Abstract
Chromosomal translocations involving the N-terminal ≈250 residues of the Ewings sarcoma (EWS) oncogene produce a group of EWS fusion proteins (EFPs) that cause several distinct human cancers. EFPs are potent transcriptional activators and interact with other proteins required for mRNA biogenesis, indicating that EFPs induce tumorigenesis by perturbing gene expression. Although EFPs were discovered more than a decade ago, molecular analysis has been greatly hindered by the repetitive EWS activation domain (EAD) structure, containing multiple degenerate hexapeptide repeats (consensus SYGQQS) with a conserved tyrosine residue. By exploiting total gene synthesis, we have been able to systematically mutagenize the EAD and determine the effect on transcriptional activation by EWS/ATF1 and cellular transformation by EWS/Fli1. In both assays, we find the following requirements for EAD function. First, multiple tyrosine residues are essential. Second, phenylalanine can effectively substitute for tyrosine, showing that an aromatic ring can confer EAD function in the absence of tyrosine phosphorylation. Third, there is little requirement for specific peptide sequences and, thus, overall sequence composition (and not the degenerate hexapeptide repeat) confers EAD activity. Consistent with the above findings, we also report that the EAD is intrinsically disordered. However, a sensitive computational predictor of natural protein disorder (PONDR VL3) identifies potential molecular recognition features that are tyrosine-dependent and that correlate well with EAD function. In summary we have uncovered several molecular features of the EAD that will impact future studies of the broader EFP family and molecular recognition by complex intrinsically disordered proteins.
Keywords: Ewings sarcoma oncogene, EWS/ATF1, intrinsically disordered proteins, EWS activation domain, molecular recognition feature
The Ewings sarcoma (EWS) protooncogene together with TLS/FUS and hTAFII68 form a subgroup (the TET family; ref. 1) within the RNP family of RNA-binding proteins. Aberrant chromosomal translocations involving EWS produce EWS fusion proteins (EFPs) that cause human cancers (2, 3) and EFPs share the following common features (Fig. 1A). First, all EFPs contain at least the N-terminal 7 exons of EWS (residues 1–264) encoding the EWS activation domain (EAD). Second, the EWS fusion partner is one of several cellular transcription factors containing a sequence-specific DNA-binding domain that specifies the tumor type. Third, EFPs are potent transcriptional activators that depend on both the EAD and the fusion partner. Gene-specific transcriptional deregulation is likely to be a primary route for tumorigenesis (2, 3). However, some biological effects of EFPs might occur by alternative mechanisms including perturbation of pre-mRNA splicing (3).
Fig. 1.
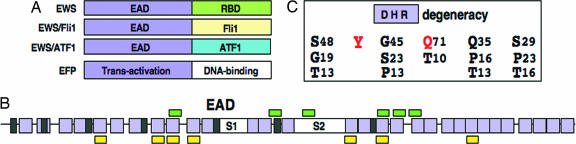
Structure of EFPs and the EAD. (A) EFPs. The normal EWS protein contains an N-terminal region (EAD) and a C-terminal RNA-binding domain (RBD). A simplified structure of the two oncogenic EFPs analyzed in the current study (EWS/Fli1 and EWS/ATF1) is shown. EWS/Fli1 and EWS/ATF1 are associated with Ewings sarcoma (EWS) (3, 4) and clear cell sarcoma (CCS) (46), respectively. The extended EFP family (2, 3) includes EWS/WT1 (desmoplastic small round cell tumor), EWS/CHOP (myxoid liposarcoma), EWS/CHN (myxoid chondrosarcoma), and EWS/ZSG (small round cell tumor). All EFPs contain the EAD (residues 1–264) and are lacking the EWS RBD. The distinct EWS fusion partners noted above are all transcription factors that in each case contribute at least a DNA-binding domain to the EFP. Intact ATF1 is a PKA-inducible activator (47), whereas, in contrast, EWS/ATF1 is a potent constitutive activator of ATF-dependent promoters (10) dependent on the EAD. (B) Primary structure of the EAD. There are multiple degenerate hexapeptide repeats (DHRs, purple boxes) with consensus sequence SYGQQS (4) and seven additional Tyr residues (dark gray boxes). The location of SH2-binding sites (YxxP, yellow boxes) and SH3-binding sites (PxxP, green boxes) are indicated. Spaces between DHRs are generally only a few residues except for S1 and S2, which are 12 and 25 residues, respectively. (C) DHRs. The sequence variation for EAD DHRs is indicated. Numbers are % occurrence and the absolutely conserved tyrosine (position 2) and well conserved glutamine (position 4) are highlighted in red.
Progress in determining structure/function relations for the EAD has been limited for several reasons. The EAD has a highly repetitive primary sequence (Fig. 1B) containing multiple copies of a degenerate hexapeptide repeat (DHR; consensus SYGQQS with Tyr absolutely conserved; ref. 4 and Fig. 1C), which has not been amenable to mutagenesis. Next, the biased amino acid composition of the EAD is characteristic of intrinsically disordered proteins (IDPs) (5–7) and determination of complex repetitive IDP structure has been confined largely to computational methods. Finally, comparative studies have yielded few pointers concerning how the EAD may work because the EAD is not obviously related to known transcriptional activation domains (TADs).
Cellular transformation by some EFPs has been directly demonstrated (8, 9). In addition, transcriptional assays have established that, in accord with the repetitive structure of the EAD, transactivation elements are dispersed throughout the EAD (8, 10, 11). The above finding and the fact that DHRs constitute 70% of the EAD (Fig. 1B) suggests that DHRs, and particularly the conserved tyrosines therein, are critical for EAD function. A small region of the EAD (residues 8–41) has been directly characterized, and Tyr residues within this region are required for transactivation (12). Whether the above finding for transactivation can be extrapolated to the intact EAD and to EFP-induced oncogenic transformation has not been addressed. Several Tyr residues within the EAD are putative phosphorylation sites, and the EAD contains many SH3/SH2 interaction motifs (13, 14). With respect to EFPs, exogenous c-Abl phosphorylates EWS/WT1 and interferes with DNA binding (15), whereas exogenous v-Src phosphorylates the EAD and modestly augments transactivation (16). Again, however, the precise tyrosine phosphorylation sites and the relationship of phosphorylation to EFP-induced changes in gene expression and cellular transformation have yet to be established.
The recent advent of commercial gene synthesis enables the study of proteins with dispersed functional elements and highly repetitive sequences via simultaneous introduction of multiple mutations. We have used the above approach to systematically alter the primary sequence of the EAD and determine the effects on transactivation by EWS/ATF1 and transformation by EWS/Fli1. Our findings show that multiple Tyr residues within a highly disordered structure are able to confer effective EAD function in the absence of Tyr phosphorylation.
Results
Mutagenesis of Highly Repetitive EAD Sequences.
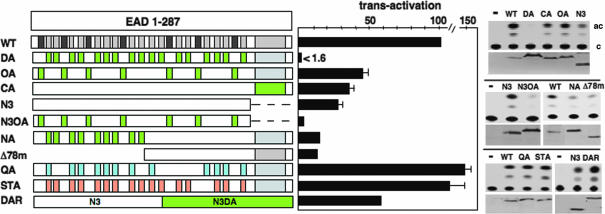
To identify transcriptional elements in the intact EAD (Fig. 2), we exploited total gene synthesis for creation of mutant proteins and a previously described assay for EWS/ATF1 (10). Briefly, JEG3 cells (EWS/ATF1 negative) were transfected with an expression vector for EWS/ATF1 (Δ287C; ref. 11) and an ATF-dependent CAT reporter [Δ(−71)SomCAT; ref. 17], resulting in an ≈250-fold EAD-dependent transactivation as shown in ref. 10. Conservation of DHR tyrosines (Fig. 1C) suggests a critical function, and we therefore made extensive Tyr to Ala substitutions and tested the effect on transactivation by the intact EAD (Fig. 2). Because the N-terminal EAD1-176 exhibits higher activity than the remaining C-terminal EAD region (10), we initially focused mutations in the former region. Changing all 17 DHR Tyr to Ala within EAD1-176 (mutant DA, DHR tyrosines changed to Ala) abolished activity [<1.6% of WT (Δ287C)] even though the DA protein is expressed at much higher levels. Changing the remaining 7 non-DHR Tyr to Ala (OA, other tyrosines changed to Ala) or the 14 remaining DHR tyrosines in the C-terminal region of the EAD (CA) had a small effect (43% and 34% of WT, respectively) when assayed in the context of the intact EAD (Fig. 2). Significantly, however, changing the seven non-DHR Tyr and testing in the context of the smaller region of the EAD (compare N3, which contains only EAD residues 1–166 and N3OA) had a much greater effect (N3OA had <11.5% of N3 activity), thus demonstrating that non-DHR tyrosines also contribute to EAD activity.
Fig. 2.
Effect of EAD mutations on transactivation by EWS/ATF1. All proteins contain the ATF1 portion of EWS/ATF1, but this is not shown. (Left) WT is an EWS/ATF1 fusion (Δ287C) containing the intact EAD (residues 1–287) and the region of ATF1(residues 66–271) present in oncogenic EWS/ATF1. DHRs (light gray boxes) and other non-DHR tyrosines (dark gray) are shown. Tyr to Ala changes (DA, OA, CA, N3OA, and NA) are indicated by green boxes. Gln to Ala changes (QA, light blue boxes) include all conserved DHR Gln residues in EAD1-176. Ser/Thr to Ala changes (STA, orange boxes) include one change in each DHR within EAD1-176 (see Materials and Methods). N3 (expressed from pΔ167C; ref. 11) contains the N-terminal 166 residues of the EAD and Δ78m is lacking the N-terminal 78 residues of the EAD. DAR contains the WT EAD region (present in N3) and the corresponding region (N3DA, green box) from DA. (Center) Quantitation of transactivation relative to 100% for WT with the SEM as error bar. In cases without error bars, the experiment was performed only twice. DA protein is expressed at much higher levels than WT, and the number indicated (1.6% of WT) overstates DA activity. (Right) Autoradiograms of representative CAT assays are shown (c, chloramphenicol; ac, acetylated chloramphenicol) together with the corresponding Western blot showing activator levels.
Because the mutational burden imposed on DA is quite high, we compared the effect of partial Tyr to Ala changes (CA and NA) with the effect of the corresponding deletions (N3 and Δ78m, respectively). In both cases, Tyr to Ala changes mimicked deletion (Fig. 2), indicating a localized effect. Next, we changed a large number of alternative residues present in EAD1-176 to Ala. QA has all conserved Gln residues at position 4 of the DHR (Fig. 1C) substituted by Ala, and STA has a total of 16 Ser/Thr residues substituted by Ala (see Materials and Methods). In contrast to DA, both QA and STA proteins retained activity, demonstrating that the effect of Tyr to Ala changes is specific. Finally, we created a protein called DAR, containing a WT EAD sequence fused to the mutated sequence present in DA (Fig. 2). DAR retained activity (225% of N3) showing that, in vivo, the gross Tyr to Ala changes present in DA do not cause a general protein malfunction or dominant inhibitory effect. Considering all of the above results, we conclude that multiple tyrosine residues in the EAD (including DHR and non-DHR) contribute to transactivation by EWS/ATF1.
Detailed Mutational Analysis of the EAD.
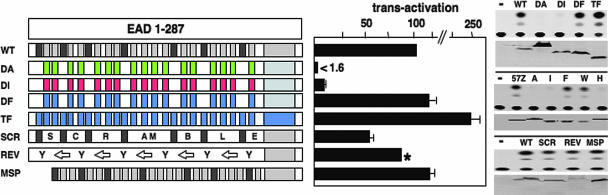
To elucidate the tyrosine side-chain requirements for transcriptional activation, we first produced a mutant protein (DF, all DHR tyrosines change to phenylalanine) that is equivalent to DA except that Tyr is changed to Phe (Fig. 3). DF protein retained activity compared with WT and was even slightly more active, although DF also is expressed at slightly higher levels than WT. Remarkably, a protein in which every Tyr residue in the entire EAD is changed to Phe (TF) also retained activity, demonstrating that the hydroxyl group of the Tyr side chain is not critical and, therefore, that effective transactivation can occur independently of Tyr phosphorylation within the EAD. Additional mutants were used to probe side-chain requirements (Fig. 3). DI protein is equivalent to DF except that Phe is replaced by Ile, which is hydrophobic but not aromatic. DI has reduced activity (≈8% of WT) although residual activity is retained compared with DA (<1.6% of WT). Trp is the only remaining aromatic amino acid, but because of poor expression of Trp-enriched proteins, we used a different assay (12) in which a smaller protein (57Z) containing EAD1-57 (including six DHRs) fused to the zta bZIP domain efficiently can activate a multisite Zta-CAT reporter (12). 57Z is the control protein (Fig. 3), whereas 57ZA, 57ZI, 57ZF, 57ZW, and 57ZH all have six DHR Tyr residues changed to Ala, Ile, Phe, Trp, and His, respectively. Significantly, proteins that contain an aromatic ring (57ZF and 57ZW) are highly active, whereas those that do not (57ZA, 57ZI, and 57ZH) are defective (Fig. 3). Overall, we conclude that the aromatic ring of multiple tyrosine side chains is critical for transactivation by EWS/ATF1, and a hydrophobic side chain (present in isoleucine) can support some residual function.
Fig. 3.
More detailed mutagenesis of the EAD. Transactivation assays, data presentation, and mutant depiction are as described for Fig. 2. Tyr to Ile (DI, red boxes) and Tyr to Phe (DF and TF, blue boxes) are indicated. Protein 57Z contains EAD1-57 (including six DHRs) fused to ATF1 and the zta bZIP domain (12). 57ZA (A), 57ZI (I), 57ZF (F), 57ZW (W), and 57ZH (H) have all six DHR tyrosines changed to Ala, Ile, Phe, Trp, and His, respectively. Transactivation assays for 57Z derivatives were performed by using a zta-CAT reporter (12). MSP protein has all spacer residues (white) between DHRs converted to two residues (see Results for details). For SCR, the relative positions of all 17 DHRs present in EAD1-176 are exchanged in a random manner. For REV, the peptide sequence between adjacent tyrosines present in EAD1-176 is inverted. ∗, REV protein level is lower than WT and normalization (under linear assay conditions) shows that REV exhibits >84% of WT activity.
DHRs constitute 70% of the EAD, prompting us to ask whether DHRs are sufficient for EAD activity. We first produced a protein called MSP (mutant spacer) in which all of the inter-DHR residues (spacers) were replaced with different dipeptides (AQ, TT, AP, or SG) while maintaining the overall composition. MSP was highly active (Fig. 3), indicating that specific spacer sequences are not critical for EAD activity. Next, we tested a protein called SCR (scrambled) in which the positions of all 17 DHRs present in EAD1-176 were scrambled. SCR also retained effective function (57% of WT, but SCR is expressed at slightly lower levels) (Fig. 3), indicating that DHRs are functionally interchangeable. The above results are consistent with a repetitive element (the DHR) in the EAD, but they also are compatible with the possibility that EAD activity is conferred by overall amino acid composition. To distinguish the above possibilities, we produced a protein called REV (reverse) in which all peptide sequences between consecutive Tyr residues present in EAD1-176 are inverted. REV also retained activity, demonstrating that there is little requirement, if any, for specific peptide sequences and, therefore, that the DHR per se is not a structural/functional determinant. This conclusion is supported by the finding that the conserved Gln residue in position 4 of the DHR is not required for EAD activity and that non-DHR Tyr residues significantly contribute to activity (Fig. 2). Together, the above experiments show that effective transactivation by EWS/ATF1 can be conferred by multiple Tyr residues presented in the context of the overall amino acid composition of the EAD.
Potential Molecular Recognition Features (MoRFs) in the EAD.
Compositional bias (lack of order promoting residues Cys, Val, Leu, Ile, and Met and enrichment in disorder promoting residues Gln, Ser, Pro, Ala, and Gly) suggested that the EAD has the characteristics of an IDP. Accordingly, circular dichroism analysis indicates that the EAD is largely unfolded (ref. 18; K.A.W.L., unpublished data). IDPs can be evaluated by various algorithms (19) including predictor of natural protein disorder regions (PONDR; ref. 5). PONDR is a set of neural network predictors based on local amino acid composition, flexibility, hydropathy, and coordination number. PONDR VL3 is highly sensitive and combines predictions of 30 neural networks for the entire protein sequence, trained by using circular dichroism, limited proteolysis, and other physical data from >150 IDPs (20). It has also been emphasized that propensity for order within IDPs can be identified by local “dips” in the PONDR curve that reflect molecular recognition elements (MoREs, (21, 22) or MoRFs (23). MoRFs (MoREs) therefore refer to subregions within IDPs that have the propensity for target-induced folding.
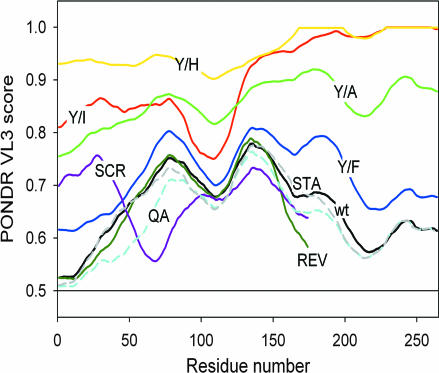
To probe for the presence of potential MoRFs in the EAD (Fig. 4), predictions were performed by using the PONDR VL3 predictor (access provided by Molecular Kinetics, Indianapolis, IN). PONDR VL3 analysis of WT and mutated EAD sequences shows that all of the proteins are generally disordered (PONDR curves mostly >0.5 threshold). However, there are several dips in the WT EAD PONDR curve, and these dips are generally reduced for Tyr/Ala, Tyr/Ile, and Tyr/His changes. In contrast, Tyr/Phe changes only marginally shifts the PONDR curve toward disorder and the PONDR curve for other active EAD mutants (QA, STA, SCR, and REV) also resembles the WT EAD. REV looks similar to WT EAD (because at a resolution of ≈10 aa they are similar), whereas SCR differs from WT EAD (because particular subregions are different) but retains overall propensity for order similar to WT. It is desirable to evaluate predictions of order propensity by using different methods (19), and we therefore applied several alternative predictors [RONN (24), IUPred (25), and FoldIndex (26)]. For the WT EAD, the above predictors are comparable with PONDR overall but show order/disorder features to different degrees [see supporting information (SI) Fig. 6]. With respect to EAD mutants, RONN and IUPred broadly agree with PONDR but are less sensitive, whereas FoldIndex appears relatively insensitive (see SI Figs. 7–9). In conclusion, predictions of order propensity by using PONDR VL3 are quite sensitive and are well correlated with activity for a range of EAD mutants, thus strongly suggesting that tyrosine-dependent MoRFs confer EAD activity.
Fig. 4.
Predictions of the presence of potential MoRFs in the EAD by using PONDR VL3. Higher PONDR scores reflect propensity for disorder (low probability for forming MoRFs), and lower scores reflect the propensity for order (higher probability for forming MoRFs). For (Y/A), (Y/I), (Y/H), and (Y/F), all EAD tyrosines are changed to Ala, Ile, His, or Phe, respectively. Color code is as follows: black, WT EAD; green, Y/A; red, Y/I; yellow, Y/H; blue, Y/F. PONDR plots derived for EAD mutants REV (dark green), SCR (purple), STA (dashed gray), and QA (dashed light blue) also are shown. Except for STA, REV, and SCR, the PONDR curve color code approximates that of the mutant proteins (Figs. 2 and 3).
Effect of EAD Mutations on Transformation by EWS/Fli1.
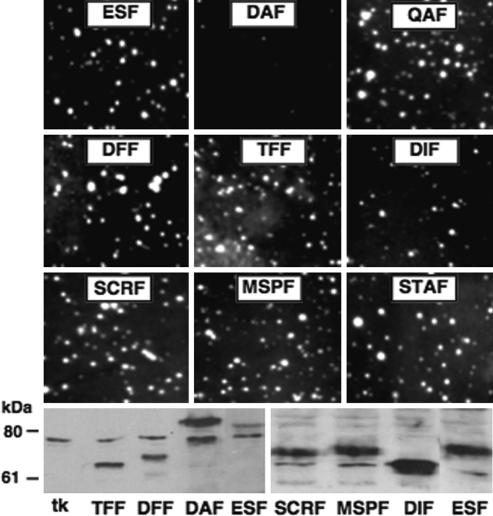
The transcription assay for EWS/ATF1 is very sensitive and robust, but it was of importance to assess EAD mutations in a more biologically significant assay. EWS/ATF1 appears to be toxic to NIH 3T3 (C.T.D. and K.A.W.L., unpublished data), thus limiting examination of EWS/ATF1 biological activity. We therefore turned to another well characterized EFP (EWS/Fli1) for which a transformation assay has been established (8). Selected EAD mutations analyzed for EWS/ATF1(TF, DF, DA, DI, QA, STA, MSP, and SCR) were introduced into EWS/Fli1 (TFF, DFF, DAF, DIF, QAF, STAF, MSPF, and SCRF, respectively), proteins expressed in NIH 3T3 cells by using a retroviral vector and anchorage-independent growth scored by soft agar colony formation (Fig. 5). All mutants tested were competent for transformation, except for DAF, which was inactive, and DIF, which exhibited reduced activity. Western blot analysis confirmed the expression of mutant proteins in transformed cells and similar to DA (Fig. 2) DAF was expressed at higher levels (Fig. 5). DIF also was expressed at higher levels, and the actual transforming capacity of DIF therefore may be less than observed. Overall, the relative activity of corresponding EWS/Fli1 and EWS/ATF1 mutants was well correlated, and we conclude that the molecular determinants of EAD activity are very similar in two assays and for two distinct EFPs. This suggests that our findings clearly can be expected to apply to the broader EFP family.
Fig. 5.
Effect of EAD mutations on transformation by EWS/Fli1. Transformation of NIH 3T3 cells was tested by using retroviral introduction of EWS/Fli1 (ESF) and selected EAD mutants, followed by soft agar colony assay. (Upper) Representative colony formation under low-serum conditions is shown. (Lower) Expression analysis was performed by Western blot with anti-Fli1 antibody. All EWS/Fli1 mutants contain EAD1-264, harboring the same mutations described for EWS/ATF1. Thus, DAF corresponds to the EWS/ATF1 mutant DA (Fig. 2) and likewise for TFF, DFF, DIF, STAF, QAF, MSPF, and SCRF.
Discussion
EAD Structure.
A close correlation between results from functional and computational approaches indicates that multiple Tyr-dependent MoRFs confer effective EAD function in the assays used. Although necessary, Tyr enrichment is insufficient for EAD function, because, for example, the EWS relative TAFII68 (1) has a similarly Tyr-rich C-terminal region that contains multiple Asp and Arg residues and fails to activate (27). This suggests that the restricted amino acid composition of the EAD creates an enabling structure (because of enrichment with Pro, Ser, Gln, and Gly) with relatively exposed tyrosine residues dispersed in a polar/neutral environment favoring hydrogen-bonding interactions and flexibility.
Repetitive structure is a common feature of IDPs (28), and modes of target recognition are beginning to emerge. First, the EAD resembles the prion domain (PD) of the Saccharomyces cerevisiae IDP Sup35p. Sup35p shows sequence similarity to the EAD (notably enrichment with Tyr) due to an imperfect reiterated peptide QGGYQQYN, and similar to the EAD, a randomized Sup35p PD retains function (29). For Sup35p, it is not known whether particular amino acids are functionally critical and potential MoRFs have not been identified. A second example of note involves the Arg/Ser-rich (R/S) domain present in splicing enhancer factors (30). An R/S domain can be reconstituted by 21 RS dipeptides, partially by 14, and not at all by 7 (30). The R/S domain might exhibit a mode of molecular recognition that broadly resembles the EAD, although the R/S domain is significantly shorter and has a less complex composition. Finally, the splicing factor SF3b155 harbors multiple copies of a degenerate motif (consensus (RK)nXRW(DE), which mediates binding to U2AF dependent on the conserved Trp residue (31). However, a single SF3b155 motif is sufficient for high-affinity U2AF binding (31), suggesting that the mechanism of molecular recognition by the EAD and SF3b155 may be quite distinct.
Whether individual Tyr residues act independently, or whether multiple Tyr residues functionally collaborate, remains to be elucidated. The spacing between tyrosines in EAD1-176 (the most active region and including 24 spacers) is quite variable and evenly distributed between two and eight residues, suggesting that spacing of tyrosines may not be critical for EAD activity. In relation to known protein recognition sites, the EAD has several SH2 (YxxP) and SH3 (PxxP) interaction motifs (Fig. 1B). Interaction of SH2 domains with YxxP motifs requires Tyr phosphorylation, and EAD/SH2 interactions therefore appear not to be critical for molecular recognition by the EAD. The PxxP motif can form a structure called the polyproline II (PPII) helix (32), but three of four PxxP motifs present in EAD1-176 can be mutated without effect (see MSP, SCR, and REV), suggesting that PPII helices do not contribute to EAD activity. Precise definition of EAD MoRFs will depend on extending the approach described here, but is also likely to require identification of critical EAD protein targets.
Transcriptional Activation by EFPs.
For the functions of EWS/ATF1 and EWS/Fli1 that we have tested, the EAD works effectively in the absence of tyrosine phosphorylation. Given the numerous tyrosine kinase interaction motifs in the EAD (Fig. 1B), it may not be surprising that tyrosine phosphorylation reportedly acts on some EFPs (15, 16), although the biological significance has not been established. c-Abl can mediate phosphorylation and inhibit DNA binding by EWS/WT1 (15), and v-Src modestly augments transactivation by EWS/WT1 (16). In relation to the current study, it must be emphasized that our results do not preclude potential biological effects of tyrosine kinases on EWS/ATF1 or EWS/Fli1. However, our findings do establish that at least some EFP functions can be mediated effectively under conditions in which the EAD is not phosphorylated on any tyrosine residues. It will be of interest to explore how broadly tyrosine phosphorylation impacts the wider EFP family and normal EWS or other TET family members (1).
A number of properties distinguish the EAD from natural TADs. First, the EAD is larger than most TADs (≈250 versus ≈40 residues). Second, although the EAD shows a resemblance to the TAD of the stage-specific activator protein (SSAP) (33), the tyrosine-rich region of SSAP requires additional elements to be effective (33). Third, the VP16C TAD has several Phe residues (34), but in contrast to the EAD, hydrophobicity rather than the aromatic ring appears to be functionally important (34). Lastly, the EAD is not normally an activation domain because the EWS RNA-binding domain represses the EAD (35). Having said the above, the EAD does resemble TADs in the following respects: (i) the presence of reiterated elements; (ii) the lack of structure (PONDR predicts TADs are 73–94% disordered (36); and (iii) flexibility engendered by enrichment in Gly, Ser, Gln, and Pro. Thus, TADs may provide instructive models for the EAD, and such models invoke multiple weak contacts between TADs and their targets, resembling a “ molecular Velcro” (37). Considering the above framework, both the size (number of potential MoRFs) and malleability of the EAD may well account for the potency of EAD-mediated transactivation.
The EAD is known to contact a limited number of transcriptional components including the coactivator CBP (38, 39), multiple TAFs (40), and the RNA polymerase II subunits rpb7 (40, 41) and rpb5 (40). rpb7 and its partner rpb4 are required for EAD-mediated transactivation in yeast (42), suggesting that the EAD might flexibly wrap around rpb7/4 via multiple contacts, a mode of recognition that is used by known IDPs (7). However, artificial promoter recruitment of rpb7 in mammalian cells does not activate transcription (K.A.W.L., unpublished data), perhaps pointing to the need for multiple EAD partners. We therefore speculate that efficient and promiscuous action by the EAD may involve recruitment of several transcriptional components via an array of related MoRFs dependent either on individual EAD Tyr residues or possibly Tyr clusters. A precedent for the above mode of action is provided by HMGA (of the HMG family of architectural TFs), which is highly unstructured (36) and can interact with at least 18 other proteins, many of which are themselves transcription factors.
Oncogenic Versus Normal EAD Function.
The essential features of the EAD described herein seem certain to impact the normal functions of EWS, whatever those functions might be. The overall structure of the EWS protein (including both the EAD and the RNA-binding domain) is highly conserved between frogs and mammals, with 70% dispersed identity. Conservation includes almost all tyrosine residues, several tyrosine phosphorylation sites, and several SH2/SH3 binding motifs. Given that the above features in EWS are likely to play important functional roles, our results indicate that certain molecular determinants (within the EAD) are finely distinguished with respect to EFP versus normal EWS function. The tumor specificity of EFPs and the fact that EFPs are implicated in tumor maintenance (43) suggest that such a distinction ultimately might allow development of EFP-specific inhibitors with therapeutic potential.
It is notable that EWS is found in several subcellular compartments and that the human proteome interaction network (44) places EWS at the hub of a disease-associated protein network via potential interactions with 20% of ≈400 different proteins (44). The malleable nature of the EAD most likely is crucial for allowing EWS to network so broadly, and the EAD mutants that we have created should provide invaluable tools for probing EWS function and identifying cognate EWS interaction partners.
Materials and Methods
Plasmids.
pΔ(−71)SomCAT (17), pΔ78 (10), pΔ287C and pΔ167C (11), and pZ7E4TCAT and p57Z (12) are as described. For construction of EAD mutants, synthetic DNA fragments containing the desired restriction sites were obtained by commercial gene synthesis (TOP Gene Technologies, Montreal, QC, Canada) and directly inserted into vectors obtained from pΔ287C (11) or p57Z (12). pSTA contains Ser- to-Ala changes for EAD residues 40, 51, 69, 87, 111, 162, 168, and 171 plus Thr-to-Ala changes for residues 8, 22, 32, 48, 64, 79, 95, and 120.
Retroviral Constructs and EWS/Fli1 Transformation Assay.
Plasmids harboring EWS/Fli1 containing EAD1-264 were constructed in pBluescript II KS and then inserted into replication-deficient retroviral construct pSRαMSVtkneo. Retroviral transduction of NIH 3T3 cells and soft agar assays were performed as described in ref. 45.
EWS/ATF1 Transactivation Assay and Western Blotting.
Transfections, CAT assays, Western blotting, and quantitation of transactivation under nonsaturating conditions were performed as described in ref. 10. In cases where mutations resulted in significant changes in protein levels, quantitation is explained in Results or in the legends to Figs. 2 and 3. Western blot detection of EWS/Fli1 and mutants was performed by using C-terminal Fli1 antibodies.
Supplementary Material
Acknowledgments
We thank Kim Li for assistance in the laboratory; Keith Dunker, Peter Tompa, and Mingjie Zhang for useful discussions; and TOP Gene Technologies Inc., Montreal, for efficient and economical gene synthesis. This work was supported by Association for International Cancer Research Grant 03-131 (to K.A.W.L.) and National Cancer Institute Grant CA087771 (to C.T.D.).
Abbreviations
- DHR
degenerate hexapeptide repeat
- EAD
EWS activation domain
- EFP
EWS fusion protein
- EWS
Ewings sarcoma
- IDP
intrinsically disordered protein
- MoRF
molecular recognition feature
- PONDR
predictor of natural protein disorder
- TAD
transcriptional activation domain.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607007104/DC1.
References
- 1.Bertolotti A, Lutz Y, Heard DJ, Chambon P, Tora L. EMBO J. 1996;15:5022–5031. [PMC free article] [PubMed] [Google Scholar]
- 2.Kim J, Pelletier J. Physiol Genomics. 1999;1:127–138. doi: 10.1152/physiolgenomics.1999.1.3.127. [DOI] [PubMed] [Google Scholar]
- 3.Arvand A, Denny CT. Oncogene. 2001;20:5747–5754. doi: 10.1038/sj.onc.1204598. [DOI] [PubMed] [Google Scholar]
- 4.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, et al. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 5.Uversky VN, Oldfield CJ, Dunker AK. J Mol Recognit. 2005;18:343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- 6.Dyson HJ, Wright PE. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 7.Tompa P. FEBS Lett. 2005;579:3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 8.Lessnick SL, Braun BS, Denny CT, May WA. Oncogene. 1995;10:423–431. [PubMed] [Google Scholar]
- 9.Kim J, Lee KAW, Pelletier J. Oncogene. 1998;16:1973–1979. doi: 10.1038/sj.onc.1201716. [DOI] [PubMed] [Google Scholar]
- 10.Brown AD, Lopez-Terrada D, Denny CT, Lee KAW. Oncogene. 1995;10:1749–1756. [PubMed] [Google Scholar]
- 11.Pan S, Ming KY, Dunn TA, Li KKC, Lee KAW. Oncogene. 1998;16:1625–1631. doi: 10.1038/sj.onc.1201671. [DOI] [PubMed] [Google Scholar]
- 12.Feng L, Lee KAW. Oncogene. 2001;20:4161–4168. doi: 10.1038/sj.onc.1204522. [DOI] [PubMed] [Google Scholar]
- 13.Felsch JS, Lane WS, Peralta EG. Curr Biol. 1999;9:485–488. doi: 10.1016/s0960-9822(99)80214-0. [DOI] [PubMed] [Google Scholar]
- 14.Guinamard R, Fougereau M, Seckinger P. Scand J Immunol. 1997;45:587–595. doi: 10.1046/j.1365-3083.1997.d01-447.x. [DOI] [PubMed] [Google Scholar]
- 15.Kim J, Lee JM, Branton PE, Pelletier J. Proc Natl Acad Sci USA. 1999;96:14300–14305. doi: 10.1073/pnas.96.25.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J, Lee JM, Branton PE, Pelletier J. FEBS Lett. 2000;474:121–128. doi: 10.1016/s0014-5793(00)01590-8. [DOI] [PubMed] [Google Scholar]
- 17.Montminy MR, Sevarino KA, Wagner JA, Mandel G, Goodman RH. Proc Natl Acad Sci USA. 1986;83:6682–6686. doi: 10.1073/pnas.83.18.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uren A, Tcherkasskaya O, Toretsky JA. Biochemistry. 2004;43:13579–13589. doi: 10.1021/bi048776q. [DOI] [PubMed] [Google Scholar]
- 19.Ferron F, Longhi S, Canard B, Karlin D. Proteins. 2006;65:1–14. doi: 10.1002/prot.21075. [DOI] [PubMed] [Google Scholar]
- 20.Obradovic Z, Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK. Proteins. 2003;53(Suppl 6):566–572. doi: 10.1002/prot.10532. [DOI] [PubMed] [Google Scholar]
- 21.Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, Dunker AK. Biochemistry. 2005;44:12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- 22.Fuxreiter M, Simon I, Friedrich P, Tompa P. J Mol Biol. 2004;338:1015–1026. doi: 10.1016/j.jmb.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 23.Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. J Mol Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 24.Yang ZR, Thomson R, McNeil P, Esnouf RM. Bioinformatics. 2005;21:3369–3376. doi: 10.1093/bioinformatics/bti534. [DOI] [PubMed] [Google Scholar]
- 25.Dosztanyi Z, Csizmok V, Tompa P, Simon I. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 26.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 27.Bertolotti A, Bell B, Tora L. Oncogene. 2000;18:8000–8010. doi: 10.1038/sj.onc.1203207. [DOI] [PubMed] [Google Scholar]
- 28.Tompa P. BioEssays. 2003;25:847–855. doi: 10.1002/bies.10324. [DOI] [PubMed] [Google Scholar]
- 29.Ross ED, Edskes HK, Terry MJ, Wickner RB. Proc Natl Acad Sci USA. 2005;102:12825–12830. doi: 10.1073/pnas.0506136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Philipps D, Celotto AM, Wang Q-Q, Tarng RS, Gravely BR. Nucleic Acids Res. 2003;31:6502–6508. doi: 10.1093/nar/gkg845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thickman KR, Swenson MC, Kabogo JM, Gryczynski Z, Kielkopf CL. J Mol Biol. 2006;356:664–683. doi: 10.1016/j.jmb.2005.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li SS. Biochem J. 2005;390:641–653. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benuck ML, Li Z, Childs G. J Biol Chem. 1999;274:25419–25425. doi: 10.1074/jbc.274.36.25419. [DOI] [PubMed] [Google Scholar]
- 34.Sullivan SM, Horn PJ, Olson VA, Koop AH, Niu W, Ebright RH, Triezenberg SJ. Nucleic Acids Res. 1998;26:4487–4496. doi: 10.1093/nar/26.19.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li KKC, Lee KAW. J Biol Chem. 2000;275:23053–23058. doi: 10.1074/jbc.M002961200. [DOI] [PubMed] [Google Scholar]
- 36.Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Biochemistry. 2006;45:6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melcher K. J Mol Biol. 2000;301:1097–1112. doi: 10.1006/jmbi.2000.4034. [DOI] [PubMed] [Google Scholar]
- 38.Fujimura Y, Siddique H, Leo L, Rao VN, Reddy ESP. Oncogene. 2001;20:6653–6659. doi: 10.1038/sj.onc.1204684. [DOI] [PubMed] [Google Scholar]
- 39.Araya N, Hirota K, Shimamoto Y, Miyagishi M, Yoshida E, Ishida J, Kaneko S, Kaneko M, Nakajima T, Fukamizu A. J Biol Chem. 2003;278:5427–5432. doi: 10.1074/jbc.M210234200. [DOI] [PubMed] [Google Scholar]
- 40.Bertolotti A, Melot T, Acker J, Vigneron M, Delattre O, Tora L. Mol Cell Biol. 1998;18:1489–1497. doi: 10.1128/mcb.18.3.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petermann R, Mossier BM, Aryee DNT, Khazak V, Golemis EA, Kovar H. Oncogene. 1998;17:603–610. doi: 10.1038/sj.onc.1201964. [DOI] [PubMed] [Google Scholar]
- 42.Zhou H, Lee KAW. Oncogene. 2001;20:1519–1524. doi: 10.1038/sj.onc.1204135. [DOI] [PubMed] [Google Scholar]
- 43.Kovar H, Aryee D, Zoubek A. Curr Opin Oncol. 1999;11:275–284. doi: 10.1097/00001622-199907000-00007. [DOI] [PubMed] [Google Scholar]
- 44.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 45.Lugo TG, Witte ON. Mol Cell Biol. 1989;9:1263–1270. doi: 10.1128/mcb.9.3.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zucman J, Delattre O, Desmaze C, Epstein AL, Stenman G, Speleman F, Fletchers CDM, Aurias A, Thomas G. Nat Genet. 1993;4:341–345. doi: 10.1038/ng0893-341. [DOI] [PubMed] [Google Scholar]
- 47.Ribeiro A, Brown AD, Lee KAW. J Biol Chem. 1994;269:31124–31128. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.