Abstract
Potassium channels share a common selectivity filter that determines the conduction characteristics of the pore. Diversity in K+ channels is given by how they are gated open. TASK-2, TALK-1, and TALK-2 are two-pore region (2P) KCNK K+ channels gated open by extracellular alkalinization. We have explored the mechanism for this alkalinization-dependent gating using molecular simulation and site-directed mutagenesis followed by functional assay. We show that the side chain of a single arginine residue (R224) near the pore senses pH in TASK-2 with an unusual pKa of 8.0, a shift likely due to its hydrophobic environment. R224 would block the channel through an electrostatic effect on the pore, a situation relieved by its deprotonation by alkalinization. A lysine residue in TALK-2 fulfills the same role but with a largely unchanged pKa, which correlates with an environment that stabilizes its positive charge. In addition to suggesting unified alkaline pH-gating mechanisms within the TALK subfamily of channels, our results illustrate in a physiological context the principle that hydrophobic environment can drastically modulate the pKa of charged amino acids within a protein.
Keywords: KCNK channels, molecular simulation, TALK-2, TASK-2
All K+ channels contain a highly conserved sequence, the P domain, which forms the selectivity filter and generally six transmembrane α-helices. The K+ channel pore is formed by four identical subunits, each comprising a P-domain and two of the six transmembrane α-helices encircling the ion conduction pathway with a 4-fold symmetry. Structures attached to the pore-forming domains are able to transduce signals, such as changes in transmembrane voltage and intra- or extracellular messages, into gating of the pore (1, 2). Potassium channels of the KCNK superfamily (3, 4) are remarkable in that they possess two P-domains and four α-helices in each subunit, form dimers, and are mostly open (“leak channels”) at resting potential. Potassium-selective leaks are fundamental to the function of various cells including nerve, muscle, and epithelia. There are 16 mammalian members to the KCNK family and their gating is variously regulated by free fatty acids, membrane tension, G protein-generated signaling, and extracellular pH. Among KCNK channels gated by extracellular pH, TASK-1 and TASK-3 form a subfamily (TASK) of channels blocked by extracellular protons (5–8). A second subfamily (TALK) of KCNK channels comprises TASK-2, TALK-1, and TALK-2¶, all activated by extracellular alkalinization. TASK-2 participates in ion fluxes necessary for cell volume regulation (10, 11), and its physiological and possible pathological importance has also been highlighted by studies in a TASK-2 knockout mouse (12) that revealed a metabolic acidosis and hypotension caused by renal loss of HCO−3. The transport of HCO−3 in the proximal tubule couples to TASK-2 activity through extracellular alkalinization, and inactivation of TASK-2 leads to a situation reminiscent of the clinical manifestation of human proximal renal tubular acidosis syndrome. TALK-1 and -2 are also activated by extracellular alkalinization and are highly expressed in the pancreas (13). Their function has not been elucidated, but they might play a role in pancreatic HCO−3 secretion.
K+ channels of the TASK subfamily are blocked by protons by titration of a histidine (N in TALK channels) residue in the first P domain (6–8), making them responsive to pH in the physiological range. The pH-sensing mechanism in the TALK subfamily is unknown. A multiple mutation in the extracellular loop neutralizing five amino acids was recently proposed as possible pH sensor in TASK-2 (14). In our hands, this mutation did not abolish the gating of the channel by extracellular pH (15), and in our opinion, the identification of the pH sensor in TASK-2 remains an unresolved problem. Rather than exploring by mutation the effect of neutralization of all putative external titratable residues to discover the pH-sensing mechanism in TASK-2, we modeled its pore using the recently published structure of Kv1.2 (16) as a template. The idea was to identify residues that might affect the electrostatic potential in the pore, its occupancy, and therefore channel gating (15). Examination of the model uncovered a hydrophobic environment around an arginine residue at the outermost portion of helix TM4 and close to the pore region. Neutralization of this arginine residue by mutation abolishes the pH dependence of TASK-2, whereas its replacement by other basic amino acids shifted its pH sensitivity in a way expected from the relative pKa values of their side chains. We propose that the single arginine residue near the pore of TASK-2 acts as a pH sensor, a function for which it has to lose its proton with a pKa of 8.0. We propose that this unusually large shift in arginine pKa is given by its hydrophobic environment. Our results also suggest that this sensing mechanism is common to the TALK subfamily of channels.
Results
We have modeled the structure of the pore of TASK-2 to try and obtain some insight into the mechanism of extracellular pH sensing. TASK-2 was assumed to assemble as a homodimer (17) with each monomer contributing two pore domains to the selectivity filter (7). Examination of the model thus derived revealed an interesting environment around an arginine residue at the outermost portion of helix TM4 and close to the pore region (R224, Fig. 1A). R224 side chain appears in a hydrophobic environment given mainly by an Ile residue (I90) and only forming a hydrogen bond interaction with a helix P1 asparagine (N87). N103, which replaces a more usual aspartate in the P1 region of the selectivity filter, is also in the vicinity. To test, at least in principle, the effect of R224 neutralization in TASK-2, the electrostatic potential profile along the pore main axis was calculated. The difference in electrostatic potential profile between the neutral minus the protonated R224 along the main axis of the permeation pathway is shown in Fig. 1B. A marked decrease in electrostatic potential, as expected, is obtained with a neutral R224. The difference reaches a minimum (−125 mV) at the position of the S2 cation binding site in the selectivity filter.
Fig. 1.
Identification of a conserved basic amino acid as extracellular pH sensor in TASK-2. (A) Model for TASK-2 based on the structure of Kv1.2 K+ channel (16). The model was generated as a homodimer, and a single monomer is shown for clarity. R224 is predicted to project toward the selectivity filter region. The positions of N87, N103, and I90 are also shown. Helices are identified, and the permeation path is shown by K+ ions at cation binding sites S1, S2, and the internal cavity. (B) Difference in the contribution to electrostatic potential (φE) calculated for a neutral minus a protonated R224 along the permeation pathway of the channel. Positions along the pore are indicated above the graph. (C) Sequence alignments for the pore regions of channel members of the TALK subfamily. The R224 sensor is highlighted red, as are basic amino acid conserved in TALK-1 and -2. Selectivity filters are also shown in red. Green identifies residues predicted to provide the environment for R224 and K242 in TASK-2 and TALK-2, respectively.
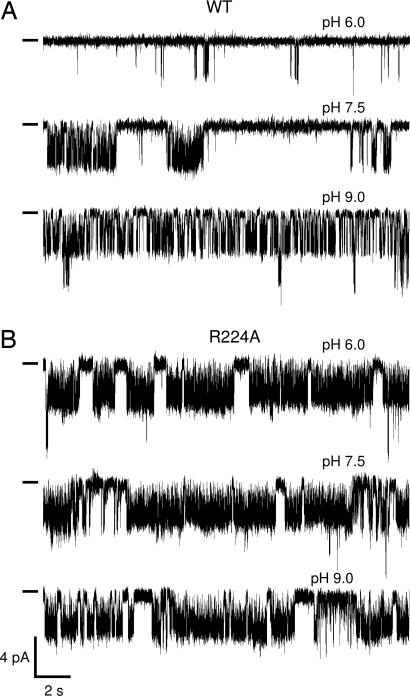
To test the hypothesis that R224 could act as the pH sensor of TASK-2, we replaced it with the nonprotonatable residues alanine or glutamine and tested for the pH dependence of the mutant channels. Mutation did not prevent expression of the channels, which presented normal time dependence for current development, selectivity of K+ over Na+, and open channel rectification (Fig. 2A–H). Testing the effect of extracellular pH on the currents, however, revealed that although alkalinization activated WT channels with a pK1/2 of 8.03 ± 0.11 and a Hill coefficient nH value of 0.85 ± 0.06 (n = 6), R224A-TASK-2 channels were essentially insensitive to extracellular protons (Fig. 2I). A similar result was obtained with R224Q-TASK-2 (data not shown). The effect of R224 neutralization on the pH dependence of TASK-2 activity was also evident in single channel recordings. Fig. 3A shows traces of WT TASK-2 single channel activity in an outside-out patch exposed to extracellular pH values of 6.0, 7.5, and 9.0. As expected, there was little activity at acid pH, but openings occurred more frequently as the medium was alkalinized to pH 7.5 and 9.0. The calculated NPo value increased from 0.1at pH 6.0 to 0.6 and 1.2 at pH 7.5 and 9.0, respectively, as expected (18). By contrast, and as shown in Fig. 3B, there was no effect of pH on the activity of R224A-TASK-2 channels. In six separate patches, average NPo values were 1.0 ± 0.27, 0.95 ± 0.29, and 0.95 ± 0.13 at pH 6.0, 7.5, and 9.0 respectively. There was no statistical difference between the groups as tested by one-way ANOVA (P = 0.754). Mutation did not alter the single channel conductance, which was 50 pS under the experimental conditions (symmetrical 140 mM K+ and holding potential of −60 mV).
Fig. 2.
Effect of mutating R224 pH sensor on TASK-2 sensitivity to extracellular protons. (C–F) Effect of mutation R224A on TASK-2 rectification, time-dependence, and selectivity. Currents recorded were elicited by transfection of TASK-2 or TASK-2-R224A into HEK-293 cells. Measurements were done in the whole-cell recording mode of the patch-clamp technique, using the voltage protocols in A (for C and E) and B (for D and F). The intracellular solution contained 140 mM K+. Results in C, D, and G correspond to WT TASK-2 and those in E, F, and H to TASK-2-R224A mutant. In C and E, the extracellular medium had 135 mM Na+ and 5 mM K+. In D and F, extracellular Na+ was replaced by an equimolar amount of K+. G shows the current–voltage relationship in WT TASK-2 under the different extracellular K+ concentrations (circles, 5 mM; triangles, 140 mM). (H) Corresponding plot for TASK-2-R224A. Results are means ± SEM of n = 6 and n = 8 for WT TASK-2 and TASK-2-R224A, respectively. The graph in I shows extracellular pH-dependence curves of WT TASK-2 (circles, n = 6) and for its mutants R224A (downward triangles, n = 8), R224H (diamonds, n = 5), and R224K (triangles, n = 5). Results are shown as means ± SEM. The lines are fits of the Hill equation and were constructed by using the average of fitted parameters of the individual experiments (10).
Fig. 3.
Effect of mutation R224A on single-channel activity of TASK-2 channels. The pH sensitivity of WT TASK-2 (A) and the mutant R224A (B) are shown at three different pH values of the bathing medium. The recording configuration was outside-out, and the short lines at the left of the recordings show the closed channel levels. Openings are downward deflections, and the patches illustrated contained at least two channels. The holding potential was −60 mV, and both pipette and bath solutions contained 140 mM K+.
Lysine and histidine are also basic amino acids whose respective pKa values differ by 2 and 3.2 pH units from that of arginine. It might be expected, therefore, that if R224 were the pH sensor of TASK-2, replacement by these amino acids might preserve the pH sensitivity of the channels but decrease its pK1/2 accordingly. The result of these experiments is also shown in Fig. 2I. Both mutants were pH-sensitive and had pK1/2 values of 6.97 ± 0.08 (n = 5) for R224K-TASK-2 and 5.60 ± 0.11 (n = 5) for R224H-TASK-2. Respective nH values were 0.80 ± 0.13 and 0.85 ± 0.09. There was a nonzero offset for R224K of 0.39 ± 0.04.
To test whether the environment of R224 affects its pKa and therefore the pK1/2 for the effect of pH on TASK-2, we replaced I90 by a tryptophan. This mutation might allow a π-cation interaction between R224 and W90 that ought to increase pK1/2 by stabilizing the protonated state of R224. Tryptophan is also an amino acid that frequently occupies that position in channels of known structures such as KcsA, Kv1.2, and KvAP. Fig. 4A shows that I90W-TASK-2 presents a pH dependence displaced to alkaline values compared with the native channel. The pK1/2 of this mutant was 9.2 ± 0.1 (n = 5) with an nH value of 0.62 ± 0.04. Similar displacements were obtained when the I90W mutation was done in the background of R224K-TASK-2 and R224H-TASK-2. As shown in Fig. 4 B and C, the pK1/2 values for I90W-R224K-TASK-2 and I90W-R224H-TASK-2 were 7.54 ± 0.08 (n = 8) and 6.22 ± 0.05 (n = 5), respectively. The corresponding nH values were 0.78 ± 0.02 and 0.88 ± 0.07.
Fig. 4.
Effect of I90W mutation on TASK-2 sensitivity to extracellular protons. The graph shows extracellular pH dependence curves of I90W-TASK-2 (A; n = 5), I90W-R224K-TASK-2 (B; n = 8), and I90W-R224H-TASK-2 (C; n = 5). Results are shown as means ± SEM. The lines are fits of the Hill equation and were constructed by using the average of fitted parameters of the individual experiments. The lines without points are taken from the fits in Fig. 2G.
Our present work demonstrates that a single arginine residue, R224, sited toward the extracellular end of TM4 in TASK-2, is capable of sensing extracellular pH and accounts for the gating of this alkali-activated K+ channel. R224 sits at what appears as a hydrophobic pocket and close enough to the pore to exert an electrostatic effect, influencing pore occupancy. Recent work by Morton et al. (14) has suggested that a series of five charged amino acids in a large extracellular loop between TM1 and TM2 could constitute the pH sensor of TASK-2. These data raise the possibility that a number of titratable residues might act as pH sensors in TASK-2 by virtue of their electrostatic effect on the pore. We do not think this is the case as can be shown by assaying the quintuple mutated TASK-2 that should abolish its pH gating, as well as by neutralizing other titratable residues near the top of the transmembrane domains and investigating the pH dependence of the mutants. In our hands, as we have reported before (15), the quintuple mutation (TASK-2–5M) purported to abolish pH dependence shifts the pK1/2 of TASK-2 in the acid direction but does not abolish gating by extracellular pH. Those experiments were performed by using the same construct as Morton et al. (14), the only difference being the expression in HEK-293 or Cos-7 cells in our published experiment (15) rather than CHO-K1 cells. We have now repeated the experiment in CHO-K1 cells as reported in Fig. 5A. Despite an acid shift in pK1/2 of 0.6 of a pH unit compared with WT and a low pH offset of 16 ± 6% (n = 3), this mutant remained quite sensitive to extracellular pH. Besides E28, the remaining four lysines of the putative pH sensor lie in an extracellular loop. Their positive charge will be, we believe, most likely shielded by the solvent and consequently unlikely to exert a direct electrostatic effect on the pore. A structural change, perhaps a partial collapse of the loop, might explain the shifted pK1/2 of TASK-2–5M by a further hampering of R224 protonation.
Fig. 5.
Effect of neutralizing various titratable residues located near the extracellular aspect of TASK-2 channels on its pH dependence. Data for WT TASK-2 are means ± SEM (n = 12) and are shown as circles in A, and the same data are repeated in panel B. Mutants also are illustrated as means ± SEM as follows: K107A (n = 3), H220N (n = 9), 5-Mut (K32N, K35N, K42N, K47N, and E28Q, n = 3), K32A (n = 3), Y190F (n = 3), Y219F (n = 7), and Y225F (n = 3). All assays were done into HEK-293 cells except those for 5-Mut, which were carried out into CHO-K1 cells. Expressing WT TASK-2 into this last cell line had no effect on its pH dependence (data not shown).
The specificity of the effect of R224 neutralization on the pH dependence of TASK-2 is also supported by the lack of effect of neutralizing a number of titratable residues predicted to be near the pore mouth [see supporting information (SI) Fig. 8]. As seen in Fig. 5 A and B, these mutants were indistinguishable from WT TASK-2 or produced only small alterations in pH dependence inconsistent with their participation in the specific pH-sensing mechanism. K32 sits toward the top of TM1 (SI Fig. 8D) and is the nearest to the membrane of the four lysines in the sensor proposed by Morton et al. (14). As shown in Fig. 5A, its neutralization in the K32A mutant, in our hands, did not alter the pK1/2 of TASK-2, but there was a significant increase in the current remaining at acid pH (12 ± 3%, n = 3 vs. 3 ± 1%, n = 12 in WT TASK-2). This residue is most probably exposed to the solvent, and its electrostatic manifestation strongly shielded as a result. Its proximity to E28 (SI Fig. 8D) might contribute to cancel an effect of the negative amino acid to increase occupancy of the pore, which might become manifest in the K32A mutant. We speculate that this mechanism is responsible for the increase of basal current at low pH. Mutations K107A and H220N affect residues located, according to our model (SI Fig. 8 A and B), toward the top of helix TM2 and TM4, respectively. In agreement with previous results (14), these mutations had no effect whatsoever on the pH gating of TASK-2 (Fig. 5A). A further series of titratable, extracellular-facing residues were assayed. These residues were tyrosines, whose phenol group can lose its proton in solution with a pK1/2 of 10.1. These residues are Y190, located at the top of P2 helix, Y225, toward the extracellular end of TM4, and Y219 at the P2-TM4 loop (SI Fig. 8C). Mutating these tyrosines to phenylalanine only produced minor effects on pK1/2 of TASK-2 of which only that of Y219F reached statistical significance (Fig. 5B).
TALK-2, a close relative of TASK-2, is a pancreatic channel gated by very strong alkalinization. A basic amino acid is conserved at the equivalent position in TALK-2 (K242). TALK-2 is closed at neutral pH, starts to show significant activity only at pH ≈9, and has a pK1/2 >10 (13). We have modeled TALK-2 pore using Kv1.2 as a template as described above for TASK-2. As seen in Fig. 6, the model shows K242 occupying a position similar to TASK-2 R224 in α-helix TM4. K242 side chain also appears stabilized through hydrogen bond interactions with N103, which is conserved in the TALK subfamily. The place of I90 of the TASK-2 model is taken by F108 in TALK-2. The charged state of K242 might be stabilized by a π-cation interaction with F108, providing an explanation for the observed pK1/2 for TALK-2, which is close to the bulk solution pKa of lysine. If K242 were the pH sensor that switches TALK-2 off at neutral pH, one should observe activity after its neutralization by mutation. This prediction was tested by mutation K242A in TALK-2. Fig. 7B shows that at neutral pH, WT TALK-2 is closed and no significant K+ currents can be evoked at pH 7.5 with the voltage protocol shown in Fig. 7A. When extracellular pH was increased to 10, robust currents were evoked as expected (Fig. 7C). The same experiments performed with TALK-2-K242A revealed already sizable currents at neutral pH (Fig. 7D) which showed no major increase at pH 10. Summary current–voltage relationships for WT TALK-2 and mutant channels are shown in Fig. 7 E and G.
Fig. 6.
Modeling TALK-2 K+ channel pore suggests structural conservation of an extracellular pH sensor. Model for TALK-2 based on the structure of Kv1.2 K+ channel (16) was generated as a homodimer. A single monomer is illustrated with K242 projecting toward the selectivity filter region. The positions of G105, N121, and F108 are also shown. Helices are identified, and the permeation path is shown by K+ ions at cation binding sites S1, S2, and the internal cavity. K+ at S1 has been made transparent to enable visualization of K242.
Fig. 7.
Neutralization of K242 of TALK-2 K+ channel switches activity on at neutral pH. (B–E) Effect of mutation K242A on TALK-2-mediated currents. Recordings were elicited by transfection of TALK-2 (B, C, and F) or TALK-2-K242A (D, E, and G) into HEK-293 cells. Measurements were done in the whole-cell recording mode of the patch-clamp technique, using the voltage protocol shown in A. The intra- and extracellular solution contained 140 mM K+. Replacing the extracellular solution by one containing 135 mM Na+ and 5 mM K+ gave the expected shift in reversal potential but no other qualitative difference. The graph in F shows the current–voltage relationships for traces in B (triangles) and C (circles). Plots in G correspond to traces as in D (triangles) and E (circles). All numbers are means ± SEM; n = 5 in each case.
Discussion
In the present paper we have used molecular simulation and site-directed mutagenesis experiments to search for the pH sensor of TASK-2, a pH-gated KCNK channel. Our results suggest that an arginine (R224) residue, so located as to be able to exert an electrostatic effect upon the channel pore, acts as the pH sensor. The hypothesis that the R224 is the sensor is supported by the loss of pH sensitivity of the channel upon its neutralization and by the modulation of the pH sensitivity of TASK-2 upon its replacement by amino acids of different pKa values or alterations in its environment. In addition, a conserved basic amino acid appears to play the same role in TALK-2, a channel that belongs to the same subfamily.
Modeling TASK-2 on the known structure of the mammalian Kv1.2 revealed that R224 is so positioned as to be a pH sensor candidate in this channel. The contention that R224 is the pH sensor is supported by the loss of pH sensitivity of TASK-2 upon neutralization of this residue by mutation to A or Q. On the other hand, changing R224 for K and H modulates the pH sensitivity in a way predicted by the shift in pKa of the amino acid side chain. R224 is close to an asparagine residue (N87) next to the selectivity filter. N103 is present at a position usually occupied by an aspartate in most K+ channels outside the KCNK family, a configuration that would stabilize a charged form of R224. A deprotonated state of Arg may seem controversial, but recent direct measurements on basic amino acids engineered into the nicotinic receptor have demonstrated marked environment-dependent shifts, of up to −7.5, in their pKa compared with that in bulk solution (19). Moreover, theoretical studies of a deprotonation reaction of a guanidine group in media of low (ε = 4.9) and high (ε = 78.6) dielectric constant suggest a considerable shift in pKa of around −7 pH units in the low dielectric environment (20).
Analysis of available crystallographic data for K+ channels discloses that a similar position to that of R224 in our TASK-2 model is occupied by Arg in KcsA (Protein Data Bank (PDB) ID code 1K4C; www.pdb.org), by Lys in Kv1.2 (PDB ID code 2A79) and KvAP (PDB ID code 1ORQ), by His in KirBac1.1 (PDB ID code 1P7B), but by Met in MthK (PDB ID code 1LNQ). R89 in KcsA is well stabilized by π-cation interaction with W67, which in turn H-bonds with selectivity filter D80 (see SI Fig. 9). A similar arrangement stabilizes the corresponding Lys in Kv1.2 and KvAP, and the His (via Phe rather than Trp) in KirBac1.1. In the case of our TASK-2 model, however, R224 sits in a hydrophobic environment, with Ile replacing the Trp and Asn taking the place of the selectivity filter Asp. This scenery would, on the one hand, predict an alkaline shift in pKa for R224 in TASK-2 due to its more hydrophobic environment compared with other channels but also a heightened relative electrostatic effect on the pore due to the absence of the conserved aspartate. To test for the effect of the environment on pKa of the putative sensor, we mutated I90 of TASK-2 to the more habitual W of other K+ channels. Whether the I90W was made in the background of WT, R224K- or R224H-TASK-2, it always led to an increase in pK1/2, consistent with the idea that providing a condition favorable to a π-cation interaction for the sensing residue makes its pKa more alkaline, stabilizing the protonated state of R224.
TALK-1 and -2 are also activated by very strong alkalinization and have low or no activity at neutral pH (18). A basic amino acid is conserved at the equivalent position to TASK-2 R224 in the K+ channels of the TALK (R333 in TALK-1 and K242 in TALK-2), but not the TASK, subfamily. It appears that deprotonation of these conserved residues might also gate the opening of these alkaline-pH-activated channels, because neutralization of K242 in TALK-2 led to channel opening at neutral pH. TASK-2 I90 is replaced by a phenylalanine residue in TALK-1 and -2. It is possible that the presence of this π-cloud-providing amino acid might contribute to the high pK1/2 of these channels.
It has recently been proposed that a group of four lysine and one glutamic acid residues, located in the extracellular loop between TM1 and P1, is the external pH sensor of TASK-2 (14). One might think that a number of titratable residues affecting the pore electrostatically could all act as pH sensors. As we report here and elsewhere, our experiments with a mutant of TASK-2 with these five residues neutralized was still gated by pH (15). We have no ready explanation for this discrepancy. The arrangement of charges proposed as the pH sensor in TASK-2 (14) is not conserved in the TALK subfamily of channels, and the hypothesis, therefore, does not provide a unified mechanism for their alkalinization-dependent gating. Although there is no structural information on this large extracellular loop, these charges are most likely exposed to, and therefore shielded by, the solvent and would not be able to exert any electrostatic perturbation in the pore. These charges might, at best, affect the local H+ and K+ concentration. On the other hand, the elimination of a total of 10 charged amino acids from the extracellular loops could cause an important structural change in the channel that might be responsible for the effect on pK1/2 that we have observed (15). Interestingly a number of mutations neutralizing extracellular-facing titratable residues had only small or no effect on TASK-2 pH gating.
Modulation of TASK-2 by extracellular pH involves changes in open probability (Po) without affecting single channel conductance (5, 18). How does neutralization of R224 affect Po? One possibility is that, in the protonated form, R224 prevents occupancy of the selectivity filter by K+ thus creating a blocked state, a situation relieved by neutralization of R224 at alkaline pH. As discussed elsewhere (15), in the absence of a full complement of aspartate residues around the mouth of the channel, two of which are N in TASK-2, the relative contribution of R224 to the electrostatic potential of the pore must be high. It is therefore conceivable that pH gating in these channels occurs at the selectivity filter in a way akin to the C-type inactivation seen in other KCNK K+ channels (21). Recent molecular dynamic simulations suggest that this situation occurs by changes in pore structure dependent upon occupancy of the S2 cation-binding site (22). Our calculations of the change in electrostatic potential upon neutralization of R224, giving a trough at S2, make occupancy-related pore gating in TASK-2 an attractive hypothesis.
Methods
TASK-2 and TALK-2 Channel Models by Molecular Simulation.
The models for the pores of TASK-2 and TALK-2 (files are available at http://www.cecs.cl/web/files/PoreModels.html) were built by using as template the crystallographic data of Kv1.2 K+ channel (PDB ID code 2A79) (16) with the program MODELLER (23). The electrostatic potential calculations were done by using the Poisson-Boltzmann equations implemented in the PBEQ module of the program CHARMm, version c31b1 (24). The choice of Kv1.2 K+ channel as a template for molecular simulation was based on the fact that Kv1.2 and MthK (PDB ID code 2A79) are the only K+ channels known in an open-channel structure. Kv1.2 was preferred over MthK because its structure contains information on the location of side chains of the pore residues. More details on the molecular simulation procedures are given in SI Text.
Electrophysiological Assays.
Mus musculus TASK-2 (KCNK-5; GenBank accession no. AF319542) plasmid had been obtained from mouse kidney. Human TALK-2 (KCNK-17; GenBank accession no. BC025726) was obtained from the Mammalian Genome Collection at the American Type Tissue Collection (Manassas, VA). Both DNAs were subcloned into the pCR3.1 vector. Transient transfections were done in HEK-293 or CHO-K1 cells as described (25). Standard whole-cell, patch-clamp recordings were performed as described elsewhere (25, 26). Currents were measured at several potentials, but data reported are those obtained at 0 mV.
Calculations.
The effect of pH on currents was evaluated by plotting current (I) measured at a membrane potential of 0 mV against extracellular [H+]. Fitting of a Hill equation to the data was done for each individual experiment. The parameters are defined in the following equation: I = Imin + (Imax − Imin)/(1+ ([H+]/K1/2)nH). For graphical representation, averages ± SEM of I/Imax values were obtained from individual experiments and plotted together with curves constructed with average parameters obtained from the individual fits. Fits were done by using the Marquardt-Levenberg algorithm as implemented in the SigmaPlot software (Systat, Erkrath, Germany).
A more detailed account of the methods used is given in SI Text.
Supplementary Material
Acknowledgments
We are grateful to Ms. Yasna Jaramillo for skilled technical support and to Dr. Marcelo Catalán for help and advice. This work is supported by Fondo Nacional de Desarrollo Científico y Tecnológico (Fondecyt) Grants 1030838 (to M.I.N.) and 1040254 (to F.D.G.-N.). Centro de Estudios Científicos is funded by an Institute grant from the Millennium Science Initiative and by Fundación Andes and Empresas CMPC. L.Z. and W.G. are supported by MECESup and CONICYT studentships.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
¶Synonyms (9) include the following: TASK-2, KCNK5, and K2P5.1; TALK-1, KCNK16, and K2P16.1; and TALK-2, KCNK17, and K2P17.1.
This article contains supporting information online at www.pnas.org/cgi/content/full/0606173104/DC1.
References
- 1.Yellen G. Nature. 2002;419:35–42. doi: 10.1038/nature00978. [DOI] [PubMed] [Google Scholar]
- 2.MacKinnon R. FEBS Lett. 2003;555:62–65. doi: 10.1016/s0014-5793(03)01104-9. [DOI] [PubMed] [Google Scholar]
- 3.Lesage F, Lazdunski M. Am J Physiol. 2000;279:F793–F801. doi: 10.1152/ajprenal.2000.279.5.F793. [DOI] [PubMed] [Google Scholar]
- 4.Goldstein SA, Bockenhauer D, O'Kelly I, Zilberberg N. Nat Rev Neurosci. 2001;2:175–184. doi: 10.1038/35058574. [DOI] [PubMed] [Google Scholar]
- 5.Reyes R, Duprat F, Lesage F, Fink M, Salinas M, Farman N, Lazdunski M. J Biol Chem. 1998;273:30863–30869. doi: 10.1074/jbc.273.47.30863. [DOI] [PubMed] [Google Scholar]
- 6.Kim Y, Bang H, Kim D. J Biol Chem. 2000;275:9340–9347. doi: 10.1074/jbc.275.13.9340. [DOI] [PubMed] [Google Scholar]
- 7.Lopes CMB, Zilberberg N, Goldstein SA. J Biol Chem. 2001;276:24449–24452. doi: 10.1074/jbc.C100184200. [DOI] [PubMed] [Google Scholar]
- 8.Rajan S, Wischmeyer E, Liu GX, Preisig-Muller R, Daut J, Karschin A, Derst C. J Biol Chem. 2000;275:16650–16657. doi: 10.1074/jbc.M000030200. [DOI] [PubMed] [Google Scholar]
- 9.Goldstein SA, Bayliss DA, Kim D, Lesage F, Plant LD, Rajan S. Pharmacol Rev. 2005;57:527–540. doi: 10.1124/pr.57.4.12. [DOI] [PubMed] [Google Scholar]
- 10.Niemeyer MI, Cid LP, Barros LF, Sepúlveda FV. J Biol Chem. 2001;276:43166–43174. doi: 10.1074/jbc.M107192200. [DOI] [PubMed] [Google Scholar]
- 11.Barrière H, Belfodil R, Rubera I, Tauc M, Lesage F, Poujeol C, Guy N, Barhanin J, Poujeol P. J Gen Physiol. 2003;122:177–190. doi: 10.1085/jgp.200308820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warth R, Barrière H, Meneton P, Bloch M, Thomas J, Tauc M, Heitzmann D, Romeo E, Verrey F, Mengual R, et al. Proc Natl Acad Sci USA. 2004;101:8215–8220. doi: 10.1073/pnas.0400081101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Girard C, Duprat F, Terrenoire C, Tinel N, Fosset M, Romey G, Lazdunski M, Lesage F. Biochem Biophys Res Commun. 2001;282:249–256. doi: 10.1006/bbrc.2001.4562. [DOI] [PubMed] [Google Scholar]
- 14.Morton MJ, Abohamed A, Sivaprasadarao A, Hunter M. Proc Natl Acad Sci USA. 2005;102:16102–16106. doi: 10.1073/pnas.0506870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niemeyer MI, González-Nilo FD, Zúñiga L, González W, Cid LP, Sepúlveda FV. Biochem Soc Trans. 2006;34:903–906. doi: 10.1042/BST0340899. [DOI] [PubMed] [Google Scholar]
- 16.Long SB, Campbell EB, MacKinnon R. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 17.Niemeyer MI, Cid LP, Valenzuela X, Paeile V, Sepúlveda FV. Mol Membr Biol. 2003;20:185–191. doi: 10.1080/0968768031000084181. [DOI] [PubMed] [Google Scholar]
- 18.Kang D, Kim D. Biochem Biophys Res Commun. 2004;315:836–844. doi: 10.1016/j.bbrc.2004.01.137. [DOI] [PubMed] [Google Scholar]
- 19.Cymes GD, Ni Y, Grosman C. Nature. 2005;438:975–980. doi: 10.1038/nature04293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norberg J, Foloppe N, Nilsson L. J Chem Theor Comp. 2005;1:986–993. doi: 10.1021/ct049849m. [DOI] [PubMed] [Google Scholar]
- 21.Zilberberg N, Ilan N, Goldstein SA. Neuron. 2001;32:635–648. doi: 10.1016/s0896-6273(01)00503-7. [DOI] [PubMed] [Google Scholar]
- 22.Bernèche S, Roux B. Structure (Cambridge) 2005;13:591–600. doi: 10.1016/j.str.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 23.Sali A, Blundell TL. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 24.Jogini V, Roux B. J Mol Biol. 2005;354:272–288. doi: 10.1016/j.jmb.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 25.Cid LP, Niemeyer MI, Ramírez A, Sepúlveda FV. Am J Physiol. 2000;279:C1198–C1210. doi: 10.1152/ajpcell.2000.279.4.C1198. [DOI] [PubMed] [Google Scholar]
- 26.Díaz M, Sepúlveda FV. Pflügers Arch. 1995;430:168–180. doi: 10.1007/BF00374647. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.