purified by using an affinity resin (RNeasy, Qiagen, Chatsworth, CA). Microarray analysis was performed according to the Affymetrix Expression Analysis Technical Manual with the following exceptions (see http://relman.stanford.edu): first-strand cDNA synthesis was performed at 42°C, and 10 μg of total RNA was used. Triton X-100 (0.01%) was used in hybridization solutions and buffers instead of Tween 20. Labeled cRNA (20 μg) was hybridized to Affymetrix HU6800 genechip microarrays (Affymetrix, Santa Clara, CA) for 18 h in buffer without EDTA. The arrays were rinsed with 6× standard saline phosphate/EDTA (SSPET, 0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA), washed in 0.1× Mes at 50°C for 30 min on a rotisserie at 60 rpm, and then rinsed in 1× Mes. Hybridized RNA was fluorescently labeled by incubation in streptavidin- R-phycoerythrin conjugate (Molecular Probes) staining buffer at 40°C for 15 min, then washed with 6× SSPET at 22°C. Quantitative analysis of hybridization patterns and intensities were performed as previously described (9) (see http://relman.stanford.edu).
Cytokine and Apoptosis Quantitation.
For cytokine analysis, 96-well plates were coated with 0.2 μg of capture mAb (MAB 279, 206, 208, R&D Systems) and then blocked with 1% BSA in PBS. One hour later, 10 ng of a biotinylated polyclonal Ab (BAF 279, 206, 208, R&D Systems) was added for 1 h. After washing, bound Ab was detected by using a streptavidin–horseradish peroxidase conjugate and tetramethylbenzadine substrate. Cytoplasmic histone-associated DNA fragments were quantitated as a measure of apoptosis by using the Cell Death Detection ELISA Plus kit per manufacturer recommendations (Roche Diagnostics).
Neutrophil Chemotaxis.
Whole human blood was mixed 1:1 with dextran in PBS for 45 min at 25°C. The top layer was overlaid on Ficoll–Paque (Amersham Pharmacia) and centrifuged at 450 × g for 45 min, then granulocytes were harvested. RBCs were lysed in hypotonic saline. Neutrophil purity was >95%. One hundred thousand neutrophils were placed on a 3-μm filter (Neuroprobe, Cabin John, MD) separating them from 29 μl of BEAS-2B supernatant in a 96-well microchamber. In some wells, anti-human IL-8 Ab was added at 10 μg/ml for the duration of the experiment (MAB208, R&D Systems). After 1 h at 37°C, only cells that had passed through the filter were quantified by using the CyQuant Cell Proliferation Kit (Molecular Probes).
Mucin Transcription and Binding.
HM3 cells (1.5–3 × 106) carrying either a MUC2 or MUC5AC promoter fusion to a luciferase reporter gene were infected with B. pertussis in DMEM without FCS in triplicate. After 2, 4, and 6 h, the cells were lysed for 30 min with 100 μl Reporter Lysis Buffer (Luciferase Assay System, Promega) and then frozen at −80°C. The luciferase assay was carried out per manufacturer recommendations.
For binding, 2.5 μg of bovine submaxillary mucin type I-S (Sigma) was desiccated onto a microtiter plate at 37°C. Mucin-treated and -untreated plates were then washed with PBS and blocked with 5% dried nonfat milk in PBS for 1 h at 37°C and again washed with PBS. A suspension of B. pertussis labeled with a 250-μg/ml solution of FITC for 20 min was then added to the plates for 1 h. Approximately 2.5 × 108 of labeled bacteria were added per well. Nonadherent bacteria were washed away with three cycles of PBS washings. Plates were then read in a 96-well fluorimeter.
Results
The Transcriptional Profile of BEAS-2B Cells Infected with B. pertussis Reveals the Basis of Inflammation in Pertussis.
Although the pathology of pertussis has been studied for decades, little is known about the response of individual cell types to infection. We examined the genome-wide transcriptional responses of a bronchial epithelial cell line, BEAS-2B (8), after infection with a wild-type B. pertussis strain (BP536) for 3 h. Preliminary analyses had shown more robust responses after 3 h than after 1 h (data not shown). Steady-state transcript abundance for approximately 6,800 human genes was analyzed in duplicate experiments. Reproducibility of the data was examined by using duplicate preparations of labeled cDNA/cRNA from RNA isolated from each of two replicate infection experiments. These analyses revealed 19.5% and 26.9% variation in mean average difference values for all probe sets in the four comparisons. The complete data sets for these (Table 2) and other experiments (Tables 3–8) are available at http://relman.stanford.edu.
Table 1 categorizes the named BEAS-2B genes that exhibited greater than a 3-fold change in steady-state transcript abundance after 3 h of infection. These results are noteworthy for the modest number of genes that responded to B. pertussis exposure, unlike the responses observed at later time points during human cytomegalovirus (10) and HIV-1 infections (J. Corbeil, D. Genini, D. Sheeter, S. Rought, L. Leoni, P. Du, M. Ferguson, D. R. Masys, J. B. Welsh, J. L. Fink, et al., personal communication). Of the 33 genes whose expression levels increased in abundance by greater than 3-fold on B. pertussis infection, the most well-represented category (8) was a group encoding proinflammatory cytokines. None of these had been previously implicated in B. pertussis infection, but their known chemoattractant activities are consistent with the cellular infiltrate seen in pertussis. Our data also revealed increased IL-1β transcription. IL-1 is known to increase growth-regulated oncogene (GRO) mRNA and has been shown to play a role in TCT-mediated respiratory epithelial cell toxicity (11, 12). Other up-regulated gene products, such as IκB, are involved in NFκB signaling and may regulate these cytokines (13).
Table 1.
Epithelial cell transcriptional profile after exposure to B. pertussis
| Down-regulated
transcripts
|
Up-regulated transcripts
|
||||
|---|---|---|---|---|---|
| Ratio | Accession no. | GDB/other names | Ratio | Accession no. | GDB/other names |
| Chemokines/Cytokines | |||||
| +22.38 | X54489 | GRO1/SCYB1/MGSA | |||
| +16.29 | M28130 | IL8 | |||
| +9.11 | Y00787 | IL8 | |||
| +6.80 | X04500 | IL1B | |||
| +6.47 | N/A | MCP1/SCYA2 | |||
| M57731 | GRO2/SCYB2/MIP-2α | ||||
| X04602 | IL6/IFNB2 | ||||
| M23178 | SCYA3/MIP-1a | ||||
| X53800 | GRO3/SCYB3/MIP-2α | ||||
| Transcription factors/Zn-finger proteins | |||||
| −3.03 | X51435 | ZNF40/HIVEP1 | +7.39 | M59465 | TNFAIP3/A20 |
| D88613 | HGCM/GCMA | X65663 | SOX6 | ||
| U10324 | ILF3/NF90 | M82882 | ELF1/E74-like factor | ||
| N/A | MYBL1/A-MYB | U69127 | FUBP3/(FUSE)-binding protein 3 | ||
| X59244 | ZNF43/zinc-finger protein | U18259 | MHC2TA/CIITA-8 | ||
| X55544 | ATF1/TREB36 | ||||
| M29581 | ZNF8/zinc-finger protein 8 | ||||
| M96843 | ID2/inhibitor DNA binding | ||||
| U78722 | ZNF165/zinc-finger protein | ||||
| U28687 | ZNF157/zinc-finger protein | ||||
| N/A | BTF3 homologue | ||||
| Secreted molecules | |||||
| U19557 | SCCA2/protease Inhibitor | +4.59 | M57730 | EFNA1/LERK1/B61/Ephrin A1 | |
| U90546 | BTN3A2/butyrophilin | M31166 | PTX3/pentaxin-related/TSG-14 | ||
| X64877 | HFL3/H factor complement | X57579 | INHBA/inhibin, β A/activin | ||
| S75256 | LCN2/lipocalin 2 | X75308 | MMP13/collagenase 3 | ||
| M21642 | AT3/antithrombin III | M31551 | PAI2/plasminogen active inhibitor | ||
| ER/Golgi associated | |||||
| X97064 | SEC23A | L10333 | RTN1/reticulon 1 | ||
| X71661 | LMAN-1/ERGIC-53/lectin | U27326 | FUT3/fucosyltransferase 3 | ||
| L41390 | acetylglucosaminyltransfer | ||||
| U14550 | STHM/sialyltransferase | ||||
| Metabolic/synthetic | |||||
| −3.44 | AC002115 | COX6B/cytochrome c oxid. | D38524 | Cytosolic purine 5′-nucleotidase | |
| M61853 | CYP2C17/P450IIC17 | X78711 | GK/glycerol kinase deficiency | ||
| M60891 | UROD/decarboxylase | M61855 | CYP2C9/cytochrome P450, IIc | ||
| D49488 | TTPA/tocopherol α transfer | Z94753 | ATP7A/Cu++ ATPase | ||
| U75362 | USP13/ubiquitin protease | ||||
| S72370 | PC/pyruvate carboxylase | ||||
| X59834 | GLUL/glutamine synthase | ||||
| M84424 | CTSE/cathepsin E | ||||
| M64082 | FMO1/monooxygenase 1 | ||||
| D28791 | PIGA/phosphate inositol glycan | ||||
| D10040 | FACL1/fatty-acid-coA ligand | ||||
| Cell surface molecules/Receptors | |||||
| −4.76 | M32315 | TNFRSF1B/TNF receptor 2 | +4.53 | Y00285 | IGF2R/IGF II receptor |
| −3.85 | N/A | PSG11/glycoprotein | +4.30 | M24283 | ICAM1/CD54 |
| L36531 | ITGA8/integrin alpha 8 | +3.27 | U92971 | F2RL2/thrombin receptor-like | |
| M23197 | CD33 | S59184 | RYK/receptor tyrosine kinase | ||
| U64871 | GPR19/G protein-coupled | K02405 | HLA-DQB1/HLA-DC-3 β | ||
| U04811 | TRO/Trophinin | U11878 | CXCR2/IL8RB/IL-8 receptor | ||
| Y09392 | TNFRSF12/APO-3/TNFR | U65011 | PRAME/antigen of melanoma | ||
| X16983 | ITGA4/CD49D/integrin | ||||
| U61500 | TMEM1/EHOC-1 | ||||
| X77753 | M1S1/TROP-2/GA733-1 | ||||
| S67798 | SPAM1/sperm adhesion | ||||
| D31784 | CDH6/cadherin-6 | ||||
| S83249 | NG-TRA/hormone pump | ||||
| U80811 | EDG2/G-protein-coupled | ||||
| Cytoskeleton | |||||
| L07261 | ADD1/α adducin 1 | L20826 | PLS1/I-plastin | ||
| U34301 | Nonmuscle myosin heavy chain | ||||
| Intracellular signaling | |||||
| U48736 | PRP4h/ser-thr protein kinase | +3.47 | L42373 | PPP2R5A/protein phosphatase | |
| M38258 | RARG/retinoic acid receptor | +3.27 | M69043 | NFKBIA/IKBA/MAD-3 | |
| U18671 | STAT2 | U77129 | MAP4K5/KHS1/kinase | ||
| M35416 | RALB/v-ral homolog B | M80563 | S100A4/CAPL calcium binding | ||
| U40371 | PDE1C/phosphodiesterase | U37546 | API2/IAP homolog C/MIHC | ||
| U77845 | hTRIP/TRAF-interacting | ||||
| X78549 | PTK6/BRK | ||||
| U20158 | LCP2/SLP-76 | ||||
| U17327 | NOS1/nitric oxide synthase | ||||
| DNA replication/transcription | |||||
| U01833 | NUBP1/nucleotide binding | +3.31 | N/A | POLE/DNA polymerase, ɛ | |
| U47414 | CCNG2/cyclin G2 | X54993 | TBP/TFIID/TATA box binding | ||
| D26528 | DDX7/RNA helicase | X95152 | BRCA2/breast cancer 2 | ||
The experiment was repeated twice independently; data from one experiment are shown. Absolute difference ratios above or below the uninfected transcription are not reported when one of the two values was less than 2-fold the background noise level.
The data in Table 1 reveal regulation of several apoptosis-related genes. TNFAIP3 and API2 are antiapoptotic factors whose genes are transcriptionally up-regulated by infection (14, 15). Furthermore, transcription of BTF3, a transcription factor associated with apoptosis, is decreased (16). Given that monocytes and neutrophils have been described to undergo apoptosis in response to B. pertussis infection (17), we looked for evidence of apoptosis in B. pertussis-infected BEAS-2B cells by using an ELISA assay for mono- and oligonucleosomes at the 3-h time point used in the microarray experiments. Representative data from duplicate experiments revealed no evidence of apoptosis [absorbance of 0.028 ± 0.005 (mean ± 95% confidence interval) for the infected cells and 0.029 ± 0.005 for the uninfected cells (P = 0.85)].
The 65 genes whose expression levels decreased by greater than 3-fold included those encoding 10 transcription factors and 6 cellular adhesion molecules. The overall transcriptional profile of these infected cells suggested a picture of relatively rapid activation and defensive host responses.
The ADP–Ribosyltransferase Activity of PT Is Responsible for Specific Transcriptional Responses.
To define the role of PT ADP–ribosylase activity at the transcriptional level, we compared the responses of BEAS-2B cells to two pairs of stimuli: (i) wild-type PT vs. a catalytically inactive but properly folded PT holotoxin (9K/129G), and (ii) wild-type B. pertussis vs. an isogenic derivative that expresses PT-9K/129G. Data sets are available as Tables 3 and 4 at http://relman.stanford.edu; Table 5 lists genes whose expression changed more than 3-fold when comparing the effects of both the purified inactive toxin with wild-type toxin and the toxin-mutant strain with its isogenic parent strain. Thus, these host cell genes represent those for which pertussis toxin enzymatic activity was both necessary and sufficient to effect a change in transcript abundance. Several of these genes are G-protein or cAMP regulated, such as those encoding matrix metalloproteinase 1, β B1-crystallin, basic fibroblast growth factor, and the Na+/Cl−-dependent serotonin transporter. Surprisingly, other genes such as the serum/glucocorticoid-regulated kinase, whose expression level decreased, and guanylate cyclase 1, whose expression level increased, showed responses in directions contrary to those suggested by the literature. The majority of the PT-response genes revealed in these experiments had not been previously associated with G-protein activity. Thirty-eight genes with discordant 3-fold changes, suggesting an effect of PT activity that depends on other bacterial factors, are highlighted (Table 6) at http://relman.stanford.edu.
The Chemokine Response of B. pertussis-Infected Epithelial Cells Overcomes the Antichemotactic Activity of PT.
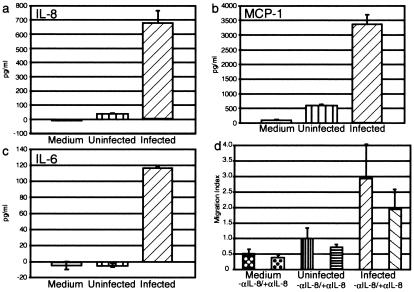
The known activities of PT and other B. pertussis virulence factors such as adenylate cyclase toxin suggest that they might be capable of blocking the chemotactic response predicted by our data (18). We first documented the predicted protein expression responses for several of the cytokine genes that were induced in BEAS-2B cells. Fig. 1a shows an increase in secreted IL-8 from 35 ± 0.8 (mean ± 95% confidence interval) to 678 ± 78 ng/ml (P = 0.0006) and MCP-1 from 580 ± 29 to 3367 ± 349 ng/ml (P = 0.004) (Fig. 1b) when the supernatants of BEAS-2B cells were harvested after 24 h of exposure to B. pertussis or mock infection. Induction of IL-6 secretion was also confirmed, with none detectable in uninfected supernatants, whereas 116 ± 2 ng/ml was detected in supernatants from infected cells (P < 2.5 × 10−7) (Fig. 1c).
Figure 1.
(a–c) Changes in secreted chemokine abundance after exposure of an epithelial cell line to B. pertussis for 24 h. Representative data from duplicate experiments are shown. Assays were performed for IL-8 (a), MCP-1 (b), and IL-6 (c). (d) Migration of neutrophils in response to the supernatants of B. pertussis-infected epithelial cells. In duplicate experiments, neutrophils migrated toward the supernatant with or without 10 μg/ml anti-IL-8 Ab. Migration index is the ratio of cells that migrated to a supernatant in comparison to the uninfected supernatant. Error bars represent the 95% confidence interval of the mean of three wells.
IL-8 and the GRO family of chemokines are strong neutrophil chemoattractants that have not been previously associated with B. pertussis infection (19, 20). We examined the chemoattractant activity of the BEAS-2B cell supernatants by using a neutrophil migration assay, in which cells are separated from BEAS-2B supernatants by a porous membrane. As shown in Fig. 1d, 2.9-fold more neutrophils were attracted to the supernatant of infected cells than to that of uninfected cells. Furthermore, cell migration could be partially inhibited (34% with the infected cell supernatant) by using an anti-IL-8 mAb, suggesting that IL-8 contributes to this chemoattractant activity and indicating a role for other chemokines as well. Although previous reports suggest that PT is capable of inhibiting neutrophil chemotaxis in vitro (21), chemoattraction was the dominant effect in this experimental system.
Antiinflammatory Drugs Suppress the Epithelial Cell Responses to B. pertussis.
The data presented above and in Table 1 reveal up-regulation of genes whose products play a role in NFκB activation as well as genes known to be regulated by NFκB. We hypothesized that pretreatment of the epithelial cells with antiinflammatory drugs that interfere with NFκB activation could help confirm mechanisms by which B. pertussis stimulates a proinflammatory cellular response. Dexamethasone was chosen because glucocorticoids have been suggested as a therapy for pertussis, despite the paucity of supportive data (22). Glucocorticoids may inhibit NFκB activity by augmenting IκB synthesis or through association of their bound receptors to NFκB (23). Sodium salicylate inhibits NFκB activation of genes by preventing phosphorylation of IκB (24).
The transcriptional responses of BEAS-2B cells pretreated with either sodium salicylate or dexamethasone and then exposed to B. pertussis BP536 differed profoundly from untreated cells (Tables 7 and 8, http://relman.stanford.edu). Pretreatment eliminated the infection-associated induction of GRO-2, GRO-1, IL-8, IL-6, IL-1β, MCP-1, MIP-1α, GRO-3, CIITA, HLA-DQ, API2/IAP, and other genes that were induced in untreated cells. However, differences were also noted in the effects of these two inhibitors. Dexamethasone failed to block a 2.9-fold infection-associated induction of the antiapoptotic molecule TNFAIP3, whereas salicylate was inhibitory. Furthermore, dexamethasone pretreatment led to a 4.8-fold infection-associated increase in expression of iNOS, which was not seen in salicylate-pretreated cells or in the absence of drug pretreatment. Transcription of IL-1, which has been implicated in the induction of iNOS activity (12), was not detected in dexamethasone-treated cells.
B. pertussis Binds Mucin and Induces Its Expression by Bronchial Epithelial Cells.
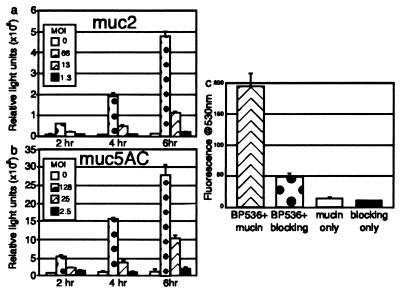
The regulated expression of mucin in the respiratory tract is one innate host defense that respiratory pathogens must overcome. Given previously published evidence of mucin gene responses to other respiratory noxious stimuli, we examined our microarray expression data for responses by the mucin gene family. MUC2, which encodes a respiratory tract mucin glycoprotein, was expressed at 1.5-fold increased levels in cells exposed to B. pertussis, whereas no response was noted for MUC3 (encodes intestinal mucin) and MUC6 (encodes gastric mucin). These data prompted further investigation by using a system that is more sensitive to small changes in mucin gene transcriptional activity. MUC2 and MUC5AC [also shown to be transcribed in respiratory epithelium (25)] were studied by using epithelial cells that harbor a stable transfected copy of the MUC2 or MUC5AC promoter fused to a luciferase reporter gene. Transcription of MUC2 and MUC5AC increased during the experiment in both a dose- and time-dependent manner (Fig. 2 a and b). At 6 h, the highest bacterial dose gave 32-fold (P = 0.002) and 17-fold (P = 0.01) increases in MUC2- and MUC5AC-luciferase activities, respectively. To our knowledge, there have been no previous descriptions of mucin gene responses to B. pertussis infection.
Figure 2.
(a and b) Induction of mucin reporter genes by B. pertussis infection. In duplicate experiments, an epithelial cell line carrying a mucin-luciferase reporter gene fusion was exposed to varying numbers of B. pertussis for increasing periods of time. Representative data from one experiment in which the fusion contained the promoter from MUC2 (a) or the promoter from MUC5AC (b) are shown. (c) Binding of B. pertussis to mucin-coated plates. One set of data from triplicate experiments that demonstrates binding of B. pertussis to mucin-coated and blocked wells vs. those coated with blocking agent alone. Background fluorescence of the mucin and nonfat milk are demonstrated by the third and fourth bars. Error bars represent the 95% confidence interval.
Given the ability of B. pertussis to induce mucin gene transcription, we hypothesized that the bacterium might be capable of binding mucin. Fig. 2c shows that B. pertussis adhered to microtiter plates at 4-fold higher levels when the wells were coated with bovine salivary mucin and then treated with a nonspecific blocking agent (nonfat dry milk) than they did to wells treated with the blocking agent alone (P < 6 × 10−12). A related species, Bordetella bronchiseptica has been shown to bind to swine nasal mucosa sialyl glycoconjugates, including mucin (26). Evaluation of several glycoconjugates revealed that a sialic acid-containing ganglioside mixture was able to inhibit B. pertussis binding to mucin by 32% (P < 0.005). N-acetyl-neuraminic acid blocked binding better than N-acetyl-d-glucosamine, but neither produced a significant decrease (22% and 12%, P values of 0.07 and 0.32, respectively). These data suggested that binding of B. pertussis to mucin is mediated by sialic acid-containing compounds.
Discussion
The use of high-density DNA microarrays to survey genome-wide transcriptional responses in host cells after exposure to microbial pathogens is a powerful approach to understanding host–pathogen interactions (10, 27, 28) (J. Corbeil, D. Genini, D. Sheeter, S. Rought, L. Leoni, P. Du, M. Ferguson, D. R. Masys, J. B. Welsh, J. L. Fink, et al., personal communication). Our data provide an overview of the interaction between B. pertussis and respiratory epithelial cells at the level of gene transcription and suggest a number of testable hypotheses. Our studies have revealed a dominant proinflammatory state in B. pertussis-exposed bronchial epithelial cells and the induction of host defenses that might be coopted or subverted by this pathogen. In addition, we have documented responses by a number of genes that were not previously known to be associated with B. pertussis or its virulence factors.
As with any model system, our experimental design and data have limitations. Transformed cell lines may respond in a manner distinct from that of primary cells by virtue of altered controls on growth and differentiation. Furthermore, we are limited by data from only a single time point and a single multiplicity of infection. Our system is also limited by the presence of only one cell type, whereas in an infected host, the responses of bronchial epithelial cells would be modified by the presence of other immune and nonimmune cells and a wide variety of soluble factors. These limitations were accepted to create an easily controlled, easily manipulated, and reproducible model of the interaction between B. pertussis and its primary target cell.
B. pertussis-Infected Bronchial Epithelial Cells Exhibit a Transcriptional Profile Dominated by a Proinflammatory Response.
Eight of the thirty-three genes that met our induction criteria on B. pertussis infection encode proinflammatory cytokines. This host response profile has not been described previously for B. pertussis infection. Although the chemokines IL-8, GRO-1, GRO-2, and GRO-3 are strong neutrophil chemoattractants, MCP-1 and SCYA3 promote monocyte infiltration (19, 20, 29). IL-8, GRO-1, and SCYA3 have also been shown to exert lymphocyte chemoattractant properties (30, 31). These results help to explain the infiltrate seen in autopsy studies of humans with pertussis and in animal models of the disease (1, 2). Furthermore, IL-8, MCP-1, and GRO-1 are known to promote expression of CD11b/CD18 (CR3) on the surface of neutrophils and monocytes (32, 33). This leukocyte integrin is a receptor for the major adhesin of B. pertussis, filamentous hemagglutinin (34). However, the binding of B. pertussis to leukocytes is not necessarily detrimental to this pathogen: the organism inhibits oxidative burst and intracellular killing and instead up-regulates its own binding to these cells and inhibits antigen-dependent T cell proliferation (34, 35). This suggests B. pertussis has adapted to the inflammatory response that it elicits from bronchial epithelial cells.
The NFκB Pathway Is Activated in B. pertussis-Infected Cells Without Evidence of Apoptosis.
Our study of human bronchial epithelial cells indicates that activation of NFκB and downstream pathways are dominant features of B. pertussis infection, as has been implicated in other infections (27, 36). The use of dexamethasone and sodium salicylate support this finding, because both inhibit NFκB activation, and they abolished the responses of genes which we propose were induced by NFκB. TRIP, which inhibits NFκB activation, is down-regulated on infection, thereby allowing NFκB activation (37). Furthermore, the transcriptional changes in TNFAIP3, API2, and BTF are suggestive of antiapoptotic signaling, despite the previous findings of B. pertussis-induced apoptosis in monocytes and neutrophils (17). In addition, our use of a phenotypic marker for apoptosis, the appearance of mono- and oligonucleosomes, revealed no evidence of apoptosis in the infected cells after 3 h. Thus, B. pertussis may have evolved specific mechanisms for killing inflammatory cells while sparing the nonciliated epithelium on which it survives.
PT-Responsive Genes Present Clues to the Pathogenesis of Pertussis.
Our use of a catalytically inactive toxin and a strain bearing this mutant toxin allowed us to investigate the role of the ADP–ribosyltransferase activity of PT without the confounding effects of other PT-associated activities. In BEAS-2B cells, two collagenases, fibroblast activation protein and matrix metalloproteinase I (MMP-1) were transcriptionally down-regulated by the enzymatic activity of PT. MMP-1 transcription is increased by other infections (38) and by basic fibroblast growth factor (39), whose transcription was increased by PT catalytic activity. Down-regulation of MMP-1 expression appears to be an active process and one that is unexpected. An examination of the responses elicited by this toxin on other cell types will likely reveal many other G-protein-linked genes.
Transcriptional analysis suggests that the quantity and quality of respiratory secretions are altered by the ADP–ribosylase activity of PT. The product of the down-regulated VIP gene increases secretion of Cl− in epithelial cells (40). In the lung, serotonin mediates airway reactivity and causes fluid transport across the epithelium into the tracheal lumen (41). Serotonin is cleared by a transporter, whose gene, SLC6A4, is up-regulated by PT. Through these regulatory events, B. pertussis may modify the physical properties of the respiratory tract mucus. This is borne out by descriptions of thick tenacious mucus at autopsy.
Inhibition of NFκB Activation by Antiinflammatory Agents Has Therapeutic Implications.
Previous studies have proposed that corticosteroids may have some benefit in the treatment of pertussis through their effect on transcription of proinflammatory effectors such as IL-1β, IL-6, and IL-8 (22). Of particular interest given the role of iNOS in epithelial cell pathology (12) was the finding that dexamethasone treatment before infection led to increased transcription of iNOS. Other studies have described a decrease in iNOS transcription when cells are treated with corticosteroids before stimulation with lipopolysaccharide and several cytokines (42). In our study, iNOS transcriptional activation occurred despite the induction of TNFAIP3 expression; in some cells, TNFAIP3 blocks transcription of iNOS (14). Thus, our data indicate the dominance of other positive regulatory factors on iNOS expression. From the perspective of disease management, these data may suggest caution in the use of corticosteroids for patients with whooping cough. These agents are predicted to worsen TCT-mediated induction of iNOS and subsequent adverse effects on neighboring ciliated epithelial cells.
B. pertussis Subverts the Mucociliary Escalator.
Although mucus is often considered a host defense; in some diseases, such as asthma, bronchitis, and cystic fibrosis, it contributes to the pathology. Our data show that B. pertussis, like many other infectious agents or irritants, is capable of inducing mucin transcription. With an intact mucociliary escalator, mucin provides an important housekeeping function. However, lipopolysaccharide, in combination with TCT of B. pertussis, paralyzes ciliated epithelial cells and causes their death and extrusion (6), leading to a buildup of respiratory secretions. As described above, PT-induced alterations in VIP and serotonin signaling could lead to thicker and even more tenacious mucus. We have demonstrated that B. pertussis is able to bind to mucin, suggesting that the pathogen may be manipulating host defenses to create a favorable microenvironment for itself.
Conclusion.
Transcriptional analysis of infected epithelial cells has helped to reveal the mechanisms by which the host responds to B. pertussis and some of the mechanisms by which this pathogen attempts to subvert host defenses. Our data help to integrate preexisting information about the pathology of pertussis, pathogen–host and receptor–ligand interactions, and host cell phenotypes, with virulence mechanisms. A more comprehensive examination of transcriptional responses of pathogens and their hosts will help define conserved and specific strategies used by both.
Acknowledgments
C.B. is a fellow of the Pediatric Scientist Development Program and is funded by National Institutes of Health Grants 7K12HD00850–13 and 5K12HD00850–14. B.K. was funded by the Walter and Idun Berry Fellowship. D.R. was supported in part by National Institute of Health/National Institute of Allergy and Infectious Diseases Grant R01 AI39587. We thank Tom Schall and Zheng Wei (Chemocentryx, San Carlos, CA) for their expertise with chemokines. We also thank Rino Rappuoli (Chiron Corporation/Istituto Ricerche Immunobiologiche, Siena, Italy) for his generous gift of B. pertussis strain BP-9K/129G and PT.
Abbreviations
- PT
pertussis toxin
- TCT
tracheal cytotoxin
- iNOS
nitric oxide synthase
- GRO
growth-regulated oncogene
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 13467.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.230262797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.230262797
References
- 1.Lapin J H. Whooping Cough. Springfield, IL: Thomas; 1943. [Google Scholar]
- 2.Khelef N, Bachelet C M, Vargaftig B B, Guiso N. Infect Immun. 1994;62:2893–2900. doi: 10.1128/iai.62.7.2893-2900.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Relman D A, Domenighini M, Tuomanen E, Rappuoli R, Falkow S. Proc Natl Acad Sci USA. 1989;86:2637–2641. doi: 10.1073/pnas.86.8.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katada T, Ui M. Proc Natl Acad Sci USA. 1982;79:3129–3133. doi: 10.1073/pnas.79.10.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hewlett E L, Weiss A A, Crane J K, Pearson R D, Anderson H J, Myers G A, Evans W S, Hantske L L, Kay H D, Cronin M J. Dev Biol Stand. 1985;61:21–26. [PubMed] [Google Scholar]
- 6.Flak T A, Heiss L N, Engle J T, Goldman W E. Infect Immun. 2000;68:1235–1242. doi: 10.1128/iai.68.3.1235-1242.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pizza M, Covacci A, Bartoloni A, Perugini M, Nencioni L, De Magistris M T, Villa L, Nucci D, Manetti R, Bugnoli M, et al. Science. 1989;246:497–500. doi: 10.1126/science.2683073. [DOI] [PubMed] [Google Scholar]
- 8.Lechner J F, LaVeck M A. J Tissue Culture Methods. 1985;9:43–48. [Google Scholar]
- 9.Wodicka L, Dong H, Mittmann M, Ho M H, Lockhart D J. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 10.Zhu H, Cong J P, Mamtora G, Gingeras T, Shenk T. Proc Natl Acad Sci USA. 1998;95:14470–14475. doi: 10.1073/pnas.95.24.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anisowicz A, Messineo M, Lee S W, Sager R. J Immunol. 1991;147:520–527. [PubMed] [Google Scholar]
- 12.Heiss L N, Lancaster J R, Jr, Corbett J A, Goldman W E. Proc Natl Acad Sci USA. 1994;91:267–270. doi: 10.1073/pnas.91.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsusaka T, Fujikawa K, Nishio Y, Mukaida N, Matsushima K, Kishimoto T, Akira S. Proc Natl Acad Sci USA. 1993;90:10193–10197. doi: 10.1073/pnas.90.21.10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grey S T, Arvelo M B, Hasenkamp W, Bach F H, Ferran C. J Exp Med. 1999;190:1135–1146. doi: 10.1084/jem.190.8.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uren A G, Pakusch M, Hawkins C J, Puls K L, Vaux D L. Proc Natl Acad Sci USA. 1996;93:4974–4978. doi: 10.1073/pnas.93.10.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brockstedt E, Otto A, Rickers A, Bommert K, Wittmann-Liebold B. J Protein Chem. 1999;18:225–231. doi: 10.1023/a:1020636308270. [DOI] [PubMed] [Google Scholar]
- 17.Khelef N, Guiso N. FEMS Microbiol Lett. 1995;134:27–32. doi: 10.1111/j.1574-6968.1995.tb07909.x. [DOI] [PubMed] [Google Scholar]
- 18.Cundell D R, Kanthakumar K, Taylor G W, Goldman W E, Flak T, Cole P J, Wilson R. Infect Immun. 1994;62:639–643. doi: 10.1128/iai.62.2.639-643.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammond M E, Lapointe G R, Feucht P H, Hilt S, Gallegos C A, Gordon C A, Giedlin M A, Mullenbach G, Tekamp-Olson P. J Immunol. 1995;155:1428–1433. [PubMed] [Google Scholar]
- 20.Wolpe S D, Sherry B, Juers D, Davatelis G, Yurt R W, Cerami A. Proc Natl Acad Sci USA. 1989;86:612–616. doi: 10.1073/pnas.86.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spangrude G J, Sacchi F, Hill H R, Van Epps D E, Daynes R A. J Immunol. 1985;135:4135–4143. [PubMed] [Google Scholar]
- 22.Roberts I, Gavin R, Lennon D. Pediatr Infect Dis J. 1992;11:982–983. [PubMed] [Google Scholar]
- 23.Scheinman R I, Cogswell P C, Lofquist A K, Baldwin A S., Jr Science. 1995;270:283–286. doi: 10.1126/science.270.5234.283. [DOI] [PubMed] [Google Scholar]
- 24.Yin M J, Yamamoto Y, Gaynor R B. Nature (London) 1998;396:77–80. doi: 10.1038/23948. [DOI] [PubMed] [Google Scholar]
- 25.Dohrman A, Miyata S, Gallup M, Li J D, Chapelin C, Coste A, Escudier E, Nadel J, Basbaum C. Biochim Biophys Acta. 1998;1406:251–259. doi: 10.1016/s0925-4439(98)00010-6. [DOI] [PubMed] [Google Scholar]
- 26.Ishikawa H, Isayama Y. Infect Immun. 1987;55:1607–1609. doi: 10.1128/iai.55.7.1607-1609.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eckmann L, Smith J R, Housley M P, Dwinell M B, Kagnoff M F. J Biol Chem. 2000;275:14084–14094. doi: 10.1074/jbc.275.19.14084. [DOI] [PubMed] [Google Scholar]
- 28.Rosenberger C M, Scott M G, Gold M R, Hancock R E, Finlay B B. J Immunol. 2000;164:5894–5904. doi: 10.4049/jimmunol.164.11.5894. [DOI] [PubMed] [Google Scholar]
- 29.Rollins B J, Walz A, Baggiolini M. Blood. 1991;78:1112–1116. [PubMed] [Google Scholar]
- 30.Gesser B, Lund M, Lohse N, Vestergaad C, Matsushima K, Sindet-Pedersen S, Jensen S L, Thestrup-Pedersen K, Larsen C G. J Leukocyte Biol. 1996;59:407–411. doi: 10.1002/jlb.59.3.407. [DOI] [PubMed] [Google Scholar]
- 31.Jinquan T, Frydenberg J, Mukaida N, Bonde J, Larsen C G, Matsushima K, Thestrup-Pedersen K. J Immunol. 1995;155:5359–5368. [PubMed] [Google Scholar]
- 32.Detmers P A, Powell D E, Walz A, Clark-Lewis I, Baggiolini M, Cohn Z A. J Immunol. 1991;147:4211–4217. [PubMed] [Google Scholar]
- 33.Vaddi K, Newton R C. J Immunol. 1994;153:4721–4732. [PubMed] [Google Scholar]
- 34.Ishibashi Y, Claus S, Relman D A. J Exp Med. 1994;180:1225–1233. doi: 10.1084/jem.180.4.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boschwitz J S, Batanghari J W, Kedem H, Relman D A. J Infect Dis. 1997;176:678–686. doi: 10.1086/514090. [DOI] [PubMed] [Google Scholar]
- 36.Ebnet K, Brown K D, Siebenlist U K, Simon M M, Shaw S. J Immunol. 1997;158:3285–3292. [PubMed] [Google Scholar]
- 37.Lee S Y, Choi Y. J Exp Med. 1997;185:1275–1285. doi: 10.1084/jem.185.7.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding Y, Uitto V J, Firth J, Salo T, Haapasalo M, Konttinen Y T, Sorsa T. Oral Dis. 1995;1:279–286. doi: 10.1111/j.1601-0825.1995.tb00194.x. [DOI] [PubMed] [Google Scholar]
- 39.Aho S, Rouda S, Kennedy S H, Qin H, Tan E M. Eur J Biochem. 1997;247:503–510. doi: 10.1111/j.1432-1033.1997.00503.x. [DOI] [PubMed] [Google Scholar]
- 40.Tokunaga T, Kiso T, Namikawa T, Ohtsubo Y. Biol Pharmacol Bull. 1999;22:745–748. doi: 10.1248/bpb.22.745. [DOI] [PubMed] [Google Scholar]
- 41.Tamaoki J, Chiyotani A, Takemura H, Konno K. Clin Exp Allergy. 1997;27:972–977. [PubMed] [Google Scholar]
- 42.Saura M, Zaragoza C, Diaz-Cazorla M, Hernandez-Perera O, Eng E, Lowenstein C J, Perez-Sala D, Lamas S. Kidney Int. 1998;53:38–49. doi: 10.1046/j.1523-1755.1998.00725.x. [DOI] [PubMed] [Google Scholar]