Abstract
Upon a dark-to-light shift, the conditional fluorescent (flu) mutant of Arabidopsis releases singlet oxygen (1O2) within the plastid compartment. Distinct sets of nuclear genes are activated that are different from those induced by superoxide (O2•−) and/or hydrogen peroxide (H2O2), suggesting that different types of reactive oxygen species activate distinct signaling pathways. It is not known whether the pathways operate separately or interact with each other. We have addressed this problem by modulating noninvasively the level of H2O2 in plastids by means of a transgenic line that overexpresses the thylakoid-bound ascorbate peroxidase (tAPX). The overexpression of the H2O2-specific scavenger reduced strongly the activation of nuclear genes in plants treated with the herbicide paraquat that in the light leads to the enhanced generation of O2•− and H2O2. In the flu mutant overexpressing tAPX, the intensity of 1O2-mediated cell death and growth inhibition was increased when compared with the flu parental line. Also, the expression of most of the nuclear genes that were rapidly activated after the release of 1O2 was significantly higher in flu plants overexpressing tAPX, whereas in wild-type plants, overexpression of tAPX did not lead to visible stress responses and had only a very minor impact on nuclear gene expression. The results suggest that H2O2 antagonizes the 1O2-mediated signaling of stress responses as seen in the flu mutant. This cross-talk between H2O2- and 1O2-dependent signaling pathways might contribute to the overall stability and robustness of wild-type plants exposed to adverse environmental stress conditions.
Keywords: ascorbate peroxidase, chloroplast, oxidative stress, reactive oxygen species
The evolution of aerobic metabolic processes such as respiration and photosynthesis led to the continuous production of reactive oxygen species (ROS) in mitochondria, chloroplasts and peroxisomes. Thereby, ground state oxygen is converted to different ROS either by energy transfer or by electron-transfer reactions. The former leads to the formation of singlet oxygen (1O2), whereas the latter results in the sequential reduction to superoxide anion radical (O2•−), hydrogen peroxide (H2O2) and hydroxyl radicals (•OH) (1). A common feature among these different ROS is their ability to react with a large variety of biomolecules such as lipids, proteins, and nucleic acids that are essential for the activity and integrity of cells. Under steady-state conditions, ROS are scavenged by various antioxidative defense mechanisms. The equilibrium between production and scavenging of ROS may be perturbed by various abiotic and biotic stress conditions, leading to a rapid and transient increase of the intracellular ROS level. Organisms under stress often suffer from damages that have been attributed to the cytotoxicity of these ROS. More recently, however, a second role has been proposed for ROS that implicates them with the signaling of genetically controlled stress-response programs (for reviews, see, for instance, refs. 2–4). Sensing changes of ROS concentrations that result from metabolic disturbances seem to be used by plants to activate stress responses that help them to cope with environmental changes.
In plants exposed to various abiotic stress conditions, a large part of the stress-induced transient increase in ROS concentration takes place within chloroplasts, when the balance between light absorption and the use of light energy is disturbed and excess light energy will lead to the inhibition of photosynthesis (5). Under these conditions, plants may activate alternative electron sinks such as photorespiration (6) and the reduction of oxygen by PSI that results in the enhanced production of O2•−, which is, in turn, dismutated to H2O2, and may help to avoid the stress-induced inhibition of photosynthesis (7, 8). Once these quenching mechanisms are no longer sufficient to maintain the acceptor site of PSII in a partially oxidized state, photoinhibition of PSII occurs and endorses the enhanced generation of 1O2. Whereas all three ROS mentioned before cause similar cytotoxic damages of photosynthetic membranes, their signaling specificities may be different from each other. 1O2, for instance, is expected to activate a stress-response program adapted to alleviate the negative impact of environmental conditions that enhance the generation of this ROS and that may be different from those that stimulate O2•−/H2O2 production. Attempts to unravel the signaling activity of each ROS separately are faced with the problem that within chloroplasts of plants under stress, enhanced levels of different ROS are produced simultaneously (9), making it very difficult to link a particular stress response to the signaling activity of a specific ROS and to separate this from its cytotoxic effect. We have tried to overcome this obstacle by using the conditional flu mutant of Arabidopsis that generates 1O2 in plastids in a noninvasive and controlled manner (10, 11). flu accumulates free protochlorophyllide in the dark, which acts as a potent photosensitizer that generates 1O2 in plastids during reillumination (10). Immediately after the release of 1O2, the growth rate of mature plants decreases, whereas seedlings bleach and die (10, 11). These two stress responses primarily are caused by the 1O2-dependent activation of genetically determined stress-response programs (12). At the same time, drastic changes in nuclear gene expression occur that affect ≈5% of the total genome of Arabidopsis. Several of these genes respond selectively to the release of 1O2 and are not affected during a treatment by paraquat, a herbicide that generates O2•−/H2O2 within chloroplasts (11, 13). These results strongly suggest that 1O2 and O2•−/H2O2 affect nuclear gene expression via distinct signaling pathways. These signaling pathways may act independently or they may interact with each other. In case of extensive cross-talk between these different signaling pathways, it would be even more difficult to define the biological activity of a given ROS and to determine its contribution to the overall response of a plant to oxidative stress than previously anticipated.
In the present work, we have used the overexpression of a thylakoid-specific ascorbate peroxidase (tAPX) to reduce in planta the level of H2O2 (14, 15). After crossing this transgenic line with the flu mutant, the possible impact of altered levels of H2O2 in plastids on 1O2-mediated stress responses could be determined noninvasively. Overexpression of tAPX in the flu mutant did not reduce the 1O2-induced stress responses, but to the contrary, the intensity of 1O2-dependent growth inhibition and cell death was even higher in the double mutant than in the parental flu line. An extensive analysis of the response of the transcriptome of Arabidopsis revealed that the expression of most of the genes that were activated after the release of 1O2 was even more enhanced in plants overexpressing tAPX, whereas overexpression of tAPX alone without the release of 1O2 seemed to have only a very limited impact. The enhancement of 1O2-induced stress responses by overexpressing a chloroplastic H2O2 scavenger suggests an antagonistic effect of these two ROS during the stress response and an intimate cross-talk between various ROS that might be essential for the fine control of adjusting antioxidants and photosynthesis to different environmental stress conditions.
Results
Overexpression of Thylakoidal Ascorbate Peroxidase Drastically Reduces Induction of Genes in Paraquat-Treated Arabidopsis.
Our previous work suggested that 1O2 and H2O2 impact nuclear gene expression via different signaling pathways (11). These pathways may operate either independently or interact with each other. We have tried to tackle this problem first by modulating the endogenous level of H2O2 in plastids by overexpressing the tAPX. Such a tAPX-overexpressing line had been shown to be more resistant to paraquat-induced photooxidative stress, whereas under normal growth conditions without exposure to paraquat, it was phenotypically indistinguishable from wild-type plants (14).
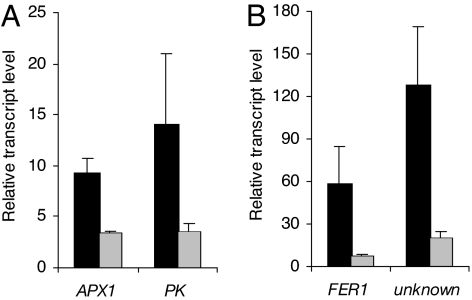
The suppressive effect of tAPX on H2O2 concentrations within the transgenic line was tested by comparing transcript levels of four genes known to be specifically activated during paraquat or H2O2 treatment (11). In tAPX-overexpressing plants that were exposed to paraquat, transcript levels of the ascorbate peroxidase 1 (APX1) gene (At1g07890), a gene encoding a pyruvate kinase (At3g49160), the ferritin 1 (FER1) gene (At5g01600), and a gene encoding an unknown protein (At3g20340) were 2.7 (APX1) to 7.4 (FER1)-fold lower than in paraquat-treated wild-type plants (Fig. 1), thus confirming the previously reported ability of the chloroplast-specific H2O2 scavenger to reduce the endogenous H2O2 level in plastids of tAPX-overexpressing plants (15). The reduced activation of these marker genes further confirms their H2O2-specific responsiveness.
Fig. 1.
The induction of genes in paraquat-treated wild-type plants (wt, black bars) and in wild-type plants overexpressing thylakoidal ascorbate peroxidase (35S::tAPX, gray bars). The transcript levels of paraquat-induced genes after 4 h of paraquat treatment (20 μM paraquat in 0.1% Tween) were expressed relative to those in mock-treated plants (Tween 0.1% only). Four genes that were reported to be induced specifically by paraquat were analyzed: the ascorbate peroxidase 1 (APX1) gene and a gene encoding a pyruvate kinase family protein (PK) (A), the ferritin 1 (FER1) gene and a gene encoding an unknown protein (unknown) (B). The results represent the average values of measurements from five independent experiments ± SE.
Overexpression of Thylakoidal Ascorbate Peroxidase Increases the Singlet Oxygen-Induced Whole-Plant Responses.
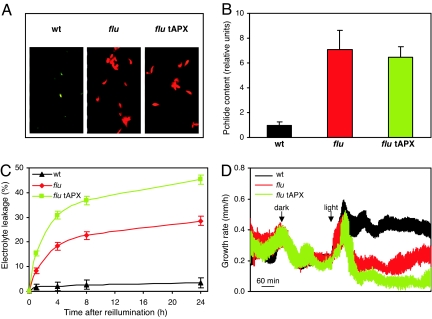
To investigate the possible impact of H2O2 in plastids on 1O2-mediated stress responses, the tAPX-overexpressing line was crossed with the flu mutant, and homozygous double mutants were selected from the segregating F2 population. In the double mutant, overexpression of tAPX did not alter the overaccumulation of protochlorophyllide in dark-treated flu plants (Fig. 2A and B) and, thus, did not interfere with the amounts of singlet oxygen that upon illumination would be generated by energy transfer from excited free protochlorophyllide to ground-state oxygen. Two 1O2-induced visible stress reactions were observed in flu plants after a dark-to-light shift: a cell-death response and the rapid inhibition of growth (11). These stress responses were compared with those of the tAPX-overexpressing flu line that was subjected to the same dark-to-light shift. The overexpression of tAPX did not reduce the intensity of the two 1O2-induced stress responses but, unexpectedly, amplified both of them.
Fig. 2.
The stimulation of singlet oxygen-mediated stress responses in tAPX-overexpressing flu. (A) The protochlorophyllide levels of etiolated seedlings of wt, flu, and tAPX-overexpressing flu (flu tAPX) were determined by comparing the red fluorescence of excited protochlorophyllide during exposure of seedlings to blue light (400–450 nm). (B) The protochlorophyllide content of rosette leaves from plants that were grown for 21 days under continuous light and transferred to the dark for 8 h. Tetrapyrroles were extracted from rosette leaves of wt, flu, and tAPX-overexpressing flu (flu tAPX) plants at the end of the dark period, separated by HPLC, and detected by their fluorescence. (C) The enhanced 1O2-mediated cell death in tAPX-overexpressing flu. The cell death was expressed as a percentage of electrolyte leakage (related to the maximum electrolyte content; see Materials and Methods). Rosette leaves from wt (black series), flu (red series), and tAPX-overexpressing flu (flu tAPX, green series) were detached under green safe light from plants transferred to the dark for 8 h and were kept on water for 30 min before reillumination. The data represent least-square means for electrolyte leakage (%) ± SE. Five to 12 plants of each line were tested in four independent experiments. (D) The enhanced 1O2-mediated growth inhibition in tAPX-overexpressing flu. The growth rates of bolting plants were recorded continuously before and after the dark/light shift. The data represent means of four to nine independent measurements ± SE.
First, the cell-death response was measured by determining the electrolyte leakage of cut leaves of flu, tAPX-overexpressing flu, and wild type that were floated on distilled water in transparent containers and before cutting had been kept in the dark for 8 h. After 24 h of reillumination, electrolyte leakage of leaves of the flu mutant reached ≈29% of total electrolyte content (Fig. 2C). In leaves of the tAPX-overexpressing flu mutant, the electrolyte leakage was almost twice as high, reaching 46% after 24 h of incubation (Fig. 2C). Least square means for flu and tAPX-overexpressing flu lines were significantly different (P < 0.001) at each tested time point after reillumination.
The second stress reaction of flu to the release of 1O2, i.e., growth inhibition, was measured by recording continuously the growth rate of the emerging stem of mature plants that are ready to bolt (Fig. 2D). The growth rate of wild type, flu, and tAPX-overexpressing flu reached ≈0.3 mm/h when these plants were kept under continuous light and was uniformly reduced in all three lines to ≈0.2 mm/h after they were transferred to the dark for 4 h (Fig. 2D). Upon reillumination the growth rates of all three lines initially increased, but after ≈30 min of reillumination, started to diverge drastically. Whereas in wild-type plants the elevated growth rate was maintained for the following hours of reillumination, the growth rate of flu plants was inhibited to slightly <0.2 mm/h and slowly recovered afterward and finally approached the rate of wild-type plants. In contrast, growth of tAPX-overexpressing flu was almost completely abolished, and the plants hardly recovered during the time of the experiment.
Similar to wild type also, the tAPX-overexpressing wild type was not inhibited in its growth and showed no increased electrolyte leakage when exposed to a dark-to-light shift (data not shown).
Overexpression of Thylakoidal Ascorbate Peroxidase Affects the Expression of Genes Induced During the Release of Singlet Oxygen.
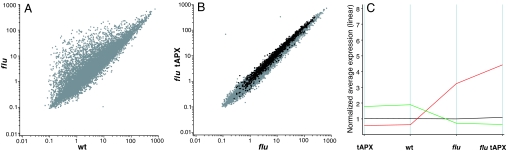
Distinct sets of genes have been shown to be activated selectively either by 1O2 alone or by O2•−/H2O2 or were stimulated by both types of ROS (11). In a first pilot experiment, marker genes from each of these three sets were used to determine by quantitative RT-PCR whether reduced levels of H2O2 in tAPX-overexpressing flu modulated their expression levels after a dark-to-light shift. The transcript levels of most of these marker genes were increased further during reillumination of flu plants that overexpressed tAPX, with the extent of stimulation being more pronounced after 2 h of reillumination than after 30 min (data not shown). Therefore, the effect of reduced levels of H2O2 on global 1O2-mediated gene expression changes in the flu mutant was analyzed comprehensively at 2 h after the dark/light shift by using Affymetrix (Santa Clara, CA) ATH1 microarrays. Two hours after the dark-to-light shift, 1,356 genes of Arabidopsis showed a >2-fold increase in their transcript levels in the flu mutant relative to wild-type plants (see Materials and Methods for the stringency of data analysis), highlighting a massive transcriptome reprogramming in response to 1O2 (Fig. 3A). A massive increase of gene expression also was seen in tAPX-overexpressing flu that is similar to the one observed in flu (Fig. 3B). However, in this line, 1,605 genes showed a >2-fold increase in their transcript levels relative to wild-type plants, indicating that the intensity of the response in tAPX-overexpressing flu is higher than in flu. When the transcript levels in flu and tAPX-overexpressing flu tAPX were compared and this comparison was restricted to the 1,356 genes initially shown to be up-regulated in flu at least 2-fold, the 1O2-mediated stimulation of these genes in tAPX-overexpressing flu was on average 1.4-fold higher than in the parental flu plants (Fig. 3 B and C). Similarly, 618 genes were down-regulated >2-fold in flu relative to wild-type as compared with the 1,004 genes in tAPX-overexpressing flu (data not shown). These 618 transcripts were on average 1.3 times more abundant in flu than in tAPX-overexpressing flu. Genes that in flu were only weakly affected by the release of 1O2, i.e., <2-fold relative to wild type, were also only very weakly stimulated in tAPX-overexpressing flu: genes that showed a < 1.1-fold difference in their transcript level to wild type were not detectably altered in their transcript level in tAPX-overexpressing flu (Fig. 3C). Thus, differences in transcript levels detected during the pairwise comparisons were not caused by the intrinsic variation of transcript measurements during the experiments (see Materials and Methods).
Fig. 3.
The impact of tAPX overexpression (tAPX) on the expression profiles of wild type (wt) and the flu mutant of Arabidopsis. (A) Scatter plot graph comparing the relative expression profiles of flu vs. wild-type plants after 2 h of reillumination. The log-intensity values for 14,088 significantly expressed genes detected as Present or Marginal in at least both replicates of one line (see Materials and Methods) were plotted (average of two replicates for each line). (B) Scatter plot graph comparing the relative expression profiles of tAPX-overexpressing flu vs. flu plants after 2 h of reillumination. The same genes as in A are plotted. Genes that, in both replicate experiments, were induced at least two-fold in flu vs. wild type are displayed as black dots, and the remaining genes are shown as gray dots. (C) Average expression profiles of wild type (wt), the flu mutant, tAPX-overexpressing wild type (tAPX), and the tAPX-overexpressing flu mutant (flu tAPX). The average expression values, derived of both replicate experiments, of 1,356 genes up-regulated at least two-fold (red line), of 618 genes down-regulated at least 2-fold (green line), and of 2,160 genes not affected (fold change between 0.9 and 1.1, black line) in flu vs. wild type are shown.
Coregulated Clusters of Genes induced by Singlet Oxygen.
1O2-activated genes were classified by using k means clustering (GeneSpring, Silicon Genetics; see Materials and Methods) to identify genes that may form coregulated clusters. To remove genes with unreliable measurements and enrich for relevant changes, the clustering of genes was restricted first to those that were detected as “Present” or “Marginal” in both replicas of all four lines, flu, tAPX-overexpressing flu, wild type, and tAPX- overexpressing wild type (see Materials and Methods). Based on these selection criteria, 839 of the 1,356 genes initially found to be activated 2-fold in flu relative to wild type, were chosen. In a second selection step aimed at enriching for early 1O2-induced genes, only those of the 839 genes were considered that were up-regulated at least 2-fold already during the first 30 min after the dark-to-light shift, according to previously reported data (11). Based on this selection of early induced genes, a total of 182 genes was retained for the final cluster analysis. Three main gene clusters could be distinguished. Cluster 1 comprised 71 genes [see supporting information (SI) Fig. 4 and SI Table 1] that were strongly up-regulated in flu by 1O2 2 h after the dark-to-light shift (average induction = 12.05-fold) and were slightly more up-regulated (in average 1.46-fold) in tAPX-overexpressing flu, but were not affected in tAPX-overexpressing wild type (average fold difference compared with wild type = 1.02). Cluster 2 contained 87 genes (see SI Fig. 4 and SI Table 1) that were moderately up-regulated in flu after the release of 1O2 (average induction = 4.46-fold), also further up-regulated in tAPX-overexpressing flu similar to genes of cluster 1 (in average 1.47) and only very weakly affected by tAPX in wild type (average fold difference compared with wild type = 1.17 fold). Cluster 3 contained 24 genes (see SI Fig. 4 and SI Table 1), whose responses to the release of 1O2 were slightly distinct from those of genes of clusters 1 and 2. These genes were up-regulated to a similar level in both tAPX-overexpressing flu and flu and were slightly up-regulated in tAPX-overexpressing wild type relative to the wild-type control (in average 1.4-fold). Although we were able to distinguish three different clusters of genes, their expression profiles were not dramatically divergent. Overall, genes that were rapidly up-regulated in flu after 1O2 release during a dark-to-light shift were up-regulated further in tAPX-overexpressing flu. The overexpression of tAPX in wild type, however, had almost no effect on gene expression during the dark-to-light shift.
Discussion
In plants placed under abiotic stress, the overall level of different ROS within chloroplasts increases rapidly. The relative contribution of each of these ROS to this general and transient increase, however, may vary depending on the actual environmental stress to which plants are exposed. For instance, in plants suffering from moderate light stress (600–700 μmol of photons·m−2·s−1) 1O2, O2•− and H2O2 are released simultaneously (9), whereas harsher light-stress conditions that lead to photoinhibition of PSII favor 1O2 production (16, 17). By using fluorescent sensors such as DanePy that is specific for 1O2 and HO-1889NH, which reacts with both 1O2 and O2•−, it has been shown that spinach leaves treated with very strong light produced 1O2 but hardly any O2•− (18). On the other hand, leaves exposed to UV light produced mainly O2•−. Leaves kept in the cold also generate little 1O2 (19), whereas the production of O2•− and H2O2 is enhanced not only under chilling stress but also under drought conditions, when CO2 reduction is restricted. The selective perception of ROS that are chemically distinct and produced preferentially under specific stress conditions (4) seems to enable plants to adjust their responses to the needs imposed by enhanced levels of a given ROS, for instance, by increasing levels of appropriate scavengers.
We have reported previously that in Arabidopsis, the release of 1O2 activates a distinct group of early stress response genes that are different from those activated by O2•−/H2O2 (11). 1O2-specific signaling pathways also have been described for mammals (20–23), Chlamydomonas (24), and the phototrophic bacterium Rhodobacter sphaeroides, in which the alternative sigma factor, sigma(E), is essential for activating a specific transcriptional response to 1O2, thereby protecting cells against this ROS (25). The present work offers previously undescribed insights into how plants may cope with oxidative stress by showing that responses to stress may not only be determined by the plant's ability to discriminate between different ROS, but that responses to a given ROS also may be modified by the concomitant perception of a second ROS. Lowering the level of H2O2 in chloroplasts by overexpressing tAPX further enhanced the 1O2-mediated cell death response and growth inhibition, suggesting that in plants under stress, enhanced levels of H2O2 may antagonize 1O2-dependent stress responses. This antagonistic interaction between 1O2 and H2O2 may explain observations published previously on the impact of different ROS in plants. For instance, Arabidopsis plants exposed to very high excess light energy (2,700 ± 300 μmol photons·m−2·s−1) that was expected to enhance primarily 1O2 production were more resistant if they were pretreated with high concentrations of exogenous H2O2 2 h before the beginning of high light stress, as indicated by a reduction of photoinhibition and the absence of visible damages of the leaves (26). It is tempting to speculate that in this case, similar to what has been reported in our present work, H2O2 protects photosynthetic membranes by modulating the signaling of 1O2 that is generated during high light stress.
The overexpression of tAPX did not confer a detectable enhanced resistance or sensitivity to plants that were challenged either with moderate high light (700 μmol of photons·m−2·s−1) that stimulates jointly the production of 1O2, O2•−, and H2O2 (9) or with low temperature combined with a mild light stress (4°C, 200 μmol of photons·m−2·s−1) (14). It is also remarkable that under no stress, the overexpression of tAPX did not affect plant fitness and only very poorly affected gene expression. The overexpression of tAPX altered the stress sensitivity of plants only under conditions that endorsed selectively the release of a particular ROS and led to a higher resistance to O2•−/H2O2 in paraquat-treated wild-type plants and to an enhanced sensitivity to 1O2 in flu plants. After a dark-to-light shift, overexpression of tAPX in the flu mutant amplified the disequilibrium between 1O2 and H2O2, thereby revealing the modulating activity of H2O2 that normally negatively regulates 1O2-mediated stress responses. The cross-talk between these two ROS also is likely to affect stress responses of wild-type plants, in particular when these are exposed to severe adverse environmental conditions.
Because the activity of thylakoidal ascorbate peroxidase is confined to the chloroplast (14) and 1O2, because of its short half-life, seems to be unable to leave the plastid compartment, the cross-talk between these two ROS is likely to take its origin from within the chloroplasts. However, it is not known yet whether this interaction is a more direct one with both ROS reacting with a common target that is shared by 1O2- and H2O2-depending signaling pathways or whether the modulating influence of different H2O2 concentrations on 1O2-mediated signaling is a more indirect one, for instance, by changing the redox state of the plastid. The tripeptide glutathione, which is essential in determining the redox state of the cell (27) and which is present in large amounts inside the plastids (27), might be a likely candidate for a factor that is involved in mediating the H2O2-dependent control of 1O2-dependent signaling. It has been shown that the size and redox state of the glutathione pool rapidly changes in response to biotic (28) and abiotic stress conditions (29) that evoke an enhanced production of ROS. Glutathione, as well as ascorbate, have been implicated with the control of stress defense-related gene expression (30–35). The impact of H2O2 and glutathione on plants, however, might be inverse, higher concentrations of H2O2 protecting plant cells against photooxidative stress and reducing the extent of photoinhibition of photosynthesis, excess amounts of glutathione having an opposite effect (26). This apparent paradox has been attributed to the reverse control of the redox status of the QA-QB-plastoquinone pools by H2O2 and glutathione. Glutathione reduces QA and lowers the electron transport efficiency in PSII, whereas treatment of chloroplast membranes with H2O2 increases the oxidation of QA and enhances the efficiency of electron transport in PSII. The former treatment would be expected to stimulate the generation of 1O2 by PSII, whereas the latter should reduce the amounts of 1O2. The next step is to investigate the effect of glutathione on 1O2-mediated stress responses and its possible role during the cross-talk between 1O2-dependent and H2O2-dependent signaling pathways.
Materials and Methods
Plant Materials, Growth Conditions, and Stress Treatments.
Arabidopsis thaliana lines used in this work were of the Columbia ecotype (Col0). They were cultivated on soil under continuous light (100 μmol·photons m−2·s−1) until they reached the rosette leaf stage. For the analysis of changes in the expression of genes after paraquat treatment, 3-week-old wild-type and thylakoidal ascorbate peroxidase-overexpressing plants were sprayed either with a solution of 20 μM paraquat (methyl viologen; Sigma, St. Louis, MO) in 0.1% Tween or with Tween alone, and rosette leaves were harvested at the indicated time point. For each sample, the rosette leaves of at least eight plants were collected for RNA extraction.
RNA Isolation and Quantitative RT-PCR.
Total RNAs were prepared as described in ref. 36, treated with RQ1 RNase-Free DNase (Promega, Madison, WI), and reverse transcribed by using random hexamers and SuperScript II RNase H− Reverse Transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations. Quantitative real-time PCR was performed with equal amounts of cDNAs by using the ABI PRISM 7700 Sequence Detection System (Applied Biosystems, Foster City, CA), a SYBR Green PCR kit from Applied Biosystems, and gene-specific primers. Relative mRNA abundance was calculated by using the comparative delta-Ct method and normalized to the profilin 1 (At2g19760) gene levels. Profilin 1 is not affected by paraquat treatment or in flu upon a dark-to-light shift, unlike the commonly used actin 2 (data not shown).
Array Hybridization and Evaluation.
Affymetrix Arabidopsis ATH1 GeneChips were used throughout the experiment. Experimental procedures are described in SI Text and according to Minimum Information about a Microarray Experiment standards for plant genomics (37, 38).
Microarray Data Analysis.
Microarray data were analyzed further with GeneSpring GX 7.3.1 software (Silicon Genetics). Raw data were preprocessed from Affymetrix CEL files by using GCRMA (39). To adjust for differences in labeling and detection efficiencies, data were normalized by using the Affymetrix standard normalization for one-color data as follows: data transformation-set measurements <0.01 to 0.01; Per Chip: Normalize to 50th percentile; and Per Gene: Normalize to median, cutoff = 10 in raw data. In addition, only those transcripts that were called present or marginal in at least both duplicates of the same line were taken into account. A cutoff value of 2-fold change was adopted to identify genes that were differentially expressed in flu compared with wild type after the dark–light shift. Only genes that showed in each duplicate of pairwise comparisons to wild type at least a 2-fold change (biological replicate A compared with A; biological replicate B compared with B) were considered as robustly regulated. This analysis identified a set of 1,356 genes showing a robust change of expression in flu after a dark–light shift.
For k means clustering analysis, only those genes were selected that were called present or marginal in both duplicates of all lines. k means was applied by GeneSpring with standard correlation as a mean to divide genes into groups based on their expression patterns.
Cell Death Measurements.
The cell death reaction was monitored by electrolyte leakage measurements. Measurements were done with a conductivity cell (TetraCon-325, Universal Pocket Multiline P4; WTW, Weilheim, Germany). Mature 3-week-old plants grown under continuous light were transferred for 8 h to the dark. At the end of the dark period, rosette leaves were cut and floated on distilled water in transparent containers. After 30 min of preincubation in the dark, the containers were transferred to the light for up to 24 h. The conductivity of the solutions was determined at different time points. The maximum electrolyte content was obtained by boiling the samples for 25 min at 100°C. The electrolyte leakage rate was compared between plant lines in four independent experiments by using the ANOVA test (SAS Software; SAS Institute, Cary, NC). Plant lines were compared by using least-square means of electrolyte leakage rate separately for each time point after reillumination.
Growth Measurements.
Growth of the primary stem was determined by using an extensometer device coupled to a laser deflection system as described in ref. 11.
Extraction and Measurement of Protochlorophyllide.
Tetrapyrroles were extracted from rosette leaves with 80% acetone supplemented with ammonia to a final concentration of 0.1% (vol/vol). Porphyrins were separated on a C18 reverse-phase silica-gel column (Nucleosil ODS 5 μm, 250 × 4.6 mm; Machery Nagel, Duren, Germany), and protochlorophyllide was detected by its fluorescence by using the 430-nm excitation and 630-nm emission wavelengths.
Supplementary Material
Acknowledgments
We thank André Imboden for his help with the plants and growth measurements and Dieter Rubli for photographs. This work was supported by grants from Eidgenössische Technische Hochschule Zurich and the Swiss National Science Foundation.
Abbreviation
- ROS
reactive oxygen species.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609063103/DC1.
References
- 1.Foyer CH, Noctor G. New Phytol. 2000;146:359–388. [Google Scholar]
- 2.Apel K, Hirt H. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 3.Dat J, Vandenabeele S, Vranova E, Van Montagu M, Inze D, Van Breusegem F. Cell Mol Life Sci. 2000;57:779–795. doi: 10.1007/s000180050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laloi C, Apel K, Danon A. Curr Opin Plant Biol. 2004;7:323–328. doi: 10.1016/j.pbi.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Asada K. Plant Physiol. 2006;141:391–396. doi: 10.1104/pp.106.082040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corpas FJ, Barroso JB, del Rio LA. Trends Plants Sci. 2001;6:145–150. doi: 10.1016/s1360-1385(01)01898-2. [DOI] [PubMed] [Google Scholar]
- 7.Asada K. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:601–639. doi: 10.1146/annurev.arplant.50.1.601. [DOI] [PubMed] [Google Scholar]
- 8.Mehler AH. Arch Biochem. 1951;33:65–77. doi: 10.1016/0003-9861(51)90082-3. [DOI] [PubMed] [Google Scholar]
- 9.Fryer MJ, Oxborough K, Mullineaux PM, Baker NR. J Exp Bot. 2002;53:1249–1254. [PubMed] [Google Scholar]
- 10.Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K. Proc Natl Acad Sci USA. 2001;98:12826–12831. doi: 10.1073/pnas.221252798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.op den Camp RG, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Gobel C, Feussner I, et al. Plant Cell. 2003;15:2320–2332. doi: 10.1105/tpc.014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner D, Przybyla D, op den Camp R, Kim C, Landgraf F, Lee KP, Wursch M, Laloi C, Nater M, Hideg E, Apel K. Science. 2004;306:1183–1185. doi: 10.1126/science.1103178. [DOI] [PubMed] [Google Scholar]
- 13.Babbs CF, Pham JA, Coolbaugh RC. Plant Physiol. 1989;90:1267–1270. doi: 10.1104/pp.90.4.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murgia I, Tarantino D, Vannini C, Bracale M, Carravieri S, Soave C. Plant J. 2004;38:940–953. doi: 10.1111/j.1365-313X.2004.02092.x. [DOI] [PubMed] [Google Scholar]
- 15.Yao N, Greenberg JT. Plant Cell. 2006;18:397–411. doi: 10.1105/tpc.105.036251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hideg E, Kalai T, Hideg K, Vass I. Biochemistry. 1998;37:11405–11411. doi: 10.1021/bi972890+. [DOI] [PubMed] [Google Scholar]
- 17.Hideg E, Spetea C, Vass I. Photosyn Res. 1994;39:191–199. doi: 10.1007/BF00029386. [DOI] [PubMed] [Google Scholar]
- 18.Hideg E, Barta C, Kalai T, Vass I, Hideg K, Asada K. Plant Cell Physiol. 2002;43:1154–1164. doi: 10.1093/pcp/pcf145. [DOI] [PubMed] [Google Scholar]
- 19.Hideg E, Kalai T, Hideg K, Vass I. Philos Trans R Soc London B. 2000;355:1511–1516. doi: 10.1098/rstb.2000.0711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klotz LO. Biol Chem. 2002;383:443–456. doi: 10.1515/BC.2002.047. [DOI] [PubMed] [Google Scholar]
- 21.Ryter SW, Tyrrell RM. Free Radic Biol Med. 1998;24:1520–1534. doi: 10.1016/s0891-5849(97)00461-9. [DOI] [PubMed] [Google Scholar]
- 22.Tyrrell RM. Methods. 1997;11:313–318. doi: 10.1006/meth.1996.0425. [DOI] [PubMed] [Google Scholar]
- 23.Valencia A, Moran J. Free Radic Biol Med. 2004;36:1112–1125. doi: 10.1016/j.freeradbiomed.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Creissen G, Firmin J, Fryer M, Kular B, Leyland N, Reynolds H, Pastori G, Wellburn F, Baker N, Wellburn A, Mullineaux P. Plant Cell. 1999;11:1277–1292. doi: 10.1105/tpc.11.7.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anthony JR, Warczak KL, Donohue TJ. Proc Natl Acad Sci USA. 2005;102:6502–6507. doi: 10.1073/pnas.0502225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karpinska B, Wingsle G, Karpinski S. IUBMB Life. 2000;50:21–26. doi: 10.1080/15216540050176548. [DOI] [PubMed] [Google Scholar]
- 27.Noctor G, Foyer CH. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- 28.Mou Z, Fan W, Dong X. Cell. 2003;113:935–944. doi: 10.1016/s0092-8674(03)00429-x. [DOI] [PubMed] [Google Scholar]
- 29.Gomez LD, Vanacker H, Buchner P, Noctor G, Foyer CH. Plant Physiol. 2004;134:1662–1671. doi: 10.1104/pp.103.033027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ball L, Accotto GP, Bechtold U, Creissen G, Funck D, Jimenez A, Kular B, Leyland N, Mejia-Carranza J, Reynolds H, et al. Plant Cell. 2004;16:2448–2462. doi: 10.1105/tpc.104.022608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomez LD, Noctor G, Knight MR, Foyer CH. J Exp Bot. 2004;55:1851–1859. doi: 10.1093/jxb/erh202. [DOI] [PubMed] [Google Scholar]
- 32.Kiddle G, Pastori GM, Bernard S, Pignocchi C, Antoniw J, Verrier PJ, Foyer CH. Antioxid Redox Signal. 2003;5:23–32. doi: 10.1089/152308603321223513. [DOI] [PubMed] [Google Scholar]
- 33.Millar AH, Mittova V, Kiddle G, Heazlewood JL, Bartoli CG, Theodoulou FL, Foyer CH. Plant Physiol. 2003;133:443–447. doi: 10.1104/pp.103.028399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pastori GM, Kiddle G, Antoniw J, Bernard S, Veljovic-Jovanovic S, Verrier PJ, Noctor G, Foyer CH. Plant Cell. 2003;15:939–951. doi: 10.1105/tpc.010538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pignocchi C, Foyer CH. Curr Opin Plant Biol. 2003;6:379–389. doi: 10.1016/s1369-5266(03)00069-4. [DOI] [PubMed] [Google Scholar]
- 36.Melzer S, Majewski DM, Apel K. Plant Cell. 1990;2:953–961. doi: 10.1105/tpc.2.10.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 38.Zimmermann P, Schildknecht B, Craigon D, Garcia-Hernandez M, Gruissem W, May S, Mukherjee G, Parkinson H, Rhee S, Wagner U, Hennig L. Plant Methods. 2006;2:1. doi: 10.1186/1746-4811-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu Z, Irizarry R, Gentleman R, Murillo F, Spencer F. Technical Report. Baltimore, MD: The John Hopkins University; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.