Abstract
RIG-I is an RNA helicase containing caspase activation and recruitment domains (CARDs). RNA binding and signaling by RIG-I are implicated in pathogen recognition and triggering of IFN-α/β immune defenses that impact cell permissiveness for hepatitis C virus (HCV). Here we evaluated the processes that control RIG-I signaling. RNA binding studies and analysis of cells lacking RIG-I, or the related MDA5 protein, demonstrated that RIG-I, but not MDA5, efficiently binds to secondary structured HCV RNA to confer induction of IFN-β expression. We also found that LGP2, a helicase related to RIG-I and MDA5 but lacking CARDs and functioning as a negative regulator of host defense, binds HCV RNA. In resting cells, RIG-I is maintained as a monomer in an autoinhibited state, but during virus infection and RNA binding it undergoes a conformation shift that promotes self-association and CARD interactions with the IPS-1 adaptor protein to signal IFN regulatory factor 3- and NF-κB-responsive genes. This reaction is governed by an internal repressor domain (RD) that controls RIG-I multimerization and IPS-1 interaction. Deletion of the RIG-I RD resulted in constitutive signaling to the IFN-β promoter, whereas RD expression alone prevented signaling and increased cellular permissiveness to HCV. We identified an analogous RD within LGP2 that interacts in trans with RIG-I to ablate self-association and signaling. Thus, RIG-I is a cytoplasmic sensor of HCV and is governed by RD interactions that are shared with LGP2 as an on/off switch controlling innate defenses. Modulation of RIG-I/LGP2 interaction dynamics may have therapeutic implications for immune regulation.
Keywords: hepatitis C virus, IFN, IPS-1, MAVS, Cardif
Virus infection of mammalian cells triggers innate immune defenses through pathogen recognition receptors (PRRs) of the host that bind to pathogen-associated molecular patterns (PAMPs) within viral products and engage intracellular signaling pathways to initiate an antiviral response. Viral RNA is a potent inducer of this host response and is recognized by specific Toll-like receptors or by the cytoplasmic RNA helicases RIG-I and MDA5 (1). RIG-I and MDA5 are unique among the helicases because they contain tandem caspase activation and recruitment domains (CARDs) (2, 3). Both RIG-I and MDA5 bind to the synthetic double-stranded (ds)RNA poly inosine:cytosine (pIC), albeit with distinct efficiencies (4, 5). Studies of human cells defective in RIG-I signaling, or of cells from mice with a targeted deletion of Rig-I or Mda5, have revealed remarkable specificity of virus recognition between each helicase that could reflect their distinctions in RNA binding and PRR function. In particular, RIG-I is essential for triggering the host response to Sendai virus (SenV) and hepatitis C virus (HCV), whereas MDA5 has been shown to be important for the response to picornaviruses (5, 6). RIG-I and MDA5 initiate the host response by binding to IPS-1 (7). IPS-1, also known as Cardif, MAVS, and VISA (reviewed in ref. 8), is a CARD family protein that mediates CARD-dependent interactions with RIG-I and MDA5. These interactions stimulate IPS-1 signaling of downstream IFN regulator factor 3 (IRF-3) and NF-κB transcription factors that induce IFN-α/β production and IFN-stimulated genes that suppress virus infection (1). Control of virus signaling is mediated in part by LGP2, a related helicase that lacks CARDs but binds dsRNA (4, 9). However, the mechanisms of this regulation are not defined. Expression of RIG-I, MDA5, and LGP2 is induced by IFN, although RIG-I is basally expressed at a low level in most tissues. RIG-I is silent in the cell until it engages dsRNA, but the signaling activity of the RIG-I CARDs is unmasked upon deletion of the C-terminal helicase domain (2, 6). These observations suggest that RIG-I is initially autoregulated by its C terminus and then subjected to transregulation by LGP2 when IFN is produced during the latter stages of the host response.
Misregulation of RIG-I could be expected to deleteriously influence the antiviral host response. An example of this comes from HCV, to which the RIG-I-dependent host response limits virus replication and spread (6, 10). HCV is a single-stranded RNA virus whose genome is punctuated with dsRNA structures within the 5′ and 3′ nontranslated region (NTR) that alone can trigger RIG-I signaling and the host response in cultured cells (6). HCV overcomes this response through viral NS3/4A protease cleavage of IPS-1 to ablate RIG-I signaling of IRF-3 activation and the host response in vivo, thus providing a foundation for chronic infection in nearly 200 million people (11, 12). Processes that control RIG-I signaling may similarly impact adaptive immunity, because α/β IFNs are essential for sustaining T cell expansion and memory formation during virus infection (13). The present study was conducted to define PRR function and regulatory processes of RIG-I and MDA5 signaling and their points of control by LGP2. We identified a repressor domain (RD) of CARD signaling whose actions impact HCV infection and immunity.
Results
RIG-I and LGP2 Bind HCV RNA.
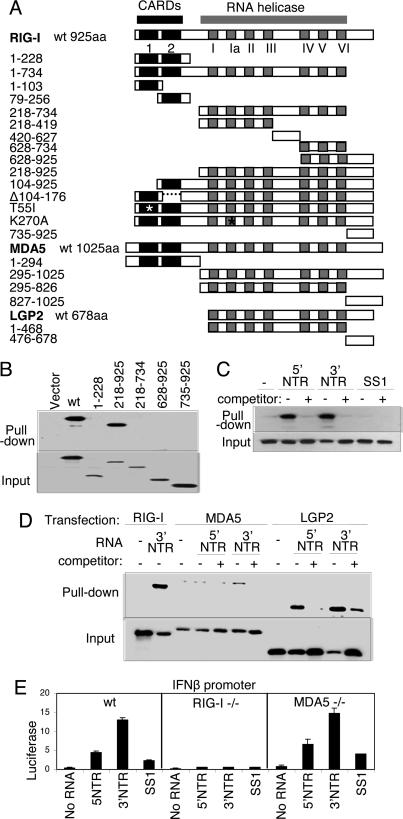
To define the functional domains of RIG-I and their relationship with homologous regions of MDA5 and LGP2, we created a panel of wild-type (WT) and mutant expression constructs, including an extensive set of RIG-I mutants (Fig. 1A). We analyzed Flag-tagged construct binding to the synthetic dsRNA, pIC, and HCV RNA by using sensitive RNA-binding/agarose bead pull-down assays to recover RIG-I from Huh7 cell extracts (6). WT RIG-I and its C-terminal region of amino acids 218–925 were recovered from cell extracts by using pIC-agarose beads (Fig. 1B). We also found that a Walker-A/ATPase-deficient mutant (RIG-I K270A) retained dsRNA binding function (data not shown). However, RIG-I constructs encoding helicase domain fragments or amino acids 735–925 alone failed to bind pIC, demonstrating that dsRNA binding requires both the RIG-I helicase domain and C terminus. RIG-I formed a specific complex with in vitro-transcribed HCV RNA encoding the highly structured 5′ or 3′ NTR but did not bind to the same length SS1 RNA (6), a defined linear, nonstructured domain of the HCV genome (14) (Fig. 1C). Similar analyses revealed efficient HCV NTR RNA binding by LGP2 and demonstrated that MDA5 binds only weakly to HCV 3′ NTR RNA (Fig. 1D). We confirmed that RIG-I and LGP2 could bind HCV genomic RNA [see supporting information (SI) Fig. 5A]. These results indicate that RIG-I binds to dsRNA or 2° structured RNA, including HCV RNA, through its helicase domain and C terminus independently of its CARDs, and binding efficiency is shared by LGP2 but not MDA5. We further evaluated the functional role of MDA5 and RIG-I in IFN-β promoter signaling by HCV RNA. When transfected into WT control mouse embryo fibroblasts (MEFs), HCV NTR RNA but not SS1 RNA triggered signaling processes that activated the IFN-β promoter. This response was absent in RIG-I-null MEFs but remained intact in MDA5-null MEFs (Fig. 1E). Similar results were obtained when MEFs were transfected with HCV genome RNA (SI Fig. 5B). These results define RIG-I as a dsRNA-binding protein and PRR for HCV RNA.
Fig. 1.
Constructs and RNA binding properties. (A) Domain structure of the RIG-I, MDA5, and LGP2 constructs showing the positions of the tandem CARDs and the RNA helicase domain and its subdomains conserved among the helicase superfamily (15). Point mutations are indicated by an asterisk. The amino acid region encoded by each construct is shown at left. (B) Extract from Huh 7 cells that were transfected with empty vector or plasmid expressing Flag-tagged RIG-I constructs encoding WT (wt) RIG-I or the indicated amino acid regions of RIG-I were mixed with pIC-agarose beads and subjected to pull-down assay for dsRNA binding. RIG-I proteins within the input material (Lower) and recovered as pull-down product (Upper) were evaluated by immunoblotting using anti-Flag antibody. (C and D) Cytoplasmic fraction (20 μg) from 293 cells transfected with plasmid encoding Flag-tagged WT RIG-I (C) or with Flag RIG-I wt, Flag-MDA5 wt, or Flag-LGP2 wt (D) were mixed with 1 μg of in vitro-transcribed biotin-UTP HCV 5′ NTR RNA, 3′ NTR RNA, or SS1 RNA alone (−) or with an excess of unlabeled homologous competitor RNA (+). RNA–protein complexes were recovered by pull-down assay using streptavidin affinity gel. Flag-tagged protein within the pull-down fraction or 25% of input material was analyzed by immunoblotting using anti-Flag antibody. (E) wt, RIG-I-null, or MDA5-null MEFs were cotransfected with plasmids encoding constitutive Renilla luciferase and firefly luciferase controlled by the murine IFN-β promoter (2). After 24 h, the cells were mock-transfected or transfected with 10 ng of in vitro-transcribed, gel-purified RNA encoding the HCV 5′ NTR, 3′ NTR, or SS1 region (6). After 24 h, the cells were harvested, and extracts were subjected to dual luciferase assay. Bars show relative luciferase values and SD.
dsRNA and ATP Analog Promote Formation of a Trypsin-Resistant 30-kDa Domain of RIG-I.
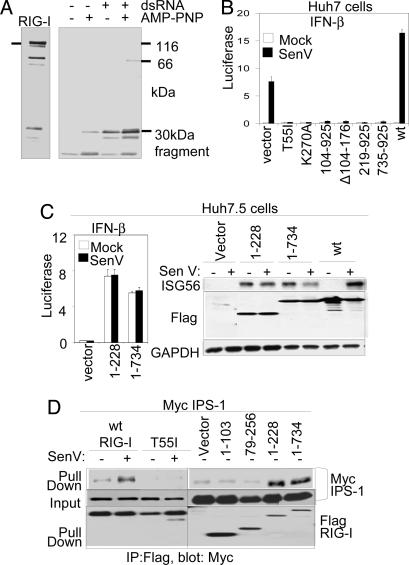
Conformational change induced by dsRNA could be a mechanism of RIG-I signaling activation. We therefore determined the trypsin sensitivity of recombinant RIG-I. Purified recombinant RIG-I produced in insect cells exhibited binding activity to 25-bp synthetic dsRNA ligand as determined by gel shift assay (SI Fig. 5C). The C-terminal portion of RIG-I, as detected by a mAb whose epitope mapped to amino acids 477–925, was highly sensitive to trypsin digestion in the absence of dsRNA ligand. However, the association of RIG-I with dsRNA and 5′-adenylyl-imidodiphosphate (AMP-PNP), a nonhydrolyzable ATP analog, conferred limited resistance to trypsin digestion and resulted in the generation of a trypsin-resistant 30-kDa polypeptide (Fig. 2A). Thus, upon binding dsRNA ligand, and in the presence of bound nucleotide, RIG-I may undergo a conformational change that displaces its C-terminal region. Because dsRNA triggers RIG-I signaling, C-terminal conformation changes induced by dsRNA could presumably confer RIG-I signaling activation during virus infection.
Fig. 2.
An RD controls RIG-I signaling. (A) Recombinant RIG-I protein was digested with trypsin in the presence or absence of dsRNA and AMP-PNP as indicated. After termination of the reaction, the mixtures were analyzed by immunoblotting using anti-RIG-I mAb, which reacts with the C-terminal portion of RIG-I. Bars indicate the undigested 115-kDa RIG-I input protein (Left) and a 30-kDa protected digestion fragment (Right). (B) Huh 7 cells were cotransfected with luciferase promoter constructs and the indicated RIG-I expression constructs. After 24 h, the cells were mock-infected or infected with SenV. Cells were harvested 20 h later for dual luciferase assay. Bars show the mean relative IFN-β promoter–luciferase levels (± SD). (C) Huh 7.5 cells were cotransfected with promoter luciferase plasmids and empty vector or the indicated RIG-I expression plasmids. Cells were mock- or SenV-infected as indicated and were processed as above for dual luciferase assay (Left) or were harvested and analyzed by immunoblot assay for ISG56, Flag-RIG-I construct (Flag), and GAPDH abundance as shown (Right). (D) Huh7.5 cells were cotransfected with plasmids encoding Myc-IPS-1 and vector control or the indicated RIG-I constructs. After 16 h, the cells were mock-infected or infected with SenV as shown, cultured for 16 h, and harvested for anti-Flag immunoprecipitation (IP) and immunoblot assays as described in ref. 10. Shown are Flag-RIG-I protein abundance in the input material (Bottom) and Myc-IPS within the IP product (Top) or input material (Middle).
RIG-I Signaling and IPS-1 Interaction Require Tandem CARDs and Are Controlled by a C-Terminal RD.
To determine the RIG-I requirements for host response signaling, we measured luciferase levels driven by the IFN-β promoter (Fig. 2B) or by the NF-κB-responsive PRDII promoter element (11, 12) during SenV infection of cells expressing RIG-I constructs (see SI Fig. 6). Ectopic WT RIG-I enhanced SenV-induced promoter activation, and RIG-I signaling required both intact CARDs in tandem (Fig. 2B and SI Fig. 6 A and B). Constructs encoding only the first or second CARD failed to signal IFN-β promoter induction (SI Fig. 6B) but lacked dominant-negative activity. The latter was a property of constructs lacking either CARD alone in the context of the full-length protein. Moreover, the RIG-I T55I and K270A point mutations respectively disrupt CARD function and ATP binding/ATPase activity (2, 6), and they each exhibit dominant-negative control of RIG-I signaling (Fig. 2B and SI Fig. 6A). RIG-I amino acids 735–925 were also dominant-negative for signaling when expressed in Huh7 (Fig. 2B) or 293 cells (SI Fig. 6A). When expressed in Huh7.5 cells, which harbor a dominant-negative mutant RIG-I allele that prevents signaling by RIG-I (6), RIG-I 1–735 alone conferred constitutive promoter signaling and expression of endogenous ISG56, in a manner similar to ectopic expression of RIG-I 1–228 encoding the tandem CARDs alone (Fig. 2C). We conclude that RIG-I mutants devoid of signaling activity but that encode an intact C terminus exhibit dominant-negative function, whereas deletion of the C terminus (amino acids 735–925) renders RIG-I constitutively active.
We conducted coimmunoprecipitation analyses to define the control points and requirements for RIG-I/IPS-1 binding. Importantly, we found that complex formation was induced upon virus infection (Fig. 2D). Although WT RIG-I formed a virus-induced complex with IPS-1, the interaction depended on the tandem RIG-I CARDs and was disrupted by the RIG-I T55I mutation. RIG-I constructs encoding amino acids 1–228 or 1–734, respectively lacking the entire helicase domain and C terminus or lacking the C terminus only (see Fig. 1A), formed a constitutive complex with IPS-1 independently of virus infection, consistent with the signaling properties of each RIG-I mutant (compare Fig. 2 C and D). Thus, both RIG-I CARDs are required for downstream signaling and interactions with IPS-1 through a process that is negatively regulated by the RIG-I C terminus. Taken together, these results define RIG-I amino acids 735–925 as an RD that controls innate defense signaling, possibly by governing RIG-I interactions.
RIG-I Signals as a Multimeric Complex Regulated by RD Interactions.
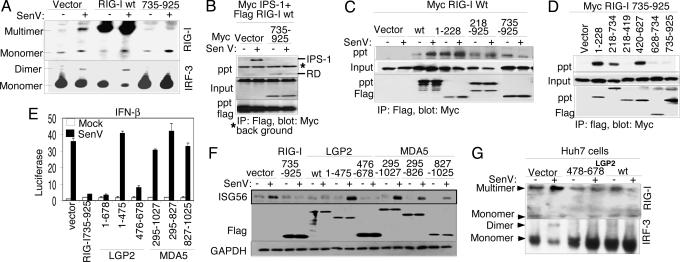
To determine the mechanisms of RD regulation of RIG-I signaling, we examined RIG-I complex formation and the influence of the RD in this process. Multimerization is a hallmark of CARD proteins (1). We therefore conducted coupled native PAGE/immunoblot analyses to determine whether RIG-I multimerizes as it signals downstream IRF-3 activation during virus infection. SenV infection of Huh7 cells induced endogenous RIG-I complex formation and concomitant dimerization of IRF-3 (Fig. 3A). RIG-I constitutively formed a complex when overexpressed but signaled IRF-3 activation and dimerization only after SenV infection. Ectopic expression of the RIG-I RD alone prevented complex formation of endogenous WT RIG-I and concomitantly blocked SenV-induced IRF-3 dimerization. Moreover, we found that when expressed alone, the RD was sufficient to prevent SenV-induced RIG-I interaction with IPS-1 in coimmunoprecipitation assays (Fig. 3B).
Fig. 3.
Mechanism of regulation by the RIG-I RD. Where indicated, cells were mock-infected or infected with SenV for 16 h before harvesting. (A) Stable Huh7 cell lines expressing vector alone, RIG-I wt, or RIG-I 735–925 were mock-infected or infected with SenV, and protein extracts were subjected to native PAGE and immunoblot analysis with anti-RIG-I antibody (Upper) or anti-IRF-3 antibody (Lower). Dimer/multimer and monomer protein forms are indicated. (B–D) Huh7 cells were cotransfected with plasmids encoding Myc-IPS-1, Flag-RIG-I wt, and vector or Myc-RIG-I 735–925 (B); Myc-RIG-I wt and the indicated Flag construct (C); or Myc-RIG-I 735–925 and the indicated Flag construct (D). Cells were infected as shown and harvested, and extracts were analyzed by immunoprecipitation (IP) and immunoblot assays. Shown are the abundance of Myc-tagged protein within anti-Flag IP products (Top), input Myc-tagged protein (Middle), and input Flag-tagged protein (Bottom). (E and F) Huh 7 cells were transfected with Renilla luciferase, IFN-β-luciferase plasmids, and plasmids encoding vector alone or the indicated Flag-tagged RIG-I, LGP2, or MDA5 constructs. After SenV infection, the cells were harvested and extracts were subjected to dual luciferase assay (E) (bars show relative luciferase and SD) and to immunoblot assay for abundance of ISG56, Flag-tagged protein (Flag), and GAPDH (F). (G) Anti-RIG-I (Upper) or anti-IRF-3 (Lower) immunoblot of Huh7 cell extracts separated by native PAGE. Protein monomer and multimer/dimer forms are indicated. Cells were transfected with vector control or expression plasmid encoding Flag-LGP2 478–378 or Flag-LGP2 wt. Extracts were prepared after SenV or mock infection.
We further assessed complex formation between epitope-tagged WT and mutant RIG-I constructs. As shown in Fig. 3C, WT Flag-RIG-I was recovered as a complex with WT Myc-RIG-I that was stimulated by SenV infection, whereas Flag-RIG-I 1–228 formed a stable complex with WT Myc-RIG-I irrespective of virus infection. In further studies we found that RIG-I 1–228 does not self-associate (SI Fig. 6CLeft) and that expression of RIG-I 1–228 induced the IFN-β promoter in WT MEFs but failed to induce promoter activity in RIG-I null MEFs (SI Fig. 6CRight), suggesting that RIG-I 1–228 constitutive signaling activity is mediated by heterocomplex with WT RIG-I independently of self CARD-CARD association. The dominant-negative Flag-RIG-I constructs encoding the helicase domain and C terminus (amino acids 218–925) or the RD alone (amino acids 735–925) each formed a constitutive complex with WT Myc-RIG-I (Fig. 3C), suggesting that these constructs prevent signaling by forming an RD trans inhibitory complex with WT RIG-I. In support of this, coimmunoprecipitation experiments demonstrated RD interactions with the RIG-I amino acids 1–228 encoding the tandem CARDs, and also demonstrated interaction with the helicase domain that mapped to within amino acids 420–627 corresponding to the linker region between helicase subdomains III and IV (15) (Figs. 3D and 1A). The expression of the RD was inhibitory for signaling by WT RIG-I but not by the tandem RIG-I CARDs alone (SI Fig. 6D). Taken together, these results indicate that (i) RIG-I signals as a self-complex whose actions are directed by tandem CARDS and induced by virus infection, and (ii) RIG-I signaling is negatively regulated through internal RD interactions that control self-association and interaction with IPS-1.
RD Function Is a Common Feature of RIG-I and LGP2 but Not MDA5.
RIG-I shares a level of sequence similarity with MDA5 and LGP2 throughout their respective C-terminal regions and has been shown to block RIG-I signaling (4). We therefore assessed signaling regulation to the IFN-β promoter (Fig. 3E) and endogenous ISG56 expression (Fig. 3F) by WT or truncation mutants of RIG-I, LGP2, and MDA5. LGP2 prevented SenV induction of the IFN-β promoter and ISG56 expression in a manner similar to the RIG-I RD, and this was attributed to LGP2 C-terminal amino acids 476–678 independently of its helicase domain. In contrast, ectopic WT MDA5 expression constitutively induced IFN-β promoter activity irrespective of its C-terminal region or SenV infection (SI Fig. 6E). Truncation constructs of MDA5 lacking the CARDs (amino acids 322–1027) or encoding amino acids 827–1025, analogous to the RIG-I RD, were inert and neither stimulated nor blocked SenV induction of the IFN-β promoter or ISG56 expression (Fig. 3 E and F). Coimmunoprecipitation experiments demonstrated that WT LGP2 or LGP2 476–678 could form a stable complex with WT RIG-I or WT MDA5 when overexpressed in Huh7 cells (SI Fig. 6F). However, when coexpressed in Huh7. 5 cells neither WT LGP2 (see SI Fig. 7) nor the LGP2 RD (data not shown) could block constitutive signaling mediated by RIG-I 1–228 or MDA5. Together, these results suggest that (i) LGP2 controls signaling by RIG-I but not MDA5 through interactions between the RIG-I helicase domain and an RD encoded by amino acids 476–678 of LGP2, (ii) the analogous region of MDA5 does not harbor RD function, and (iii) the LGP2 interaction does not block signaling by MDA5. When expressed alone in Huh7 cells, the LGP2 RD functioned in a manner similar to WT LGP2 to block both SenV induction of endogenous RIG-I self-association and subsequent IRF-3 dimer formation (Fig. 3G)
RIG-I Signaling and Cell Permissiveness for HCV Are Governed by the RD.
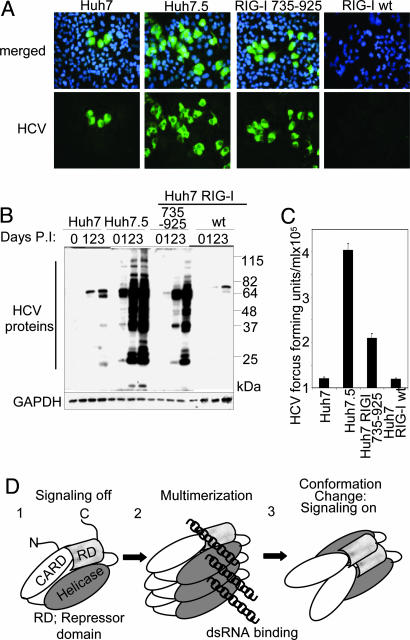
To determine the functional role of the RIG-I RD in regulating PRR signaling and host control of virus infection, we analyzed a set of Huh7 cell lines that constitutively express the RIG-I RD (Huh7-RIG-I-735–925) or WT RIG-I (Huh7-RIG-I-wt). We compared their host response and virus infection phenotype with Huh7 control cells and with Huh7.5 cells. The latter are highly permissive to HCV (6, 10, 16). SenV infection triggered the IFN-β promoter and ISG56 expression in Huh7 cells and Huh7-RIG-I-wt cells but stimulated neither IFN-β nor ISG56 expression in Huh7.5 cells or Huh7-RIG-I-735–925 cells (see SI Fig. 8). We also assessed the relative permissiveness of each cell line to HCV infection, with the JFH1 strain genotype 2a HCV infectious clone (17). HCV infection triggers the host response through a RIG-I-dependent process that limits initial permissiveness of Huh7 cells for infection (6, 10). Analysis of infected cultures demonstrated marked differences in infected cell numbers, viral protein abundance, and infectious virus production. Huh7 cells were less permissive for HCV infection than Huh7.5 cells. When compared with Huh7 control cells, expression of the RD in Huh7-RIG-I-735–925 cells conferred enhanced permissiveness for infection, increased viral protein abundance, and higher levels of infectious virus production (Fig. 4 A–C, respectively). In contrast, Huh7-RIG-I-wt cells exhibited reduced HCV permissiveness and virus production, consistent with RIG-I enhancement of host defenses. We note that Huh7.5 cells overall showed the highest permissiveness to HCV infection, implying that additional features act with the RIG-I deficiency to effect high permissiveness for HCV. We conclude that RD control of the RIG-I pathway and host defense signaling is an important determinant of cellular permissiveness to HCV infection.
Fig. 4.
The RD regulates cell permissiveness to HCV. Huh7, Huh7.5, Huh RIG-I wt, or Huh7-RIG-I-735–925 cells were infected with JFH-1 HCV 2A at a multiplicity of infection of 0.5. (A) After 48 h, cells were immunostained with HCV 2A antiserum (green). Nuclei were visualized by staining the cells with DAPI (blue). (B) Anti-HCV (Upper) and GAPDH immunoblot (Lower) of extracts from mock-infected cells (0) or from cells infected with JFH1 for 1, 2, or 3 days as indicated. The positions of HCV proteins are shown and have been defined previously (10). (C) Titer of HCV in supernatants collected from the indicated cell cultures 48 h after infection. (D) Model of RIG-I autoregulation and activation by virus infection. The RIG-I CARDs and domains encoding the helicase region and the RD are indicated.
Discussion
In this study, structure–function analyses of RIG-I revealed dsRNA PAMP binding and an induced conformation shift that associates with RIG-I PRR signaling actions. We found that RIG-I signaling requires the full tandem CARD arrangement, that it likely signals as a multimeric complex, and that an RD in RIG-I and LGP2 tightly regulates CARD signaling actions. Native gel analyses showed that, although RIG-I resides as a monomer in resting cells, virus infection or high levels of expression promote its self-association. Thus, RIG-I signals through its tandem CARDs as a multimeric complex. Further support for this idea comes from two sets of observations. First, deletion of either CARD alone ablated RIG-I signaling actions. Second, the RIG-I T55I mutant formed a complex with WT RIG-I that ablated IPS-1 binding and downstream signaling (see Fig. 2 and SI Fig. 6). RIG-I constructs lacking either CARD were also dominant-negative for signaling. Together, this indicates that RIG-I signals at least as a dimeric unit in which residue T55, located within the first CARD, might participate in IPS-1 interaction. In addition, we found that MDA5 also forms a homocomplex concomitant with signaling action when expressed in Huh7 cells (T.S. and M.G., unpublished observations). Thus, multimerization is a common feature for RIG-I and MDA5 signaling.
RIG-I has been shown to discriminate between RNA substrates wherein RNA containing a free 5′ triphosphate end, including purified influenza virus RNA, is selectively bound over RNA lacking free 5′ phosphates (18, 19). RNA transcribed in vitro, including the HCV RNA used in our study, contains 5′ triphosphate. It is notable that the HCV ss1 RNA is nonstructured (14) and did not bind efficiently to RIG-I, whereas 5′ or 3′ HCV NTR RNA did bind. Because the latter contains extensive secondary structure (14), it is likely that RIG-I discriminates self from nonself RNA by a combination of 5′ triphosphate and dsRNA motif recognition. We found that the RIG-I helicase domain and RD together form the functional unit that is sufficient to bind RNA, and that RIG-I and LGP2 bind HCV RNA. Relevance to this is supported by crystal structure analyses of DExD/H-box RNA helicase family members, in which interdependent interactions between helicase motifs I, II, and VI are necessary for ATP binding and hydrolysis, whereas motifs Ia and IV participate in oligonucleotide substrate binding (15). Thus, during virus infection, binding of dsRNA ligand could be expected to impose conformation changes that reposition the RD. This idea is supported by our trypsin digestion studies that identified a protease-resistant conformation change of the C terminus when RIG-I was bound to dsRNA and AMP-PNP. Our data indicate that the RD alone is a functional domain that, when expressed in trans, can mediate additional interdependent interactions with the RIG-I CARDs and the linker region between helicase motifs III and IV (amino acids 420–627) in which the latter interaction is essential for signaling inhibition. These results imply that the RD autoregulates RIG-I through internal interactions that control self-association.
Our results support a model of RIG-I autoregulation and signaling (Fig. 4D). This model predicts that, in resting cells, the RD mediates conformational constraints that, through internal interactions, maintain RIG-I in a orientation that masks the CARDs from signaling. During virus infection, RIG-I activation and signaling will then occur in steps involving dsRNA binding and conformation changes that stimulate self-association and that possibly involve ATP hydrolysis to displace the RD and unmask the CARDs for signaling through IPS-1 interaction. During the IFN response, increased levels of RIG-I may further promote its self-association and potentiate signaling to drive an IFN amplification loop. This model reflects the observed complex between the RD with RIG-I amino acids 1–228 encoding the tandem CARDs and is consistent with autoregulation of other CARD proteins, including Apaf-1 and NOD1. In each case, autoregulation by the C-terminal WD-40 and leucine-rich region (LRR) controls CARD signaling of caspase 9 by Apaf-1 and NF-κB activation by NOD1, respectively. Thus, the WD-40 or LRR function as the RD equivalent to control self-association and signaling, and deletion of each renders a constitutively active molecule (20, 21). This model reveals a common theme in CARD protein regulation, in which protein function is regulated by ligand binding-induced conformation changes that alter the RD or C-terminal motif to control oligmerization and signaling.
We also identified an RD within the C terminus of LGP2. The LGP2 RD was necessary and sufficient for inhibition of RIG-I, but not MDA5, signaling despite being able to form a complex with either protein. Thus, LGP2 complex formation with MDA5 is not sufficient for signaling inhibition, possibly reflecting a unique cofactor or sequence differences of RIG-I and MDA5 control. We note that the MDA5 C-terminal region does not function as an RD. This lack of RD function and inhibitory control by LGP2 allows MDA5 to signal constitutively when expressed in abundance and could reflect an important role for MDA5 in amplifying IFN production and in the host response. We found that LGP2 blocked signaling by WT RIG-I but not RIG-I 1–228 in Huh7 cells, and the former occurred concomitant with disruption of WT RIG-I complex formation. In other work, LGP2 blocked signaling by RIG-I 1–228 when expressed in HEK 293 cells (22). This discrepancy could reflect cell-specific differences in the RIG-I pathways or disparate assay conditions among studies. Our results indicate that LGP2 inhibits RIG-I through RD interactions that block RIG-I self-association, possibly by disrupting homotypic CARD/helicase domain and/or C terminus interactions. We also found that LGP2 binds to dsRNA and HCV RNA, thus implicating HCV dsRNA motifs as PAMP ligands of RIG-I and LGP2. Our studies indicate that RIG-I signaling inhibition is mediated directly by the LGP2 RD and possibly indirectly through sequestration of RNA substrates (2, 23). Moreover, LGP2 has been shown to block RIG-I signaling by disrupting assembly of a signaling complex on IPS-1 (22). Thus, LGP2 may influence the RIG-I pathway at multiple levels. As an IFN-stimulated gene, LGP2 expression is induced as a result of RIG-I signaling, and inhibition of RIG-I by LGP2 may provide a mechanism of feedback control to overall limit host response toxicity.
Huh7 cells expressing the RIG-I RD alone failed to induce the IFN-β promoter upon SenV infection, and they exhibited enhanced permissiveness for HCV. Our results affirm the role of RIG-I as a PRR of HCV and define the RD as a key modulator of host defenses that control HCV infection and production. Viral and therapeutic regulation of RD function could have implications for the modulation of immunity by directly regulating RIG-I signaling actions and IFN defenses.
Materials and Methods
Cell Culture and Viruses.
Huh7 cells, Huh7.5 cells, and MEFs from WT, RIG-I-null, or MDA5-null cells, and their culture methods, have been described in refs. 6, 16, 24, and 25. Stable Huh7 cell lines harboring vector alone (Huh7v), or expressing the RIG-I RD (Huh7-RIG-I-735–925) or WT RIG-I (Huh7-RIG-I-wt), were produced by transfecting Huh7 cells with pCDNA3.1-hygro (Invitrogen, Carlsbad, CA) alone or in combination with pEFTak encoding RIG-I 735–925 or WT RIG-I, respectively. Cell clones were selected for resistance to hygromycine. Sendai virus (SenV; Cantell strain) was obtained from Charles River Laboratories (Wilmington, MA). HCV was produced from the RNA made from the pJFH-1 HCV 2a infectious clone, exactly as described in ref. 10. Virus infections and titrations were conducted and quantified as described in ref. 10.
Plasmids, Transfection, and Protein Analysis.
For protein expression, we prepared modified pEF and pcDNA3.1 expression vectors (Invitrogen) pEFTak-Flag and pcDNA3.1-Myc, encoding amino-terminal tandem Flag or Myc epitopes, respectively. cDNA encoding the complete ORF of RIG-I, MDA5, or LGP2 was isolated by PCR from total cellular RNA and cloned into the pEFTak-Flag or pcDNA3.1-Myc by using standard methods. Mutant RIG-I, MDA5, or LGP2 expression constructs were prepared by using a PCR strategy or the Quickchange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Primer sequences are available upon request. pIFN-β-luc, pCMV-Renilla-luc, and pPRDII-luc have been described previously (11, 12). Transfection, promoter-luciferase assay, immunoblot assay, and immunostaining and microscopy were conducted exactly as described in ref. 11.
Recombinant RIG-I was produced as a GST-RIG-I fusion by using baculovirus and High Five cells. GST-RIG-I was bound to glutathione Sepharose (Amersham Pharmacia Biosciences, Piscataway, NJ), then RIG-I was eluted by thrombin digestion and excess thrombin was eluted by passing through a benzamidine Sepharose column. RIG-I was then purified by Q Sepharose chromatography.
RNA Methods.
RNA was synthesized from plasmids by using the T7 Megascript kit (Ambion, Austin, TX) in accordance with the manufacturer's protocol. JFH1 RNA, and HCV 5′ and 3′ NTR RNA, were produced from PCR products, made from the pJFH1 clone or HCV-N cDNA, respectively (26), by using a 5′ primer containing the T7 promoter. HCV ss1 RNA was transcribed from pCDNA3.1 HCV 1b NS3/4A (11). Biotinylated HCV RNA was transcribed and purified by using the AmpliScribe Flash transcription kit (Epicentre, Madison, WI) and Biotin-16-uridine-5′-triphosphate (Roche Diagnostics, Indianapolis, IN). For RNA-binding assay, biotinylated RNAs (1 μg) were incubated for 1 h at 25°C with 10 μg of protein from the cytoplasmic fraction of cells that were transfected with pEFTak-Flag expressing RIG-I, MDA5, or LGP2. The mixture was transferred into 400 μl of dialysis buffer containing 25 μl of streptavidin agarose affinity gel (Sigma, St. Louis, MO), rocked at 4°C for 2 h, collected by centrifugation, washed three times, resuspended in SDS sample buffer, incubated in a boiling bath for 5 min, and analyzed by SDS/PAGE and immunoblotting. For pIC, agarose pull-down assays were conducted as described in ref. 6.
Other Materials and Methods.
Antibodies and additional molecular biology reagents are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank G. Sen (Cleveland Clinic Foundation, Cleveland, OH), M. David (University of California at San Diego, La Jolla, CA), T. Wakita (National Institutes of Infectious Diseases, Tokyo, Japan), and L. Gitlin and M. Colona (Washington University, St. Louis, MO) for reagents. This work was supported by National Institutes of Health Grants R01AI060389 and U19AI040035 Project 4, the Burroughs–Wellcome Fund, and a gift from Mr. and Mrs. R. Batcheldor (to M.G.).
Abbreviations
- CARD
caspase activation and recruitment domain
- HCV
hepatitis C virus
- IRF-3
IFN regulator factor 3
- MEFs
mouse embryo fibroblasts
- NTR
nontranslated region
- PAMP
pathogen-associated molecular pattern
- pIC
poly inosine:cytosine
- PRR
pathogen recognition receptor
- RD
repressor domain
- SenV
Sendai virus.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0606699104/DC1.
References
- 1.Meylan E, Tschopp J, Karin M. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 2.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 3.Kang DC, Gopalkrishnan RV, Wu Q, Jankowsky E, Pyle AM, Fisher PB. Proc Natl Acad Sci USA. 2002;99:637–642. doi: 10.1073/pnas.022637199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr, Akira S, et al. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 5.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 6.Sumpter R, Loo Y-M, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M., Jr J Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 8.Johnson CL, Gale M., Jr Trends Immunol. 2006;27:1–4. doi: 10.1016/j.it.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Cui Y, Li M, Walton KD, Sun K, Hanover JA, Furth PA, Hennighausen L. Genomics. 2001;78:129–134. doi: 10.1006/geno.2001.6661. [DOI] [PubMed] [Google Scholar]
- 10.Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida H, Yoneyama M, et al. Proc Natl Acad Sci USA. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foy E, Li K, Wang C, Sumpter R, Ikeda M, Lemon SM, Gale M., Jr Science. 2003;300:1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- 12.Foy E, Li K, Sumpter R, Jr, Loo YM, Johnson CL, Wang C, Fish PM, Yoneyama M, Fujita T, Lemon SM, Gale M., Jr Proc Natl Acad Sci USA. 2005;102:2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolumam GA, Thomas S, Thompson LJ, Sprent J, Murali-Krishna K. J Exp Med. 2005;202:637–650. doi: 10.1084/jem.20050821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simmonds P, Tuplin A, Evans DJ. RNA. 2004;10:1337–1351. doi: 10.1261/rna.7640104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caruthers JM, McKay DB. Curr Opin Struct Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 16.Blight KJ, McKeating JA, Rice CM. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, et al. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 19.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 20.Inohara N, Koseki T, del Peso L, Hu Y, Yee C, Chen S, Carrio R, Merino J, Liu D, Ni J, Nunez G. J Biol Chem. 1999;274:14560–14567. doi: 10.1074/jbc.274.21.14560. [DOI] [PubMed] [Google Scholar]
- 21.Hu Y, Ding L, Spencer DM, Nunez G. J Biol Chem. 1998;273:33489–33494. doi: 10.1074/jbc.273.50.33489. [DOI] [PubMed] [Google Scholar]
- 22.Komuro A, Horvath CM. J Virol. 2006;80:12332–12342. doi: 10.1128/JVI.01325-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, Yamamoto M, Akira S, Fitzgerald KA. J Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 24.Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 25.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Proc Natl Acad Sci USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beard MR, Abell G, Honda M, Carroll A, Gartland M, Clarke B, Suzuki K, Lanford R, Sangar DV, Lemon SM. Hepatology. 1999;30:316–324. doi: 10.1002/hep.510300137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.