Abstract
Mammalian centromere function depends upon a specialized chromatin organization where distinct domains of CENP-A and dimethyl K4 histone H3, forming centric chromatin, are uniquely positioned on or near the surface of the chromosome. These distinct domains are embedded in pericentric heterochromatin (characterized by H3 methylated at K9). The mechanisms that underpin this complex spatial organization are unknown. Here, we identify the essential histone variant H2A.Z as a new structural component of the centromere. Along linear chromatin fibers H2A.Z is distributed nonuniformly throughout heterochromatin, and centric chromatin where regions of nucleosomes containing H2A.Z and dimethylated K4 H3 are interspersed between subdomains of CENP-A. At metaphase, using the inactive X chromosome centromere as a model, complex folding of this fiber produces spatially positioned domains where H2A.Z/dimethylated K4 H3 chromatin juxtaposes one side of CENP-A chromatin, whereas a region of H2A/trimethyl K9 H3 borders the other side. A second region of H2A.Z is found, with trimethyl K9 H3 at the inner centromere. We therefore propose that H2A.Z plays an integral role in organizing centromere structure.
Keywords: centromere organization, chromosome structure, histone variants
All active centromeres contain an evolutionary conserved variant of histone H3 (CENP-A in mammals). Although poorly understood, fundamental to centromere function is the 3D organization of CENP-A containing chromatin, so that it is presented on the poleward face of a chromosome while being embedded in pericentric heterochromatin. This heterochromatin has a role in the cohesion of sister chromatids (1). The complex nature of this organization was demonstrated by showing that, whereas blocks of human CENP-A (and the Drosophila counterpart CID) and H3 nucleosomes (≈15- 40 kb) are interspersed along an extended chromatin fiber, the folding of this fiber in mitotic chromosomes yields separate CENP-A- and H3-containing domains, with the H3 domain located toward the inner chromatid region (together, these domains have been referred to as centric chromatin; ref. 2). Most interestingly, this H3-containing domain is enriched with a euchromatic mark, H3 dimethylated at K4 and not K9 H3 methylation associated with the flanking heterochromatin (3). Therefore, it was postulated that this distinct histone modification pattern might contribute to centromere structure and/or identity (3).
Our recent studies have demonstrated that another histone variant, H2A.Z, has a role in mitosis (4). When H2A.Z is depleted in mammalian cells, major defects in chromosome segregation (and cytokinesis) are observed with a corresponding loss of HP1α from pericentric heterochromatin, indicating this domain is severely disrupted. The mechanism responsible for this defect remains unresolved. Although others (5) and we (6) have shown that the concentration of H2A.Z can dynamically increase at pericentric heterochromatin under different physiological contexts, whether H2A.Z is a core component of this domain in mammals is not clear. If it is a core component, then the issue arises as to where it is located in this large megabase domain. In addition, the possibility that H2A.Z is a constituent of centric chromatin has not previously been considered.
In this study, we investigated: (i) whether H2A.Z is a core constituent of pericentric heterochromatin; (ii) the interplay between H2A.Z and CENP-A, including whether H2A.Z is a component of centric chromatin; (iii) the 3D spatial organization of H2A.Z chromatin at metaphase and its relationship to the other marks of the centromere; and (iv) whether the histone modifications that define centric and pericentric chromatin in flies, humans (3), and fission yeast (7) are conserved in mice.
Results
H2A.Z Is Present in Pericentric Heterochromatin at the Inner Centromere.
Previously, we engineered several stable mammalian cell lines to express H2A.Z siRNA in response to ponasterone A, an analogue of ecdysone, by utilizing a mammalian ecdysone-inducible hsp70 expression vector (4). Using this system, we demonstrated that H2A.Z is essential for chromosome segregation and cytokinesis. One of several major defects resulting from H2A.Z depletion was the formation of chromosome bridges between sister chromatids as they separated and moved toward opposing spindle poles (4). The mechanism responsible for this defect is not known. The hypothesis being tested here is that H2A.Z is a key structural component of pericentric heterochromatin, and the formation of these bridges, which occurs in its absence, is a direct result of the entanglement of disrupted pericentric heterochromatin between sister chromatids.
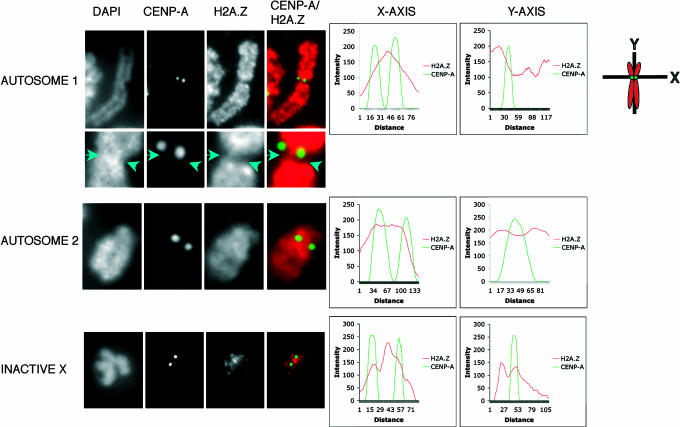
The pericentric heterochromatin found beneath centric chromatin (the inner centromere) plays a critical role in the cohesion of sister chromatids at metaphase and their subsequent faithful separation at anaphase (1). To investigate whether H2A.Z is located within this region, human HEK293 metaphase chromosomes were stained with DAPI and indirectly fluorescently labeled with H2A.Z and CENP-A antibodies. Shown are representative autosomes (autosomes 1 and 2) and an inactive X chromosome with 2D intensity plots following the x axis between the two-sister CENP-A-containing domains (Fig. 1). The 2D plot of the intensity of H2A.Z immunostaining clearly shows that H2A.Z is present at the inner centromere between the two sister chromatid peaks of CENP-A-containing chromatin for autosomes 1 and 2. All chromosomes examined (n = 3 metaphase spreads) showed this distribution of H2A.Z (data not shown).
Fig. 1.
H2A.Z is located between sister centromeres on metaphase chromosomes. Human HEK 293 metaphase chromosomes (n = 3 metaphase spreads) were stained with DAPI and subjected to indirect immunofluorescence by using antibodies to H2A.Z and CENPA. The intensity of fluorescence for each antibody following the shown x and y axes was determined and plotted for two autosomes and the inactive X chromosome. The metaphase spreads showing these analyzed chromosomes are displayed in SI Fig. 9A.
Previously, we noted that the inactive X chromosome, although containing macroH2A, is depleted in H2A.Z in somatic cells (6). However, if H2A.Z does play a fundamental role at pericentric heterochromatin, we would expect it to be retained at this domain. Indeed, this is the case; very little H2A.Z can be detected on the arms of the inactive X-chromosome, but it is clearly enriched at the inner centromere (Fig. 1).
Next, we analyzed the 2D location of H2A.Z across the centromere region along the chromosome arm (the y axis passing through one sister chromatid centromere; Fig. 1). Most interestingly, we identified at least two types of H2A.Z distribution patterns. For the inactive X chromosome, there was an unequal distribution of H2A.Z, with it being present on one side of the CENP-A domain while being relatively depleted on the other side. Such an arrangement of H2A.Z was seen on autosomes (autosome 1) irrespective of the concentration of DNA (compare the DAPI-staining pattern of the centromere with the distribution of H2A.Z, Fig. 1). An enlargement of the autosome centromere highlighting one side of the CENP-A region depleted in H2A.Z (along the y axis) is shown in Fig. 1. However, this arrangement of H2A.Z is not universal (autosome 2), indicating that individual chromosomes can differ in their chromatin organization around the centromere (see Discussion). An extensive 3D analysis of the inactive X chromosome centromere examining the precise location and spatial organization of H2A.Z-containing chromatin confirms our interpretation and will be described below (Fig. 3).
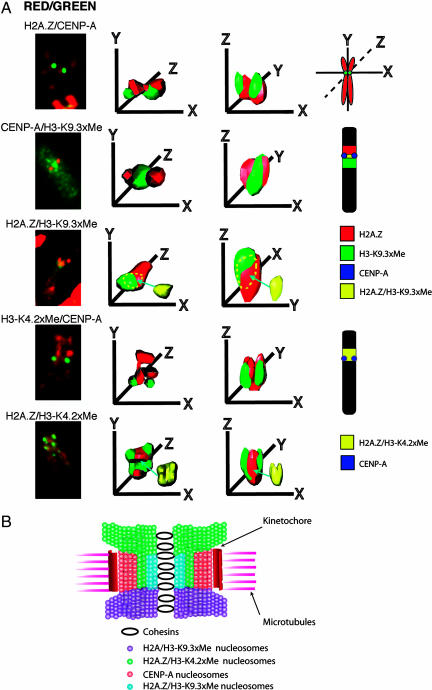
Fig. 3.
3D deconvolution analysis of the human inactive X chromosome centromere. (A) Human inactive X metaphase chromosomes in HEK 293 cells were immunostained with different pairwise combination of antibodies that recognize H2A.Z, CENP-A, trimethyl K9 H3, or dimethyl K4 H3, analyzed by 3D deconvolution microscopy and modeled to determine the spatial position of domains containing H2A.Z, CENP-A, trimethyl K9 H3, and dimethyl K4 H3. For each combination of antibodies shown, five to eight metaphase chromosomes were analyzed and modeled. (B) A model for the folding of pericentric and centric chromatin fibers into the unique 3D organization of the human inactive X chromosome centromere highlighting the spatial position of H2A.Z containing chromatin. This diagram has been modified from Sullivan and Karpen (3).
H2A.Z Is Present Discontinuously Along Extended Pericentric Heterochromatin Fibers.
In the mouse, it is possible to examine the histone composition and histone modification status of pericentric heterochromatin, because it is associated with A/T-rich major satellite DNA (megabases of a repeated 234-bp unit), which differs from CENP-A-containing centric chromatin (comprised of 120-bp minor satellite repeats). To investigate whether H2A.Z is a core component of pericentric heterochromatin in mammals, we analyzed the chromatin composition of major satellite DNA by using the extended chromatin fiber technique. Extended fibers (≈100 times their normal interphase length) were analyzed by DNA FISH by using a specific major satellite probe and by indirect immunofluorescence by using affinity-purified antibodies against H2A, H2A.Z, dimethyl K4 H3, and trimethyl K9 H3.
H2A.Z is associated with major satellite DNA but, unlike H2A, its distribution is nonuniform; large regions (≥50 μm in length) either contain or are depleted of H2A.Z [see supporting information (SI) Fig. 4A]. The length of the extended fibers (>500 μm in length) made analysis along the entire domain difficult, but it appeared that subdomains of H2A.Z-containing chromatin were present throughout the extended pericentric fiber. Based on the extent of overlap, we estimate that H2A.Z and H2A cover 44 ± 13% and 79 ± 2% of major satellite DNA, respectively (n = 6). As expected, the heterochromatic mark trimethylated K9 H3 is enriched along the major satellite chromatin fiber (98 ± 1%; n = 4), whereas the euchromatic and centric mark dimethylated K4 H3 is not, although it is not totally excluded (15 ± 4%).
To confirm the presence of H2A.Z on major satellite DNA, we performed the ChIP assay. Chromatin fragments were precipitated with the same antibodies used in the analysis of chromatin fibers as well as an antibody against the heterochromatin-binding protein HP1α for an additional control. The purified DNA was analyzed by semiquantitative PCR (16 cycles) using specific primers for major satellite DNA as previously described (8). Supporting the fiber analysis, H2A.Z-containing nucleosomes associate with major satellite DNA but to a lesser extent than H2A (see SI Fig. 4B). Taken together, we conclude that H2A.Z is a core component of constitutive heterochromatin in mammalian cell lines. It is also worth noting that H2A.Z is relatively more abundant on the chromosome arms compared with pericentric heterochromatin, which may explain our previous inability to detect H2A.Z at chromocenters in mouse L929 interphase cells (4).
Chromosome Bridges Are Composed of Major Satellite DNA.
Accurate chromosome segregation requires that once a chromosome has replicated, the resulting sister chromatids remain attached until all sister centromeres capture spindle microtubules originating from opposite poles. Pericentric heterochromatin and HP1 mediate this cohesion between sister chromatids at metaphase, enabling them to separate accurately at anaphase (1). Given that H2A.Z is a component of this domain, we would predict that the chromosome bridges that form between separating sister chromatids in H2A.Z siRNA cells would be composed of entangled defective heterochromatin. To test this proposal, under the same experimental conditions that we previously used (4), H2A.Z siRNA was expressed for 24 or 48 h in mouse L929 cells, and the integrity of pericentric heterochromatin was examined at interphase (see SI Fig. 5B) and metaphase (see SI Fig. 6A), and the composition of chromosome bridges was investigated at telophase (see SI Fig. 6A) by using DNA FISH, with major and minor satellite probes.
After the expression of H2A.Z siRNA for 48 h, the endogenous level of H2A.Z mRNA was reduced by 65 ± 5% (see SI Fig. 5A). It is worth highlighting that, despite defects in the chromosome segregation process, H2A.Z RNAi-treated cells continue to divide, eventually dying in a process unrelated to apoptosis (4). At interphase, major satellite DNA exists as large foci, which colocalize with DAPI-dense spots (see SI Fig. 5B). These chromocenters are the result of clustering of pericentric heterochromatin domains from different chromosomes. As previously noted, minor satellites are located at the periphery of the chromocenters (9).
When H2A.Z is depleted, there is a gradual decompaction of chromocenters at 24 h (see arrow in SI Fig. 5B) in 25% of cells (see SI Fig. 5C) and, by 48 h, this decompaction process continues with regions of major satellite DNA becoming dispersed throughout the nucleus (see SI Fig. 5B) in 50% of cells (see SI Fig. 5C), clearly indicating the loss of pericentric heterochromatin integrity at interphase. We also noted that in some affected cells (25%), this loss of heterochromatin structure leads to an aggregation of major and minor satellite DNA, which probably reflects the severely disrupted nature of the pericentric heterochromatin.
At metaphase, the integrity of pericentric heterochromatin and the tight interaction between sister chromatids can be visualized by a single spot of major satellite DNA, and inside this single spot are two closely positioned foci of minor satellite DNA which are the two sister centromeres (see SI Fig. 6A). A recent study demonstrated that sister chromatid cohesion was lost between sister major satellite regions in cells lacking Suv 39h, because a double, rather than a single, major satellite spot was observed at metaphase (9). In contrast to all control chromosomes, where only a single focus of major satellite was observed, in H2A.Z RNAi-treated cells, 47% of chromosomes displayed double foci of major satellite DNA separated by 0.83 ± 0.20 μm (see SI Fig. 6A). Significantly, at anaphase/telophase, the generation of chromosome bridges is the consequence of this disrupted heterochromatin, because all bridges observed (15% of cells) are comprised of major satellite DNA (see SI Fig. 6A). Interestingly, these chromosome bridges lost HP1α but appeared to retain trimethyl K9 H3 (see SI Fig. 6C).
Based on the finding that chromosome bridges form between separating sister chromatid heterochromatic regions, it would be expected that centromeres would be located on or at the base of the chromosome bridge. Indeed, this was the case. All chromosome bridges analyzed had either CENP-A foci located at the base (67%) or on the bridge (33%) (see SI Fig. 6B). Consistent with this, minor satellite DNA had a similar location (see SI Fig. 6A). Interestingly, often double CENP-A spots were located only at one end of the chromosome bridge, suggesting that some sister chromatid centromeres did not separate properly at anaphase despite prematurely separating at metaphase (see SI Fig. 6B; compare both ends, R and L, of the same chromosome bridge). One possible explanation for this observation is that the two separating sister chromatid centromeres become trapped because of the entanglement of their heterochromatic regions. This finding supports our observation that chromosomes can segregate unequally when H2A.Z is depleted (4). Taken together, we conclude that H2A.Z is an important structural component of pericentric heterochromatin, because without it, proper heterochromatic interactions between sister chromatids are disrupted leading to the formation of chromosome bridges.
Centric Chromatin Is Composed of Regions of CENP-A Interspersed with H2A.Z/Dimethyl K4 H3-Containing Nucleosomes.
The possibility that H2A.Z is a component of centric chromatin has not been explored. To investigate this, we examined the composition of extended minor satellite centric chromatin fibers by combining DNA FISH (using a specific minor satellite DNA probe) with indirect immunofluorescence (using the same antibodies as used above). Minor satellite chromatin was extended to ≈60 μm, which ensured little overlap with major satellite DNA (see SI Fig. 7A). Significantly, this analysis shows that minor satellite chromatin is associated with H2A.Z (see SI Fig. 7A). We estimate that H2A.Z and H2A cover 21 ± 2% and 86 ± 6% of minor satellite DNA, respectively (n = 5). Like flies and humans, minor satellite DNA is associated with dimethylated K4 H3 (65 ± 20%) but with very little trimethylated K9 H3 (3 ± 2%) (n = 5). ChIP assays followed by semiquantitative PCR (18 cycles) support the linear fiber analysis (see SI Fig. 7B). In addition, real-time quantitative PCR revealed that 28 ± 4% and 71 ± 7% of H2A.Z and H2A, respectively, are associated with minor satellite DNA (data not shown).
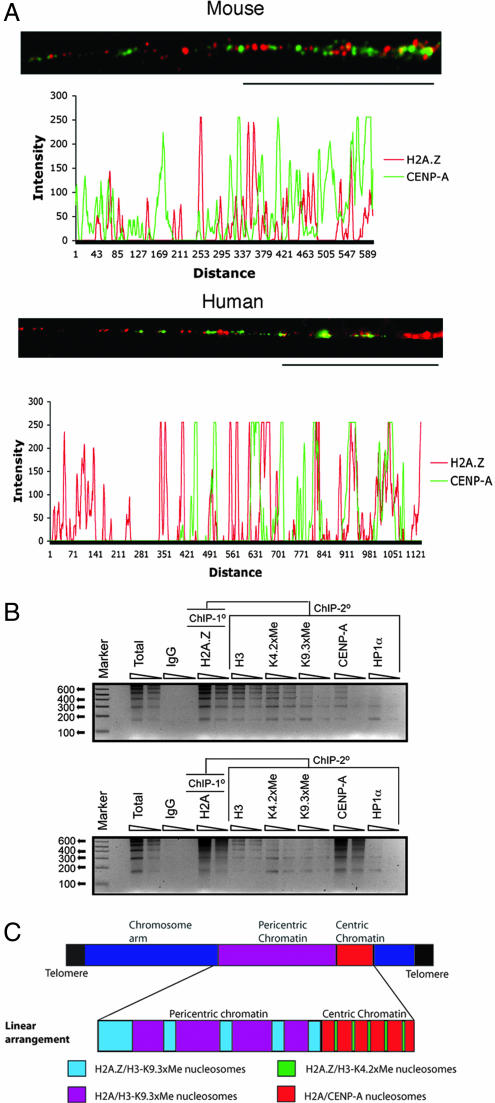
Next, we investigated whether the H2A.Z located at minor satellite DNA is coincident with blocks of CENP-A or located with the previously reported dimethyl K4 H3 interspersed subdomains (2). Analysis of linear mouse centric chromatin (≈40–50 μm) demonstrates that H2A.Z does not colocalize with CENP-A but is found between the subdomains of CENP-A (n = 5) (Fig. 2A); each block of CENP-A occupies ≈10–20 kb of DNA (assuming 1 μm ≈ 12 kb of minor satellite DNA). Such a distribution is also seen in human HEK293 cells (Fig. 2A). We believe that the occasional overlap (<5%) observed is because of incomplete unfolding of the centric fiber.
Fig. 2.
H2A.Z is a component of centric chromatin and is associated with dimethyl K4 H3. (A) Extended centric chromatin fibers from mouse L929 and human HEK 293 cells were analyzed by immunofluorescence by using H2A.Z and CENP-A antibodies. (Scale bar, 20 μm.) The intensity of fluorescence for each antibody following the linear path of each extended fiber is shown. The scales are in arbitrary units. (B) ChIP-ReChIP experiments were performed. The primary ChIP was carried out with antibodies raised against H2A.Z or H2A, and the secondary ChIP was performed with the antibodies shown. Ten and 5 ng of precipitated DNA, respectively, were analyzed by PCR by using specific primers for minor satellite DNA. (C) A schematic diagram summarizing the discontinuous linear distribution of H2A.Z throughout extended pericentric and centric chromatin.
To verify that H2A.Z and CENP-A are exclusively localized, we performed ChIP-ReChIP experiments. The primary ChIP was carried out with antibodies to H2A.Z or H2A, and the secondary ChIP was performed with antibodies, as shown in Fig. 2B. Purified ChIP DNA was analyzed by PCR using specific primers for minor satellite DNA. Consistent with our analysis of extended chromatin fibers, CENP-A nucleosomes contain H2A and essentially no H2A.Z. On the other hand, H2A.Z preferentially associates with dimethyl K4 H3 on minor satellite DNA. We conclude that H2A.Z is a component of centric chromatin and is associated with dimethyl K4 H3 and not CENP-A. This observation also shows that H2A.Z can cross epigenetic boundaries associated with both pericentric and centric chromatin (our interpretation of these results is summarized in Fig. 2C).
The 3D Organization of H2A.Z Chromatin at the Inactive X Chromosome Centromere.
Given that H2A.Z is present discontinuously along extended pericentric and centric chromatin fibers that are associated with different H3 modifications, the next issue to be addressed is how these regions of H2A.Z/trimethyl K9 H3 and H2A.Z/dimethyl K4 H3 are spatially organized to produce the specialized structure of the centromere. Of particular interest is the organization of this epigenetic information at asymmetric H2A.Z centromere structures shown in Fig. 1. To carry out this aim, we used indirect immunofluorescence and 3D deconvolution microscopy. We performed this analysis on the inactive X chromosome in human HEK293 cells, taking advantage of our finding that the highest concentration of H2A.Z observed on this chromosome is in and around the centromere (Fig. 1), thus removing any potential interference from H2A.Z located on adjacent chromosomal arms. Shown in SI Fig. 8B are the metaphase chromosome spreads and the inactive X chromosomes used in this investigation.
The 3D deconvolution analysis of the inactive X metaphase chromosome immunostained with antibodies to H2A.Z and CENP-A revealed that there is no overlap between H2A.Z and CENP-A and, most interestingly, as our 2D analysis indicated (Fig. 1), H2A.Z forms a single domain that surrounds only one side of each sister X chromosome centromere along the y axis (Fig. 3A; see also SI Movie 1). As reported, CENP-A is present in a cylinder-like structure (2). Significantly, this H2A.Z domain extends between the sister CENP-A regions in the inner centromere region (along the x axis), also supporting the results of Fig. 1.
Next, we repeated this analysis to investigate the 3D relationship between pericentric heterochromatin enriched with trimethyl K9 H3 and CENP-A (Fig. 3A). Significantly, similar to the domain of H2A.Z, trimethyl K9 H3 juxtaposes only one side of the pair of CENP-A cylinders and is also found in the inner centromere at the inactive X chromosome. To determine whether trimethyl K9 H3 and H2A.Z colocalize or are found at the opposite sides of the CENP-A domain (along the y axis), we repeated this analysis using metaphase chromosomes immunostained with H2A.Z and trimethyl K9 H3 antibodies. Significantly, the bulk of H2A.Z and trimethyl K9 H3 do not overlap, showing that H2A.Z must juxtapose one side of the pair of CENP-A cylinders while a region of trimethyl K9 H3/H2A borders the opposite side. However, H2A.Z and trimethyl K9 H3 do partially overlap at the inner centromere (≈20%; the region of overlap between H2A.Z and trimethyl K9 H3 is shown separately in yellow, and the location of this overlap in the 3D reconstruction is indicated by the yellow dashed line). A schematic diagram summarizing the location of H2A.Z and trimethyl K9 on the inactive X metaphase chromosome is shown (Fig. 3A). Note that the domain of H2A.Z is located on the short arm of the X chromosome.
We repeated this analysis, examining the spatial location of CENP-A and dimethyl K4 H3, and H2A.Z and dimethyl K4 H3. Most interestingly, unlike trimethyl K9 H3, the bulk of dimethyl K4 H3 and H2A.Z do spatially colocalize (≈60% overlap shown in yellow), being located together on the same side of the CENP-A cylinders and in the inner centromere region (summarized in Fig. 3A).
Finally, we performed 3D deconvolution analysis on metaphase autosomes using antibodies to CENP-A and H2A.Z. Despite the high abundance of H2A.Z on the chromosomal arms, it is clear that certain autosome centromeres can also display an unequal distribution of H2A.Z, with one side of the CENP-A “cylinder-like” structures being relatively depleted in H2A.Z (see SI Fig. 9 and SI Movie 2), confirming the findings of Fig. 1 (autosome 1). Taken together, we conclude that H2A.Z contributes to the unique architecture of a centromere by participating with both trimethyl K9 H3 and dimethyl K4 H3 to generate distinct spatially positioned domains.
Discussion
This investigation has provided insights into centromere structure and function by studying how the essential variant H2A.Z contributes to the unique organization of the centromere. A major finding is the identification of H2A.Z as a component of centric chromatin. The importance of this centromeric H2A.Z is illustrated by our observation that a major location of H2A.Z on the inactive X chromosome is at its centromere. Despite being less abundant than H2A, H2A.Z is also a key structural component of pericentric heterochromatin. Using the inactive X chromosome centromere as a model, we find that H2A.Z/trimethyl K9 H3 and H2A.Z/dimethyl K4 H3 occupy spatially distinct regions, thereby contributing to the unique 3D architecture of a centromere.
Our previous ChIP analysis revealed that chromatin regions of up to 20 kb in length contain H2A.Z plus trimethyl K9 H3 and HP1α and no detectable H2A (10). Based on our 2D immunofluorescence analysis (Fig. 1) and 3D reconstruction experiments (Fig. 3), we propose that H2A.Z with trimethyl K9 H3 can form a distinct domain at the inner centromere where sister chromatids pair. Because our recent biophysical analysis of in vitro reconstructed nucleosomal arrays demonstrated that H2A.Z, compared with H2A, can generate condensed chromatin secondary structures, and this level of compaction is dramatically enhanced with HP1α (10), it is attractive to propose that this condensed conformation generated by H2A.Z is important for heterochromatin integrity and sister chromatid interactions. Interestingly, the disruption to heterochromatin in response to depleting cellular H2A.Z causes the loss of HP1α binding (4), even though trimethylation of K9 H3 at chromosome bridges appears to be unaffected. This finding supports our previous proposal that the conformation of heterochromatin also plays a role in the stable binding of HP1α (10).
Our 3D deconvolution studies on the inactive X chromosome centromere allows us to propose a model for the organization of a centromere at metaphase, taking into account our discovery of previously unidentified H2A.Z at the centromere (Fig. 3B). We propose that a compacted domain of H2A.Z/dimethyl K4 H3 juxtaposes on one side of the pair of CENP-A cylinders (along the y axis), whereas a heterochromatic region of H2A/trimethyl K9 H3 borders the opposite side (with distinct regions of H2A.Z/dimethyl K4 H3 and H2A.Z/trimethyl K9 H3 in the inner centromere) on the X chromosome. These results suggest that H2A.Z may functionally substitute for trimethyl K9 H3 (5) and/or assemble a specialized compacted domain with the euchromatic dimethyl K4 H3 mark. These findings may have important future implications in understanding the role of histone posttranslational modifications, because the composition of the underlying nucleosome may have a role in determining their ultimate function. In the future, it may also be of interest to determine whether, in addition to satellite DNA sequences, the domain of H2A.Z/ dimethyl K4 H3 located next to CENP-A chromatin on the short-arm side of the inactive X chromosome is associated with unique DNA sequences (see below).
The presence of H2A.Z on one side of CENP-A chromatin observed for the inactive X chromosome is seen for some (Fig. 1 and SI Fig. 9) but not all autosomes (Fig. 1). We do not know the reason for this difference, but it is clear that centromere organization can vary in a chromosome-dependent manner in human cells, as has been reported for the location of di- and tri-K9 H3 methylation adjacent to the centromere (3). Consistent with our above hypothesis, H2A.Z may assemble dimethyl K4 H3 containing euchromatin into highly compacted chromatin structures needed for centromere function on those chromosomes, that are relatively deficient in pericentric heterochromatin. We have observed that, compared with the differentiated state, H2A.Z is expressed at a low level in different types of embryonic stem cells (4, 11), but we believe that this basal expression provides the stem cell with centromeric H2A.Z. In undifferentiated mouse ES cells, we have identified H2A.Z at the centromere on different chromosomes (data not shown). The centromeric function of H2A.Z appears to be evolutionary conserved as a genetic interaction between H2A.Z and components of the kinetochore occurs in yeast (12).
A previously unresolved issue was whether K9 H3 is trimethylated in centric chromatin (minor satellite) in mice (3). Our results show that, like flies, humans (3), and fission yeast (7), trimethylated K9 H3 and dimethylated K4 H3 are generally restricted to pericentric and centric chromatin, respectively. In conclusion, this study shows that the organization of the centromere involves the interplay of two essential histone variants, H2A.Z and CENP-A.
Materials and Methods
Inducible Expression of H2A.Z siRNA.
Mouse L929 cell lines stably expressing pIND.H2A.Z siRNA and the pVgEcR/RXR regulator vector and the induction of siRNA synthesis by ponasterone A have been described (4).
ChIP and ChIP-ReChIP Assays.
These assays, which produced chromatin fragments averaging 400–800 bp in size, were carried out according to our previous study (ref. 10; see SI Text).
Preparation of Extended Chromatin Fibers.
Extended chromatin fibers were prepared from mouse L929 cells and human HEK 293 cells and analyzed, as described (2), with the exception that the NaCl concentration was reduced to 200 mM to avoid any possibility that H2A/H2B is removed.
Cytological Preparations.
Immunofluorescence analysis of asynchronized and synchronized cells was performed as described (4, 6). Extended chromatin fibers were analyzed by immunofluorescence (3) and DNA FISH (13) (see SI Text).
Microscopy and Image Analysis.
All images were collected by using an Olympus (Melville, NY) IX81 inverted microscope by using a DP70 CCD camera along with DP70 image software (see SI Text).
Supplementary Material
Acknowledgments
We thank Michael Devoy for excellent technical assistance in growing and analyzing the different cell lines used here; Cathy Gillespie for help in the 3D deconvolution analysis; Andy Choo, Jeff Craig, and Richard Saffery for continued support of this work; and especially Paul Kalitsis for providing the anti-mouse CENPA antibodies. This work was supported by National Health and Medical Research Council grants (to D.R. and D.J.T. and to P.R. and D.J.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607870104/DC1.
References
- 1.Bernard P, Allshire R. Trends Cell Biol. 2002;12:419–424. doi: 10.1016/s0962-8924(02)02344-9. [DOI] [PubMed] [Google Scholar]
- 2.Blower MD, Sullivan BA, Karpen GH. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sullivan BA, Karpen GH. Nat Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rangasamy D, Greaves I, Tremethick DJ. Nat Struct Mol Biol. 2004;11:650–655. doi: 10.1038/nsmb786. [DOI] [PubMed] [Google Scholar]
- 5.Bulynko YA, Hsing LC, Mason RW, Tremethick DJ, Grigoryev SA. Mol Cell Biol. 2006;26:4172–4184. doi: 10.1128/MCB.00135-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rangasamy D, Berven L, Ridgway P, Tremethick DJ. EMBO J. 2003;22:1599–1607. doi: 10.1093/emboj/cdg160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jia S, Kobayashi R, Grewal SI. Nat Cell Biol. 2005;7:1007–1013. doi: 10.1038/ncb1300. [DOI] [PubMed] [Google Scholar]
- 8.Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 9.Guenatri M, Bailly D, Maison C, Almouzni G. J Cell Biol. 2004;166:493–505. doi: 10.1083/jcb.200403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan JY, Rangasamy D, Luger K, Tremethick DJ. Mol Cell. 2004;16:655–661. doi: 10.1016/j.molcel.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 11.Greaves IK, Rangasamy D, Devoy M, Marshall Graves JA, Tremethick DJ. Mol Cell Biol. 2006;26:5394–5405. doi: 10.1128/MCB.00519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krogan NJ, Baetz K, Keogh MC, Datta N, Sawa C, Kwok TC, Thompson NJ, Davey MG, Pootoolal J, Hughes TR, et al. Proc Natl Acad Sci USA. 2004;101:13513–13518. doi: 10.1073/pnas.0405753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greaves IK, Svartman M, Wakefield M, Taggart D, De Leo A, Ferguson-Smith MA, Rens W, O'Brien PC, Voullaire L, Westerman M, Graves JA. Chromosome Res. 2001;9:251–259. doi: 10.1023/a:1016656722134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.