Abstract
The molecular mechanism of neurodegeneration in transmissible spongiform encephalopathies remains uncertain. In this study, it was demonstrated that prion-infected hypothalamic neuronal GT1 cells displayed a higher sensitivity to induced oxidative stress over noninfected cells. In addition, the infected cells presented an increased lipid peroxidation and signs of apoptosis associated with a dramatic reduction in the activities of the glutathione-dependent and superoxide dismutase antioxidant systems. This study indicates for the first time that prion infection results in an alteration of the molecular mechanisms promoting cellular resistance to reactive oxygen species. This finding is vital for future therapeutic approaches in transmissible spongiform encephalopathies and the understanding of the function of the prion protein.
The cellular form of the prion protein (PrPC) is a highly conserved cell surface glycoprotein expressed by a broad range of cells and in particular by neuronal cells. In transmissible spongiform encephalopathies (TSEs), this molecule is converted into a conformationally modified protease-resistant isoform called PrPSc (1). The relationship between the PrPSc presence and neurodegeneration is not absolute. In fact, it was shown in vivo that PrPSc deposition in neuronal tissue not expressing PrPC has no pathological consequence (2). Moreover, in both infectious and genetic models of TSEs, neurodegeneration has been detected in the absence of observable protease-resistant PrPSc (3, 4). Recently, it was proposed that a putative transmembrane form of the prion protein (PrP), and doppel, a PrP-related molecule, could be responsible for neurodegeneration in transgenic mice (4, 5), but it is not clear how these two molecules may be implicated in authentic TSEs.
Although advances have been made in understanding prion diseases, the function of PrPC remains elusive. Studies based on structural homology (6) have revealed limited information concerning the function of the protein. However, it was shown that the octapeptide repeat region of the molecule binds Cu (7) and it has been proposed that PrPC may play a role in the oxidative state of the cell through a regulation of the copper transport (8) and/or through a modification of Cu/Zn superoxide dismutase activity (7). Because increasing data suggests that oxidative stress plays a role in other neurodegenerative diseases, such as Parkinson's and Alzheimer's disease and amyotrophic lateral sclerosis (9), it appeared to be important to examine the role of PrPC and the consequences of its conversion into PrPSc in the defense of the cell against oxidative stress. By using infected cell lines recently developed in the laboratory (10), we were able to demonstrate that prion infection renders neuronal cells more susceptible to oxidative stress and impairs their free radical metabolism.
Materials and Methods
Reagents and Antibodies.
Pefabloc and proteinase K were purchased from Boehringer Mannheim. DMEM was from Life Technologies (Grand Island, NY), and FCS from BioWhittaker. 3-Morpholinosydnonimine (SIN-1) was from Molecular Probes, buthionine sulfoximine (BSO), N-acetylcysteine, and 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) were from Sigma. Tris hydroxymethyl-aminomethane, thiobarbituric acid, azide, 70% perchloric acid, and potassium cyanide were from Merck. NADPH was from Randox (Crumlin, U.K.) and 1-butanol from Prolabo (Paris). All other reagents were from Sigma.
Rabbit polyclonal antibody P45–66, raised against synthetic peptide encompassing mouse PrP residues 45–66, has been described earlier (11). Scrapie-associated fibril (SAF) 60, SAF 69, and SAF 70 are three mAb produced by J. Grassi and coworkers (Commissariat à l'Energie Atomique-Saclay, Gif sur Yvette, France). They were obtained by using as immunogen SAF that were prepared from infected hamster brains. In enzyme immunometric assays, SAF 60, SAF 69, and SAF 70 were characterized as recognizing the peptide epitope 142–160 of hamster PrP (J. Grassi, personal communication). A mixture consisting of an equal volume of ascitis of SAF 60, SAF 69, and SAF 70 antibodies was used to improve PrP detection. Cu/Zn superoxide dismutase (Cu/Zn SOD) polyclonal antibody was from Calbiochem. Secondary antibodies were from Jackson ImmunoResearch.
Cell Culture.
Infection of GT1–7 cell lines with Chandler (GT1Chl), 22L (GT122L), and 87V (GT187V) strains is reported elsewhere (10). To generate the two other GT1 cell lines used in this work, GT1Mock and GT1FK, 10% brain homogenate (in PBS containing 5% glucose) of control uninfected C57/Bl6 mice and Fukuoka-1-infected mice were used. Briefly, about 1 × 105 GT1–7 cells in a 35-mm culture dish were incubated with 1 ml of the diluted homogenate at 0.2% in serum-free DMEM for 5 h, to which was added 1 ml of DMEM/10%FCS. Once the cells had become confluent, they were split into a 25-cm2 culture flask. PrPSc was tested after 5 and 10 passages by immunoblotting as described below. PrPSc was not detected in the control GT1Mock cells, whereas the infection of GT1FK was successful. These ex vivo transmission experiments have been repeated three times with the same outcome.
The cell lines were routinely cultured in DMEM supplemented with 10% heat-inactivated FCS and penicillin-streptomycin and were maintained at 37°C in 5% CO2 in the biohazard P3 laboratory of our institute.
Western Blotting.
Cells at 90% confluence were washed two times into PBS and lysed for 30 min at 4°C in Triton/deoxycholate lysis buffer [150 mM NaCl/0.5% Triton X-100/0.5% sodium deoxycholate/50 mM Tris⋅HCl (pH 7.5)], plus protease inhibitors (1 μg/ml pepstatin and leupeptin/2 mM EDTA). After 1 min of centrifugation at 10,000 × g, the supernatant was collected and its total protein concentration measured by using a bicinchoninic acid protein assay (Pierce). Protein concentration was adjusted with lysis buffer. For detection of PrPC and Cu/Zn SOD, an equivalent volume of the samples were directly mixed with the same volume of 2X SDS loading buffer.
For detection of PrPSc, equivalent volumes of the samples were digested with 16 μg of proteinase K per mg of total protein at 37°C for 30 min, and the digestion was stopped by incubating with 1 mM Pefabloc for 5 min on ice. The samples were centrifuged at 20,000 × g for 45 min at 4°C, and the pellet resuspended in 30 μl of SDS loading buffer.
Samples were applied onto 12% SDS/PAGE and the proteins were transferred onto a membrane (Immobilon-P, Millipore) in 3-(cyclohexylamino)-1-propanesulfonic acid buffer containing 10% methanol at 400 mA for 1 h. The membrane was blocked with 5% nonfat dry milk in TBST [0.1% Tween 20/100 mM NaCl/10 mM Tris⋅HCl (pH 7.8)] for 1 h at room temperature. Nondigested mouse PrPC was detected by immunoblotting by using P45–66 as described (11) and mouse PrPSc was detected by using a mixture of three mAb, SAF 60, SAF 69, and SAF 70 (mixture of ascitic fluids diluted 1/200 in TBST). The anti-Cu/Zn SOD polyclonal antibody was used at 1/500 in 3% nonfat dry milk in TBST.
DNA Fragmentation Assay.
DNA fragmentation was demonstrated in infected cells by the DNA-laddering technique modified from Schatzl et al. (12). Briefly, cells from a 125-cm2 flask were washed twice with cold PBS and lysed in 500 μl of hypotonic lysis buffer for 5 min [5 mM Tris⋅HCl (pH 7.5)/20 mM EDTA/0.5% Triton-X100]. The lysates were centrifuged at 14,000 rpm for 30 min. The supernatants were deproteinized by digestion with 0.3 mg/ml proteinase K for 30 min at 60°C and extracted once in phenol/chloroform and once in chloroform/isoamylalcohol (24:1), and then precipitated in 2.5 volumes of 100% ethanol and 0.1 volume of 3 M sodium acetate. After centrifugation at 12,000 rpm at 4°C for 30 min, the pellets were washed in 70% ethanol and resuspended in 25 μl of Tris/EDTA buffer (pH 7.5) and 60 μg/ml RNase A for 1 h at 37°C; 20 μl were supplemented with gel-loading buffer, subjected to electrophoresis on a 1.5% agarose gel, and stained with ethidium bromide.
Cell Viability Assay.
Cell viability was assessed by using a modified MTT assay from Hansen et al. (13). MTT is a sensitive first indicator of mitochondrial damage induced by oxidative stressors (14). Briefly, around 10,000 GT1 cells per well were plated in 96-well microtiter plates with 100 μl of medium. The next day, the medium was changed and the cells were challenged for 24 h with the indicated concentration of BSO or SIN-1. The medium was then changed and the cells were incubated for an additional 24 h without the drugs. For the MTT assay, 10 μl of MTT (5 mg/ml stock in PBS) was added to each well for 1 h at 37°C; 100 μl of 50% DMSO/5% SDS solubilization solution was then added, and adsorption readings were performed at 570 nm with reference at 630 nm.
Lipid Peroxidation.
The lipid peroxidation was evaluated by using an assay based on fluorescence of a malondialdehyde thiobarbituric acid adduct measured after extraction with 1-butanol (15). Subconfluent cells were trypsinized in 75-cm2 flasks, washed three times by 10 ml of isotonic, trace element-free 400 mM Tris⋅HCl (pH 7.3), and then lysed in hypotonic 20 mM Tris⋅HCl buffer by five freeze-defrost cycles; 750 μl of a mixture of thiobarbituric acid at 8 g/7% perchloric acid (2/1) were added to 100 μl of sample. After agitation, the mixture was placed in a 95°C water bath for 60 min and then cooled in an ice bath. The fluorescent compound was extracted by mixing with 1-butanol for 2 min. After centrifugation, the fluorescence in the 1-butanol phase was determined with an Aminco-Bowman fluorimeter with excitation at 532 nm and emission at 553 nm. A blank was run for each sample. The calibration curve was created with a stock solution of 1,1,3,3-tetraethoxypropane prepared in alcohol. The results were expressed as μmol malondialdehyde/g protein.
SOD Activity.
For SOD activity, the method from Oberley et al. (16) was first used. After two washes in PBS, the cells were sonicated in 0.015 M sodium phosphate buffer (pH 7.8) and centrifuged at 4,000 rpm for 4 min. The supernatant was then tested for total SOD activity. To determine the Mn SOD activity, the supernatant was incubated with 4 mM potassium cyanide for 20 min before testing to inhibit the Cu/Zn SOD activity. One unit of SOD is given as that level allowing for a 50% inhibition of formazan production. The specific activity was determined as units/mg protein.
As a control, an alternative method was used as follows. Subconfluent cells in 75 cm2 flasks, were washed three times and collected in 10 ml of isotonic, trace element-free 400 mM Tris⋅HCl (pH 7.3), and then lysed in hypotonic 20 mM Tris⋅HCl buffer by five freeze-defrost cycles. After 10 min of centrifugation at 4,000 rpm and 4°C, the lysate was assayed for metalloenzyme activities and soluble protein content. Total SOD, Mn SOD, and Cu/Zn SOD were determined by using the pyrogallol assay following the procedure described by Marklund and Marklund (17), based on the competition between pyrogallol oxidation by superoxide radicals and superoxide dismutation by SOD, spectrophotometrically read at 420 nm. Briefly, 50 μl of the sample was added with 1,870 μl of 50 mM Tris/1 mM diethylenetriaminepentaacetic acid/20% cacodylic acid buffer (pH 8.3) and with 80 μl of 10 mM pyrogallol to induce an absorbance change of 0.02 in the absence of SOD. The amount of SOD inhibiting the reaction rate by 50% in the given assay conditions was defined as one SOD unit. The specific Cu/Zn SOD inhibition by KCN (60 μl of 54 mM KCN added to 300 μl of lysate) allows the Mn SOD determination in the same condition. Each sample was assayed twice, and results were expressed as SOD units and normalized to the cell protein content.
Analysis of Glutathione (GSH)-Dependent Antioxidant System.
For the determination of total GSH levels, after washing twice in PBS, confluent 25-cm2 cells were scraped into 1 ml of PBS, centrifuged at 7,000 rpm for 5 min, the pellet was resuspended in 100 μl of cold H2O, and vortexed for 5 min; 5 μl was then removed for protein determination (bicinchoninic acid) and 95 μl of cold 10% trichloroacetic acid (wt/vol) was added. After 10 min at 4°C, the solution was spun at 3,000 rpm for 15 min and the supernatant was then assayed for total GSH content according to the method of Akerboom and Sies (18).
The glutathione peroxidase (GPX) activity was assayed by the method of Gunzler et al. (19). GPX was measured in a coupled reaction with glutathione reductase (GR). Tert-butyl hydroperoxide was the substrate. Briefly, 25 μl of the sample was added with 900 μl of 50 mM Tris/1 mM Na2 EDTA/4 mM azide buffer (pH 7.6) (azide was included in the assay mixture to inhibit interference of catalase) and with 20 μl of 0.15 M GSH/20 μl of 200 units/ml GR/20 μl of 8.4 mM NADPH2 in order and left 1 min for mixture equilibrium. Tert-butyl hydroperoxide (20 μl) was then added and the decrease in absorbance was monitored for 200 s. The difference in absorbance per min was used to calculate the enzyme activity and results were expressed as GPX units/g protein.
GR activity was determined by following the oxidation of NADPH to NADP+ during the reduction of oxidized GSH (20). The main reagent was prepared by combining 18 ml of 139 mM KH2PO4/0.76 mM EDTA (pH 7.4)/2 ml of 2.5 mM NADPH2. Sample (20 μl) was added with 220 μl of the main reagent, and then 30 μl of 22 mM oxidized GSH plus 10 μl of KH2PO4 were added to start the reaction. The absorbance was followed at 340 nm for 175 s. The difference in absorbance per min was used to calculate the activity of the enzyme. The results were expressed as units/g protein GR.
Results and Discussion
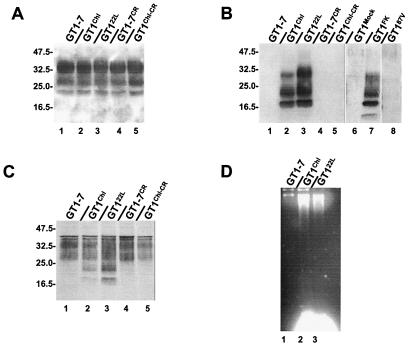
In an attempt to evaluate the role of prion generation in neurodegeneration, the sensitivity to oxidative stress of prion-infected cultures was tested. GT1–7 cells, a subcloned cell line of immortalized hypothalamic GT1 cells and a kind gift from D. Holtzman (Washington University, St. Louis), infected with two prion strains, Chandler (GT1Chl) and 22L (GT122L), were recently developed in the laboratory (10). These cell lines were not transfected, or subcloned after infection and therefore represented a good culture model avoiding clonal differences (21). Although the total amount of full-length PrPC was not modified between control and infected cells (Fig. 1A), infection was associated with the accumulation of an amino terminally truncated molecule (Fig. 1B) (22) that may be generated by an endogenous metaloprotease (23), and by the detection of the protease-resistant PrP isoform, PrPSc (Fig. 1C). As it is shown by us (Fig. 1D) and by another group (12) that infected GT1 cells exhibited signs of apoptosis, it indicated that such infected cells would represent a suitable in vitro model to study prion infection. In this study, Congo red (CR)-treated GT1Chl cells (GT1Chl-CR) were used as a control because treatment with CR (1 μg/ml) allowed for a cessation and removal of PrPSc in GT1Chl cells (24) (Fig. 1B). In addition, we also used three new GT1 cell lines, GT1Mock, GT187V, and GT1FK, with only the latter being successfully infected and producing PrPSc (Fig. 1B).
Figure 1.
Cell lysates of GT1 lines were loaded onto 12% polyacrylamide gel, transferred onto poly(vinylidene difluoride) membrane, and PrPC detected with antibody P45–66 raised against the amino terminus of the protein (A) or with a mixture of mAb (SAF 60, SAF 69, and SAF 70) raised against the carboxy terminus of the protein (C). PrPSc was detected with the same mix of mAbs but after proteinase K digestion (B). Molecular mass markers are indicated on the left in kDa. In D, DNA fragmentation was demonstrated in infected cells by the DNA laddering technique.
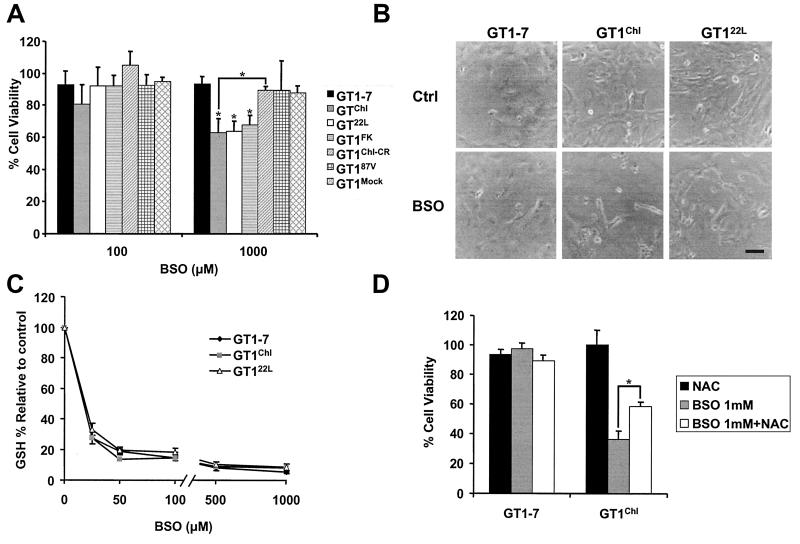
In a first attempt to evaluate the variation in cellular redox state between infected and control cells, the lipid peroxydation was measured by determining the formation of malondialdehyde (25). Cellular damages were significantly higher in GT1Chl cells when compared with GT1–7 (Table 1), indicating that the oxidative state was modified in infected cells. Interestingly, GT1 cells have previously been used to investigate neuronal cell death, Bcl-2 function, and reactive oxygen species (ROS)-induced stress (14, 26). These cells have been shown to display high sensitivity to the toxicity from BSO, a γ-glutamylcysteine synthetase inhibitor that results in the depletion of GSH in the cytosol and mitochondria of cells (14). To test the consequences of prion replication, infected and control GT1 cells were treated with BSO (100 μM or 1,000 μM) for 24 h and their viability assessed after 48 h by a modified tetrazolium salt (MTT) assay, an assay of mitochondrial activity proportional to cell viability (13). Each of GT1Chl, GT122L, and GT1FK displayed a concentration-dependent sensitivity to BSO that was significantly higher than that of the noninfected and the control cell lines (GT1Chl-CR, GT1Mock, and GT187V) (Fig. 2A). These results were confirmed by using as additional cell survival test, a Trypan blue exclusion assay (data not shown), and it is noteworthy that our results have been reproduced in several independently cultured cell lines. A visual illustration of BSO-induced neuronal death in infected cells was obtained in phase contrast microscopy (Fig. 2B). Because BSO sensitivity differed between the infected and noninfected cells, it was important to measure GSH levels before and after the treatment (18). GT1–7, GT122L, and GT1Chl all had similar GSH content and exposure to increasing concentrations of BSO had an equivalent effect on the depletion of GSH between the cell types (Fig. 2C). This indicated that differences among cell lines were not related to various levels of available GSH. Importantly, N-acetylcysteine, a known precursor of GSH and a ROS scavenger, allowed for protection against BSO-induced toxicity in our model (Fig. 2D). Taken together, our results suggest that prion infection increases sensitivity of neuronal cells to oxidative stress and ROS are an important death factor in TSEs.
Table 1.
Determination of lipid peroxidation and SODs, GPX, GR activities
| Parameter | GT1-7 | GT1Chl |
|---|---|---|
| MDA | 69.3 ± 13.9 | 113.3 ± 8.0** |
| Total SOD | 15.2 ± 0.4 | 9.9 ± 1.6* |
| Mn SOD | 6.4 ± 0.6 | 4.1 ± 04* |
| GPX | 40.4 ± 2.6 | 20.1 ± 4.6** |
| GR | 368.4 ± 97.1 | 174.1 ± 57.7* |
Data are expressed as the mean ± SD for three independent determinations. MDA is in nmol/g of protein; SOD activities are in unit/mg of protein; and GPX and GR activities are in unit/g of protein. Significantly different from GT1-7 value (*, P < 0.05; **, P < 0.01, Student's t test).
Figure 2.
(A) Cell viability evaluated by a modified MTT assay was measured in GT1 cell lines after BSO challenge. Each of the infected lines, GT1Chl, GT122L, and GT1FK, presented an increased sensitivity to 1,000 μM BSO when compared with control GT1–7, GT187V, and GT1Mock. CR-treated cells, GT1Chl-CR, recovered a normal phenotype when compared with infected GT1Chl cells. [Bars represent means ± SD; and *, P < 0.01 (Student's t test)]. (B) The phase contrast microscopy of BSO-challenged GT1 lines showed an intense cell death in the infected cell lines GT1Chl and GT122L when compared with control GT1–7. (C) Total glutathione levels (GSH) were similar in all cell lines and decreased alike on treatment with various concentrations of BSO. (D) N-acetylcysteine preincubation (1 h; 1 mM) showed a significant protective effect on cell viability in the presence of 1 mM BSO. [Bars represent means ± SD; and *, P < 0.01 (Student's t test)].
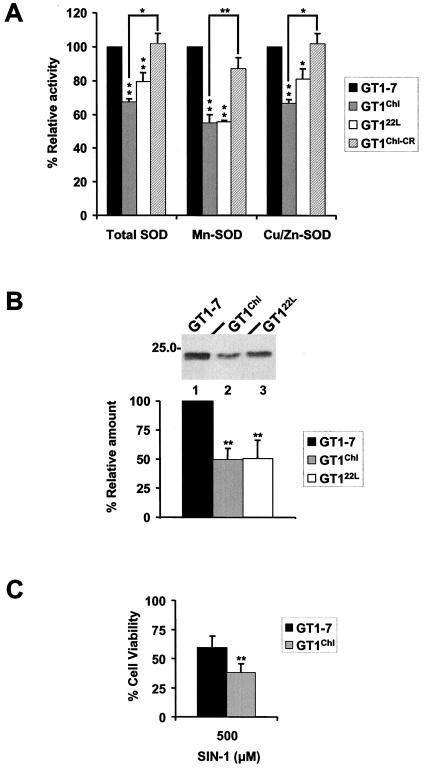
ROS toxicity has been implicated in several other neurological disorders. Such toxicity could result from an imbalance between antioxidant enzymes which normally have an appropriate relationship to each other (27). Among the multileveled interdependent antioxidant systems, which aerobic cells have evolved to protect against oxidant injury, is the family of SODs, which catalyzes the conversion of the superoxide ion (O2⨪) into hydrogen peroxide (H2O2). It has been suggested that PrPC itself may play a role in the response of the cell to oxidative stress possibly through its involvement in copper metabolism and/or in SOD activity. Indeed, it was shown in various experimental models that cells derived from PrP°/° mice were more sensitive to oxidative stress, copper toxicity, and had a reduced SOD activity when compared with control cells (7, 28, 29). Finally, modulation of SOD activity by PrP has been reported to result from an alteration in the incorporation of copper in this enzyme (7). However, these latter data were not confirmed recently (30) and analyses in affected brains have not permitted a verification of these data (31). To investigate further whether increased sensitivity to stress of infected cells may be linked to an alteration in their enzymatic defenses, SOD activities present in the cultures were measured by using the xanthine and Nitro Blue Tetrazolium technique (16). A significant reduction in both Cu/Zn and Mn SOD activities was observed in both GT122L and GT1Chl when compared with GT1–7 and GT1Chl-CR control cells (Fig. 3A). The latter results were also reproduced by using an alternative method of SOD detection, that of autooxidation of pyrogallol (25) (Table 1). Interestingly, Western blot detection of Cu/Zn SOD in GT1 lines indicated that the total level of the protein was significantly reduced in infected cells (Fig. 3B). This may either reflect a down-regulation of expression of the enzyme or a degradation because of fragmentation under conditions of higher oxidative stress (32). Either way, the consequences of lowered SOD activity is an increased O2⨪ that combined with nitric oxide could produce peroxynitrite the toxicity of which is well recognized (33). Interestingly, SIN-1, which generates nitric oxide (14) induced in GT1 cells a rapid and reproducible neuronal cell death (Fig. 3C) that was higher in infected cells. This confirmed the higher sensitivity of infected cells to oxidative stress and suggests that peroxynitrite may play an important role in the neuropathology of TSEs.
Figure 3.
(A) SOD activities present in the cultures were measured by using the xanthine and Nitro Blue Tetrazolium technique. Total, Mn, and Cu/Zn SOD activities are all significantly lower in both infected lines, GT1Chl and GT122L, when compared with control lines GT1–7 and GT1Chl-CR lines. [Bars represents means ± SD; *, P < 0.05; and **, P < 0.01 (Student's t test)]. (B) Cell lysates of GT1 lines were prepared as in Fig. 1A and the total protein concentration measured by using the BCA Protein Assay Kit (Pierce). Equal amounts of protein were Western blotted by using an anti-Cu/Zn SOD sheep polyclonal antibody from Calbiochem (Upper). Specific Cu/Zn SOD bands (Upper) and from three other experiments were quantitated by densitometry, and plotted as a percentage of the total amount in GT1–7 cells. Infected cells presented a significant decrease in Cu/Zn SOD signal. [Bars represent means ± SD; and **, P < 0.01 (Student's t test)]. (C) Cell viability evaluated by a modified MTT assay was measured in GT1Chl and GT1–7 cells after SIN-1 challenge. GT1Chl cells presented an increased sensitivity to 500 μM SIN-1 when compared with control GT1–7 cells. [Bars represents means ± SD; and **, P < 0.01 (Student's t test)].
In addition, on further examining the interdependent oxidative enzymes, although GSH levels were equivalent between cells, it was observed that both the GPX and GR activities were significantly reduced in our infected cells (Table 1) (25). We recently confirmed by using N2a cell lines infected with the Chandler prion strain (10) that SOD as well as GPX activities were also affected by prion replication in these cells. Values for control vs. infected N2a cells, calculated as described in Table 1, were for SOD: 24.8 ± 3.7 and 10.8 ± 1.9 (P < 0.01, Student's t test) and for GPX: 454.6 ± 16.3 and 299.1 ± 33.8 (P < 0.01, Student's t test), respectively. Previous results obtained with infected hamsters revealed an increase of the activities of these two enzymes in the brain (34). This discrepancy may be related to the fact that in the latter study, the enzymatic activities included those of glial and microglial cells that are known to be activated in TSEs. In our case, the lowering of GSH enzymes could be explained by a cellular response to a lowered SOD activity (27). However, because the infected cells displayed a higher sensitivity to SIN-1, it is possible that the lowered activities may be caused by an increased protein nitration (35). The latter could also explain the reduced Mn SOD activity (36). Interestingly, it was reported that PrP°/° cells possessed a higher sensitivity to H2O2, which was attributed to a lowered GR altering the cellular redox state (37). Our observation that both GPX and GR were lowered during prion infection suggests, therefore, that prion diseases may evolve from a disturbance of PrPC function by PrPSc.
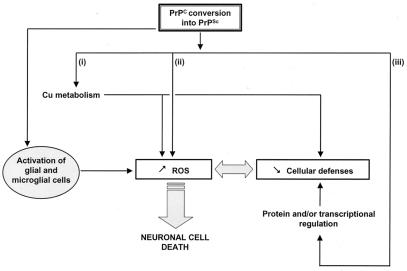
The results presented here demonstrate for the first time that prion-infected cells are more susceptible to imposed oxidative stress caused by disturbance of oxidative defense and suggest that ROS play an important role in TSEs. Three main hypotheses can now be proposed to explain neurodegeneration in prion diseases (Fig. 4): (i) It is possible that the pathological conversion of PrPC to PrPSc may lead to an aberrant copper metabolism altering the cellular redox state and the activities of important oxidative enzymes. However, knowing the recent results (30) obtained in transgenic animals, this may be unlikely and studies on the uptake of Cu by infected cells could test this hypothesis; (ii) The conversion of PrPC to PrPSc may lead to a general increase in ROS levels that would weaken cellular defenses and finally trigger cell death. Recent data showed that PrP itself could have a SOD-like activity (38), conversion into PrPSc could result in loss of activity leading to increased ROS or toxic gain of function as in amyotrophic lateral sclerosis (39); and (iii) on the other hand, modification of the PrP function may result in a modulation of oxidative defenses possibly involving protein regulation or (metal-regulated) transcription factors. This idea is supported by the fact that GPX, GR, Cu/Zn SOD, and Mn SOD partially share common signaling and biochemical pathways. In this case, PrP would function as a specific stress sensor leading to resistance of cells to oxidative stress through a yet unknown mechanism. This exciting idea is reminiscent of “the π receptor hypothesis” formulated by others (40) and we are now looking for a possible signaling pathway in which PrP could be involved.
Figure 4.
Conversion of PrPC into PrPSc increases the sensitivity of neuronal cells to oxidative stress through one or several of the following mechanisms: (i) Cu metabolism could be modified influencing the redox status or the oxidative enzyme systems of the cell; (ii) the production of ROS could be directly modified through, for example, the SOD-like activity of PrP; and (iii) the conversion could act on protein regulation and/or transcription factors involved in response to oxidative stress. Moreover, prion infection activates glial and microglial cells, which likely results in increased ROS production, triggering the death of already impaired neuronal cells.
In conclusion, it is becoming more and more evident that although the etiological factors of neurodegenerative diseases may differ, the neurodegeneration process present in such disorders share some common mechanisms. The implication of ROS and oxidative stress in TSEs allows now the possibility of new therapeutic approaches to substitute and balance antioxidant deficiencies in the disease state.
Acknowledgments
We thank David Harris (Washington University, St. Louis) for antibody P45–66; Jacques Grassi and Yveline Frobert (Commissariat à l'Energie Atomique-Saclay, Gif sur Yvette, France) for SAF 60, SAF 69, and SAF 70 antibodies; and Dr. R. Carp and J. Tateishi for providing infected mouse brains. This work was supported by grants from the Fondation de la Recherche Médicale, Cellule de Coordination Interorganismes sur les Prions, Centre National de la Recherche Scientifique, and the European Community Biotech BIO4CT98–6055 and BIO4CT98–6064. W.R. is supported by the Lebanese National Council for Scientific Research. O.M. is a MNESER fellow.
Abbreviations
- BSO
buthionine sulfoximine
- CR
Congo red
- GPX
glutathione peroxidase
- GR
glutathione reductase
- MTT
3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide
- PrP
prion protein
- PrPC
cellular isoform of PrP
- PrPSc
scrapie isoform of PrP
- ROS
reactive oxygen species
- SIN-1
3-morpholinosydnonimine
- SOD
superoxide dismutase
- TSEs
transmissible spongiform encephalopathies
- SAF
scrapie-associated fibril
- GSH
glutathione
- GT1Chl
GT1 cells infected with Chandler
- GT122L
GT1 cells infected with 22L
- GT187V
GT1 cells infected with 87V
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250289197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250289197
References
- 1.Prusiner S B, Scott M R, Dearmond S J, Cohen F E. Cell. 1998;93:337–348. doi: 10.1016/s0092-8674(00)81163-0. [DOI] [PubMed] [Google Scholar]
- 2.Brandner S, Isenmann S, Raeber A, Fischer M, Sailer A, Kobayashi Y, Marino S, Weissmann C, Aguzzi A. Nature (London) 1996;379:339–343. doi: 10.1038/379339a0. [DOI] [PubMed] [Google Scholar]
- 3.Lasmezas C I, Deslys J P, Robain O, Jaegly A, Beringue V, Peyrin J M, Fournier J G, Hauw J J, Rossier J, Dormont D. Science. 1997;275:402–405. doi: 10.1126/science.275.5298.402. [DOI] [PubMed] [Google Scholar]
- 4.Hegde R S, Mastrianni J A, Scott M R, DeFea K A, Tremblay P, Torchia M, Dearmond S J, Prusiner S B, Lingappa V R. Science. 1998;279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- 5.Moore R C, Lee I Y, Silverman G L, Harrison P M, Strome R, Heinrich C, Karunaratne A, Pasternak S H, Chishti M A, Liang Y, et al. J Mol Biol. 1999;292:797–817. doi: 10.1006/jmbi.1999.3108. [DOI] [PubMed] [Google Scholar]
- 6.Glockshuber R, Hornemann S, Billeter M, Riek R, Wider G, Wuthrich K. FEBS Lett. 1998;426:291–296. doi: 10.1016/s0014-5793(98)00372-x. [DOI] [PubMed] [Google Scholar]
- 7.Brown D R, Besinger A. Biochem J. 1998;334:423–429. doi: 10.1042/bj3340423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pauly P C, Harris D A. J Biol Chem. 1998;273:33107–33110. doi: 10.1074/jbc.273.50.33107. [DOI] [PubMed] [Google Scholar]
- 9.Simonian N A, Coyle J T. Annu Rev Pharmacol Toxicol. 1996;36:83–106. doi: 10.1146/annurev.pa.36.040196.000503. [DOI] [PubMed] [Google Scholar]
- 10.Nishida N, Harris D A, Vilette D, Laude H, Frobert Y, Grassi J, Casanova D, Milhavet O, Lehmann S. J Virol. 2000;74:320–325. doi: 10.1128/jvi.74.1.320-325.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehmann S, Harris D A. J Biol Chem. 1995;270:24589–24597. doi: 10.1074/jbc.270.41.24589. [DOI] [PubMed] [Google Scholar]
- 12.Schatzl H M, Laszlo L, Holtzman D M, Tatzelt J, Dearmond S J, Weiner R I, Mobley W C, Prusiner S B. J Virol. 1997;71:8821–8831. doi: 10.1128/jvi.71.11.8821-8831.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen M B, Nielsen S E, Berg K. J Immunol Methods. 1989;119:203–210. doi: 10.1016/0022-1759(89)90397-9. [DOI] [PubMed] [Google Scholar]
- 14.Kane D J, Sarafian T A, Anton R, Hahn H, Gralla E B, Valentine J S, Ord T, Bredesen D E. Science. 1993;262:1274–1277. doi: 10.1126/science.8235659. [DOI] [PubMed] [Google Scholar]
- 15.Richard M J, Portal B, Meo J, Coudray C, Hadjian A, Favier A. Clin Chem. 1992;38:704–709. [PubMed] [Google Scholar]
- 16.Oberley L W, Spitz D R. Methods Enzymol. 1984;105:457–464. doi: 10.1016/s0076-6879(84)05064-3. [DOI] [PubMed] [Google Scholar]
- 17.Marklund S, Marklund G. Eur J Biochem. 1974;47:469–474. doi: 10.1111/j.1432-1033.1974.tb03714.x. [DOI] [PubMed] [Google Scholar]
- 18.Akerboom T P, Sies H. Methods Enzymol. 1981;77:373–382. doi: 10.1016/s0076-6879(81)77050-2. [DOI] [PubMed] [Google Scholar]
- 19.Gunzler W A, Kremers H, Flohe L. Z Klin Chem Klin Biochem. 1974;12:444–448. doi: 10.1515/cclm.1974.12.10.444. [DOI] [PubMed] [Google Scholar]
- 20.Goldberg D M, Spooner R J. Glutathion Reductase. Basel: Birkhauser; 1983. pp. 258–265. [Google Scholar]
- 21.Bosque P J, Prusiner S B. J Virol. 2000;74:4377–4386. doi: 10.1128/jvi.74.9.4377-4386.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caughey B, Raymond G J, Ernst D, Race R E. J Virol. 1991;65:6597–6603. doi: 10.1128/jvi.65.12.6597-6603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jimenez-Huete A, Lievens P M, Vidal R, Piccardo P, Ghetti B, Tagliavini F, Frangione B, Prelli F. Am J Pathol. 1998;153:1561–1572. doi: 10.1016/S0002-9440(10)65744-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mange A, Nishida N, Milhavet O, McMahon H E M, Casanova D, Lehmann S. J Virol. 2000;74:3135–3140. doi: 10.1128/jvi.74.7.3135-3140.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parat M O, Richard M J, Beani J C, Favier A. Biol Trace Element Res. 1997;60:187–204. doi: 10.1007/BF02784439. [DOI] [PubMed] [Google Scholar]
- 26.Bonfoco E, Zhivotovsky B, Rossi A D, Aguilar-Santelises M, Orrenius S, Lipton S A, Nicotera P. NeuroReport. 1996;8:273–276. doi: 10.1097/00001756-199612200-00054. [DOI] [PubMed] [Google Scholar]
- 27.Amstad P, Moret R, Cerutti P. J Biol Chem. 1994;269:1606–1609. [PubMed] [Google Scholar]
- 28.Brown D R, Schulz-Schaeffer W J, Schmidt B, Kretzschmar H A. Exp Neurol. 1997;146:104–112. doi: 10.1006/exnr.1997.6505. [DOI] [PubMed] [Google Scholar]
- 29.Kuwahara C, Takeuchi A M, Nishimura T, Haraguchi K, Kubosaki A, Matsumoto Y, Saeki K, Matsumoto Y, Yokoyama T, Itohara S, et al. Nature (London) 1999;400:225–226. doi: 10.1038/22241. [DOI] [PubMed] [Google Scholar]
- 30.Waggoner D J, Drisaldi B, Bartnikas T B, Casareno R L, Prohaska J R, Gitlin J D, Harris D A. J Biol Chem. 2000;275:7455–7458. doi: 10.1074/jbc.275.11.7455. [DOI] [PubMed] [Google Scholar]
- 31.Lee D W, Sohn H O, Lim H B, Lee Y G, Kim Y S, Carp R I, Wisniewski H M. Free Radical Res. 1999;30:499–507. doi: 10.1080/10715769900300541. [DOI] [PubMed] [Google Scholar]
- 32.Ookawara T, Kawamura N, Kitagawa Y, Taniguchi N. J Biol Chem. 1992;267:18505–18510. [PubMed] [Google Scholar]
- 33.Troy C M, Derossi D, Prochiantz A, Greene L A, Shelanski M L. J Neurosci. 1996;16:253–261. doi: 10.1523/JNEUROSCI.16-01-00253.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi S I, Ju W K, Choi E K, Kim J, Lea H Z, Carp R I, Wisniewski H M, Kim Y S. Acta Neuropathol. 1998;96:279–286. doi: 10.1007/s004010050895. [DOI] [PubMed] [Google Scholar]
- 35.Asahi M, Fujii J, Suzuki K, Seo H G, Kuzuya T, Hori M, Tada M, Fujii S, Taniguchi N. J Biol Chem. 1995;270:21035–21039. doi: 10.1074/jbc.270.36.21035. [DOI] [PubMed] [Google Scholar]
- 36.Yamakura F, Taka H, Fujimura T, Murayama K. J Biol Chem. 1998;273:14085–14089. doi: 10.1074/jbc.273.23.14085. [DOI] [PubMed] [Google Scholar]
- 37.White A R, Collins S J, Maher F, Jobling M F, Stewart L R, Thyer J M, Beyreuther K, Masters C L, Cappai R. Am J Pathol. 1999;155:1723–1730. doi: 10.1016/S0002-9440(10)65487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown D R, Wong B S, Hafiz F, Clive C, Haswell S J, Jones I M. Biochem J. 1999;344:1–5. [PMC free article] [PubMed] [Google Scholar]
- 39.Ghadge G D, Lee J P, Bindokas V P, Jordan J, Ma L, Miller R J, Roos R P. J Neurosci. 1997;17:8756–8766. doi: 10.1523/JNEUROSCI.17-22-08756.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shmerling D, Hegyi I, Fischer M, Blattler T, Brandner S, Gotz J, Rulicke T, Flechsig E, Cozzio A, von Mering C, et al. Cell. 1998;93:203–214. doi: 10.1016/s0092-8674(00)81572-x. [DOI] [PubMed] [Google Scholar]