Abstract
Syntaxin is a key presynaptic protein that binds to N- and P/Q-type Ca2+ channels in biochemical studies and affects gating of these Ca2+ channels in expression systems and in synaptosomes. The present study was aimed at understanding the molecular basis of syntaxin modulation of N-type channel gating. Mutagenesis of either syntaxin 1A or the pore-forming α1B subunit of N-type Ca2+ channels was combined with functional assays of N-type channel gating in a Xenopus oocyte coexpression system and in biochemical binding experiments in vitro. Our analysis showed that the transmembrane region of syntaxin and a short region within the H3 helical cytoplasmic domain of syntaxin, containing residues Ala-240 and Val-244, appeared critical for the channel modulation but not for biochemical association with the “synprint site” in the II/III loop of α1B. These results suggest that syntaxin and the α1B subunit engage in two kinds of interactions: an anchoring interaction via the II/III loop synprint site and a modulatory interaction via another site located elsewhere in the channel sequence. The segment of syntaxin H3 found to be involved in the modulatory interaction would lie hidden within the four-helix structure of the SNARE complex, supporting the hypothesis that syntaxin's ability to regulate N-type Ca2+ channels would be enabled after SNARE complex disassembly after synaptic vesicle exocytosis.
Syntaxin (1, 2), a key component of the SNARE core complex involved in synaptic vesicle docking or fusion (3, 4), binds to multiple synaptic proteins (5) and functions as an organizing center for exocytosis (6). Activation of exocytosis at presynaptic terminals in brain is dominated by Ca2+ influx through N- and P/Q-type Ca2+ channels (7–10). Syntaxin binds to N- and P/Q-type Ca2+ channels in biochemical studies (1, 2, 11–16). At fast synapses, the simultaneous association of syntaxin with synaptic vesicle proteins and voltage-gated Ca2+ channels helps anchor synaptic vesicles near Ca2+ entry sites (14, 16, 17), thereby safeguarding the speed and efficiency of Ca2+-triggered exocytosis (18).
In addition to serving an anchoring role, interactions between syntaxin and Ca2+ channels may also cause inhibition of Ca2+ channels in expression systems (6, 19–23) as well as in isolated nerve terminals (24). Syntaxin acts by promoting the slow inactivation of Ca2+ channels (25). The modulatory action of syntaxin is affected by synaptotagmin, SNAP-25 (21, 26, 27), and possibly cysteine string protein (28, 29). Thus, syntaxin–channel interactions not only serve an anchoring function but may also facilitate reverse communication from the secretory machinery to Ca2+ channels (6, 24, 29). Recent reports indicate that syntaxin exerts similar inhibitory effects on the cystic fibrosis transmembrane regulator (30) and influences epithelial sodium channels (31), whereas SNAP-25 modulates P/Q-type Ca2+ channels (32). Thus, the ability of SNARE proteins to influence plasma membrane ion channels may be a general phenomenon.
The structural determinants responsible for interactions between syntaxin and α1B, the pore-forming subunit of N-type Ca2+ channels, have already been partially characterized. Syntaxin 1A (35 kDa) consists of four α-helical regions in its cytosolic N-terminal domain (HA, HB, HC, and H3) and a short C-terminal transmembrane domain (1, 2, 33, 34). The H3 domain seems most critical for interactions with other synaptic proteins, including α1B (1, 2, 11, 12). Within the α1B subunit (262 kDa), an important structural feature is the syntaxin-interaction (“synprint”) site in the large cytosolic loop between repeats II and III, first identified by Catterall's group in biochemical experiments with recombinant proteins in vitro (15). There are clear functional advantages to a structural arrangement that would allow modulatory effects of syntaxin on Ca2+ entry to vary with vesicular status while leaving the colocalization of syntaxin and α1B undisturbed. However, little is known about the structural basis of the modulatory interaction. Are the anchoring and modulatory interactions between syntaxin and Ca2+ channels mediated by the same molecular determinants? What is the role of syntaxin's transmembrane region? How might other syntaxin binding partners influence syntaxin inhibition? Here we combine site-directed mutagenesis with functional expression in Xenopus oocytes to address such questions.
Materials and Methods
Expression in Xenopus Oocytes.
N-type Ca2+ channels (α1B, β3, and α2/δ) were coexpressed in Xenopus oocytes with rat syntaxin 1A or its mutants (19). Properties of expressed N-type Ca2+ channels were evaluated in two-electrode voltage clamp experiments with 5 mM Ba2+ as charge carrier (19). The recordings were carried out 2–4 days after the second cRNA injection. pclamp6 software (Axon Instruments, Foster City, CA) was used to digitize, store, and analyze current traces. Leak and capacitance currents were subtracted on line with a P/6 protocol.
Syntaxin 1A Mutagenesis.
Rat syntaxin 1A (1) mutants were generated by PCR, subcloned into pGEMHE (35), and confirmed by sequencing. The following mutants were generated: s1A-N (M1-K189), s1A-M267X (M1-I266), s1A-K265X (M1-K264), s1A-C (M168-G288), M267 (M267-G288), s1A-V248 M (V→M248-G288), s1A-M215 (deletion of M215-I266 fragment), and s1A-I195 (deletion of I195-I266 fragment). For terminal transmembrane region (TMR) swap mutants, the PCR gene-fusion method was used to replace the carboxy terminus of syn1A (I270-G288) with the carboxy terminus from other syntaxin isoforms as follows: s1A-TMR2 (syntaxin 2), s1A-TMR2′ (syntaxin 2′), s1A-TMR2′′ (syntaxin 2′′), s1A-TMR3 (syntaxin 3), s1A-TMRScr (sequence 270TGCIFIGILVCISIGIIAG288, a random rearrangement of the 19 amino acids of s1A-TMR). S1A-H3 point mutants were generated by using the cytoplasmic domain syntaxin mutant constructs (syn1A11 series) (5) as a template in PCR reaction and cloned into the full-length s1A-pGMHE. The following mutants were generated: s1A-YK12 (R198A, I202A, L205A, I209A), s1A-YK13 (R198A, I202A, L212A, F216A), s1A-YK34 (L212A, F216A, V223A, Q226A), s1A-YK45 (V223A, Q226A, I230A, I233A), s1A-YK6 (A240V, V244A), and s1A-YK7 (A247V, T251A).
In Vivo 35S-Met Labeling and HPC-1 Immunoprecipitations.
Freshly isolated Xenopus oocytes were injected with cRNA encoding syntaxin 1A and syntaxin mutants as described above. Oocytes were incubated for 48 h at room temperature in ND96 with 35S-Met (250 μCi/ml) added to the medium. After incubation, 10 healthy oocytes were washed 3 times with ice-cold ND96 containing 10 mM unlabeled methionine and homogenized on ice in 1 ml of the homogenization buffer (100 mM NaCl/1% Triton X-100/20 mM Tris⋅HCl, pH 7.6/10 mM methionine/0.1% PMSF/5 μg/ml leupeptin/2 μg/ml aprotinin). Membrane proteins were extracted on ice for 30 min, then nonsolubilized material was pelleted by a 15-min spin at 12,000 rpm (Bedaman J2-21). The supernatant was incubated overnight in the presence of monoclonal HPC-1 antibodies (1:1,000) (36) and 0.1% SDS in an end-over-end shaker at 4°C, followed by incubation in the presence of protein G-agarose (Pharmacia) for 2 h. Protein G–agarose complexes were pelleted by 5-min centrifugation at 1,000 × g, washed three times with 1 ml of TENT-Triton 1% (50 mM Tris⋅HCl, pH 7.6/5 mM EDTA/150 mM NaCl/1% Triton X-100) and once with 10 mM Tris⋅HCl, pH 7.6/2 mM EDTA/0.1% SDS, transferred to the SDS-gel loading buffer, heated to 90°C, analyzed by PAGE in 12% acrylamide, and quantified by phosphoimaging.
N-Type Ca2+ Channel Mutagenesis.
The following N-type Ca2+ channel II/III loop truncation mutants were generated by PCR on the basis of the human α1B chimeric construct (37) and verified by sequencing: Nd2 (deletion of L780-G844), Nd5 (deletion of A735-A871), Nd6 (deletion of A735-L1029), and Nd7 (deletion of A735-A1105).
Scintillation Proximity Assay for Protein–Protein Interaction.
A fragment of the cytosolic II/III loop of α1B (N730-P879) that encompasses the syntaxin-binding site (A772-N858) (15) was amplified by PCR and subcloned into the pGEX-KG vector (Pharmacia) for bacterial expression. GST fusion proteins were purified on a glutathione–agarose column, diluted in 50 mM Tris buffer (pH 8.0), and used to coat the wells of a Scintistrip microtiter plate by overnight incubation at 4°C. The remaining nonspecific sites were blocked by incubation in 0.01% BSA for 1 h at 4°C. An [35S]methionine-labeled syntaxin 1A fragment (s1A-M267X) was generated through in vitro translation, diluted in binding buffer (120 mM potassium acetate/2 mM EDTA/0.05% Tween-20/20 mM Hepes, pH 7.4), and added in aliquots to the precoated and preblocked wells. The binding incubation was performed in the presence of a soluble competitor at room temperature for 1.5 h. The microtiter plate was washed in cold buffer (Tween-20 in the binding buffer replaced by 5% glycerol), air dried, and used for determination of bound 35S-s1A-M267X by liquid scintillation counting (Wallac, Gaithersburg, MD).
Results
Assessment of Syntaxin 1A Effects on N-Type Ca2+ Channel Gating.
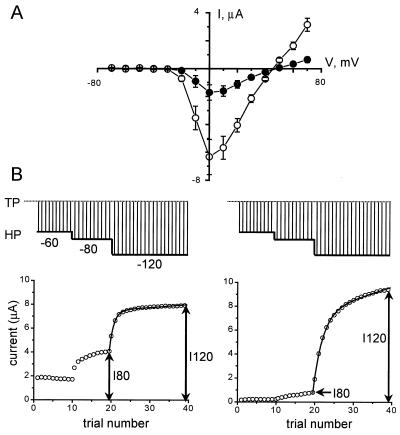
Coexpression of syntaxin 1A with N-type (α1Bβ3α2) Ca2+ channels in Xenopus oocytes reduced the size of the current without significant effects on the shape and position of the peak current–voltage relationship (Fig. 1A). The decrease in current amplitude resulted from a pronounced change in the balance between resting and inactivated states of the N-type channels (19, 25). The shift in gating states was readily apparent when oocytes were subjected to a “descending staircase” voltage stimulation protocol (Fig. 1B) (19). In this procedure, the oocyte membrane was initially maintained at a holding potential of −60 mV. Availability of the expressed channels was assessed by brief (50 ms) depolarizing pulses to a test potential corresponding to the peak of the current–voltage relationship (0 mV). After 10 test pulses, the holding potential was changed to −80 mV, and 10 more test pulses were applied. Finally, the holding potential was changed to −120 mV, and 20 additional test pulses were applied. The interpulse interval (10 s) and the test potential (0 mV) were kept constant throughout the experiment. As illustrated in Fig. 1B, the size of the test current increased progressively as the holding potential became more negative, corresponding to recovery of N-type channels from the inactivated state. The ratio of the peak currents evoked by the last test pulse from holding potential (HP) = −80 mV (I80) and the last test pulse from HP = −120 mV (I120) provided a convenient index of recovery from inactivation. The I80/I120 ratio averaged 0.47 ± 0.02 (n = 38) in control oocytes expressing N-type channels without coexpression of syntaxin but only 0.13 ± 0.01 (n = 41) when syntaxin 1A was coexpressed. The sharp difference in I80/I120 reflected the ability of syntaxin to promote slow inactivation of N-type channels (25). The descending staircase protocol was convenient for structure–function studies, because it could be executed more rapidly than conventional prepulse protocols, allowing examination of a larger number of oocytes within a particular time window after successive injections of Ca2+ channel and syntaxin cRNAs.
Figure 1.
Coexpression of syntaxin 1A with N-type Ca2+ channels in Xenopus oocytes inhibited channel activity. (A) Peak current–voltage relationships of N-type Ca2+ channels (α1Bβ3α2) expressed in Xenopus oocytes, in isolation (open circles, n = 4) or with coexpressed syntaxin 1A (filled circles, n = 4). Symbols and error bars display mean ± SEM. (B) N-type Ca2+ channel inactivation properties analyzed by use of a “descending staircase” stimulation protocol. Comparison between behavior of channels expressed in isolation (Left) and in the presence of syntaxin 1A (Right). Inward Ba2+ currents were evoked by 50-ms depolarizing pulses to 0 mV from a holding potential of −60 mV. After 10 test pulses, holding potential was changed to −80 mV for 10 more test pulses, then further changed to −120 mV for 20 additional test pulses. The interpulse interval (10 seconds) and the test pulse level (0 mV) were kept constant throughout the experiment.
The Importance of the Transmembrane Domain of Syntaxin 1A for Functional Interactions.
Fig. 2 compares the ability of different syntaxin 1A mutants to affect N-type channel gating. Coexpression with a syntaxin construct (s1A-K265X, amino acids 1–264) comprising all but the carboxyl TMR failed to affect N-type channel gating (Fig. 2). Similar results were found with other TMR-lacking constructs, either a shorter N-terminal construct (s1A-N, amino acids 1–189) (19) or a slightly longer one (s1A-M267X, amino acids 1–266) (Fig. 2). The ineffectiveness of TMR-lacking syntaxin fragments must be interpreted cautiously because of their relatively low expression levels, 10-fold less than wild-type syntaxin 1A as determined by [35S]methionine labeling of cellular protein in Xenopus oocytes and immunoprecipitation with anti-syntaxin antibody (see Materials and Methods). Nevertheless, these results point to the importance of TMR region for functional interactions of syntaxin with N-type Ca2+ channels. Two general hypotheses arise. The TMR could be critical for increasing the concentration of syntaxin in the membrane, or the TMR might contain unique sequence information important for interactions with N-type channels. Along these lines, the observed failure of syntaxin 2 to inhibit N-type channels in oocyte experiments has been attributed to sequence divergence from syntaxin 1A within the TMR (21).
Figure 2.
The carboxyl-terminal TMR of syntaxin 1A is essential for N-type Ca2+ channel modulation. The mutant constructs shown (Left) are described in Results and Materials and Methods. Striped areas denote various TMRs taken from syntaxin 2, syntaxin 2′, syntaxin 3, and the carboxyl-terminal region of syntaxin 2′′. Crosshatched area denotes a scrambled version of the TMR of s1A. Bar graph displays mean ± SEM values of the I80/I120 ratio for each construct (number of oocytes shown in parentheses).
To test for TMR-based specificity, we generated a series of chimeric constructs in which the syntaxin 1A TMR was replaced with the corresponding region from other syntaxins. The carboxyl-terminal donors included syntaxin 2 (s1A-TMR2), syntaxin 2′ (s1A-TMR-2′), syntaxin 2′′ (s1A-TMR2′′), and syntaxin 3 (s1A-TMR3) (38). As shown in Fig. 2, these chimeras generally resembled wild-type syntaxin in their ability to promote N-type channel inactivation in coexpression experiments. The single exception was s1A-TMR2′′, which contained a carboxyl-terminal domain that is much shorter and less hydrophobic than that of the other syntaxins and is not thought to be a TMR (38). The findings obtained with constructs incorporating full-length TMRs were consistent in showing no TMR specificity. As a further test, we made another chimeric construct where the amino acid sequence of the s1A TMR was scrambled (s1A-TMRScr). Here, once again, the effect on channel gating was very similar to that of wild-type syntaxin 1A.
Because large variations in TMR sequence can be tolerated without loss of the functional effect on N-type channels, it appears the TMR does not determine the specificity of the channel interaction. This leaves open the possibilities that the TMR may influence N-type channels through sequence-independent hydrophobic interactions or simply by increasing the concentration of syntaxin in the membrane, thereby facilitating separate interactions of low to moderate affinity.
Importance of a Helical Cytoplasmic Domain for Syntaxin Modulation of Gating.
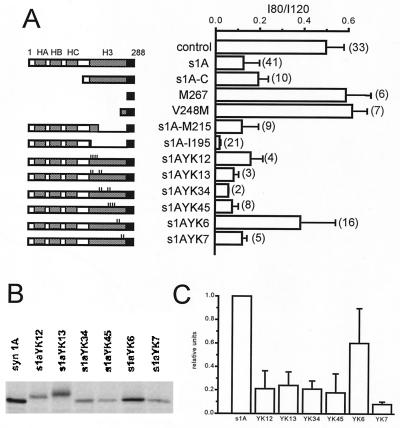
Additional constructs were made to narrow down the structural determinants on syntaxin 1A that support its functional interaction with N-type channels (Fig. 3). The s1A-C construct (amino acids 168–288) lacked helices HA, HB, and HC but produced nearly the same degree of N-type channel inhibition as full-length syntaxin (Fig. 3A). This led to further analysis of structural determinants within the carboxyl-terminal segment, which consists of a helical domain H3 (amino acids 191–265) (5, 33) along with the TMR (amino acids 266–288). We tested whether this helical region is important for functional interaction with N-type channels, or whether the TMR is sufficient. We examined additional constructs consisting of the TMR together with a short fragment of H3 (construct V248 M; amino acids 249–288; valine-248 mutated to methionine to initiate translation) or the TMR alone (amino acids 267–288; construct M267). A Kozak sequence was included at the 5′ end of both clones to increase translation efficiency. Injection of cRNA for either of these constructs gave no discernable effect on channel gating in oocytes (Fig. 3A). These negative results need to be interpreted carefully, as we cannot rule out the possibility that the oocytes simply failed to express these short peptides in significant quantity. Nonetheless, the simplest interpretation is that a cytoplasmic helical region is needed along with the TMR to support modulation of N-type channel gating. Further analysis was carried out with syntaxin constructs that lacked part or nearly all of the H3 domain (Fig. 3A). The ability to modify channel properties was retained with partial omission of H3 (s1AM215, deleted amino acids 215–266) or even near-complete removal of H3 (s1AI195, deleted amino acids 195–266). The s1AI195 mutant produced a clear functional effect on N-type channels (I80/I120 = 0.04 ± 0.01 (n = 21) no less severe than that of wild-type syntaxin 1A.
Figure 3.
The helical H3 domain of syntaxin 1A contains structural determinants critical for the modulatory interaction with N-type Ca2+ channels. (A) Bar graph displays mean ± SEM values of the I80/I120 ratio for each mutant construct (number of oocytes shown in parentheses). The mutant constructs are described in Materials and Methods. Domain deletions in s1A-C, M267, V248 M, s1A-M215, and s1A-I195 are as indicated. The small vertical lines denote point mutations in s1A-H3 point mutants. Of the various point mutant constructs, only s1A-YK6 shows an I80/I120 ratio significantly greater than wild-type s1A. For further details, see text. (B) In vivo 35S-Met labeling of s1A constructs expressed in Xenopus oocytes and immunoprecipitation with HPC-1 antibody. (C) Levels of 35S-Met labeling, normalized by expression level of wild-type syntaxin 1A in each experiment, shown as mean ± SEM.
A more refined test of the possible involvement of H3 was carried out with an extensive series of syntaxin constructs containing various point mutations along the H3 region (5). In each case, a small subset of residues along the hydrophobic face of H3 was converted to alanine (or valine if the wild-type residue was alanine). We found that one mutant (s1A-YK6) was much less effective than wild-type syntaxin 1A [I80/I120 = 0.38 ± 0.05 (n = 16)]. This construct differed structurally from wild type at only two positions (Ala240→Val, Val244→Ala). The behavior of s1A-YK6 stood out in sharp contrast to constructs that contained point mutations to the N-terminal side of positions 240 and 244 (s1A-YK45) and to the carboxyl-terminal side (s1A-YK7). Thus, it appeared that some of the residues critical for functional interaction between syntaxin and N-type channels could be localized to a small portion of H3 domain, bounded by Ile-233 and Ala-247.
We carried out control experiments to be sure that the relative ineffectiveness of s1AYK6 was not simply because of lack of expression or instability of this particular mutant in Xenopus oocytes. [35S]methionine labeling and HPC-1 (36) immunoprecipitation were performed as described in Materials and Methods. The relative abundance of expressed syntaxin and point mutants was analyzed by SDS/PAGE and autoradiography (Fig. 3B). Fig. 3C shows pooled data from three independent phosphoimaging experiments. Indeed, point mutations caused a general reduction in the amount of expressed protein in oocytes relative to wild-type syntaxin 1A. However, the s1A-YK6 mutant was expressed more efficiently than any of the other functionally active point mutants. Because the inability of s1A-YK6 to affect channel gating cannot be explained by deficient expression in oocytes, we interpret the electrophysiological results as compelling evidence for involvement of H3 in Ca2+ channel modulation. This moves the analysis beyond the contradictory data with the s1A-C construct containing the H3 domain and little else in the cytosolic segment and other constructs lacking H3 (Fig. 3A). Finding modulation in both cases is reminiscent of puzzling results obtained with deletion mutagenesis in other systems. As a tentative explanation, we hypothesize that the modulation of Ca2+ channel gating normally involves H3, but that another hydrophobic helix in syntaxin (HA, HB, or HC) may substitute for H3 in its absence.
Is the II/III Loop of N-Type Ca2+ Channels Critical for Functional Responsiveness to Syntaxin 1A?
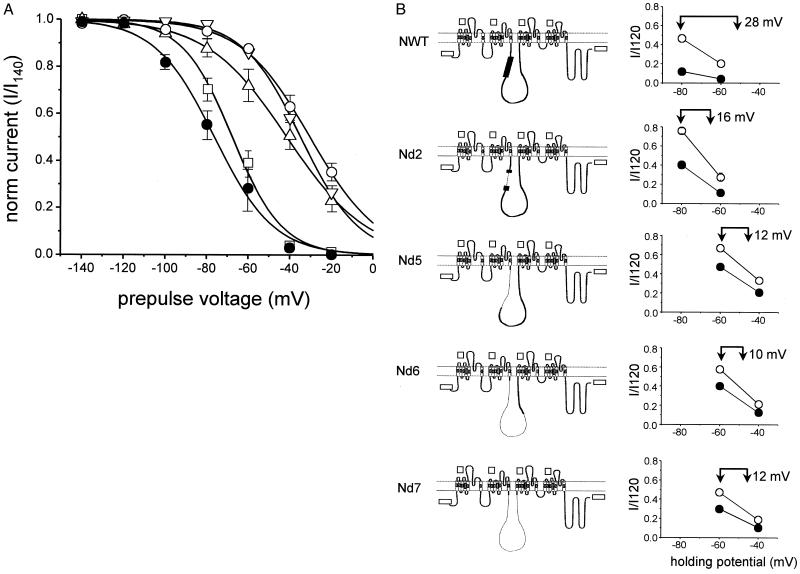
A biochemical interaction of syntaxin 1A has been reported with N-type and P/Q-type Ca2+ channels (11, 13). In agreement with these data, syntaxin 1A affects N- and P/Q- but not L-type Ca2+ channels in the oocyte expression system (ref. 19 but see refs. 21 and 39). Sequence alignment of the corresponding α1 subunits indicates that the II/III loops of α1B (N-type) and α1A (P/Q-type) are almost three times longer than that of α1C (L-type). In vitro biochemical experiments have identified a portion of the cytosolic II/III loop of α1B (amino acids 772–858) as a site responsible for N-type channel interaction with syntaxin (15). We set out to clarify the possible role of the extended II/III loop of the α1B subunit in influencing N-type Ca2+ channel function and its modulation by syntaxin. To this end, we generated a series of partial deletion mutants, depicted schematically in Fig. 4B. The smallest alteration of the II/III loop, a deletion of 65 amino acids (Nd2, Δ 780–844), resulted in removal of the main portion of the biochemically identified syntaxin-binding site (15). Increasingly severe deletions in the II/III loop were made in three additional α1B constructs, the fractions omitted ranging from one-third (Nd5, Δ735–871) to two-thirds (Nd6, Δ 735-1029) to nearly all of the II/III loop (Nd7, Δ 735-1105). All four deletion mutants were successfully expressed in oocytes. Each of the constructs gave a peak current–voltage relationship similar to that of wild-type α1B subunits, with maximal inward current evoked at 0 mV (data not shown). Strikingly, the curves describing the voltage dependence of inactivation were displaced toward depolarized voltages relative to wild type, by ≈10 mV for Nd2 and by as much as 40 mV for mutants Nd5, Nd6, and Nd7 (Fig. 4A). These data provide evidence that the II/III loop of voltage-gated Ca2+ channels may influence their inactivation properties.
Figure 4.
Effect of deletions from the II/III loop of α1B influence N-type channel inactivation properties and affect responsiveness of N-type channels to syntaxin 1A modulation. (A) Voltage-dependent availability of wild-type N-type channels (filled circles, V1/2 = −76 mV), Nd2 (open squares, V1/2 = −68 mV), Nd5 (open circles, V1/2 = −36 mV), Nd6 (downward triangles, V1/2 = −35 mV), Nd7 (upward triangles, V1/2 = −41 mV). N-type channel mutant constructs are described in Materials and Methods. Data points are averages derived from at least three independent experiments for each channel mutant (error bars indicate ± SEM). (B) Analysis of midpoint shifts (ΔV1/2) produced by coexpression of syntaxin. (Left) Wild-type N-type channels (with synprint site shown as thick line), shown schematically above a series of II/III loop deletion constructs Nd2, Nd5, Nd6, Nd7 (extent of deletion indicated by thin dotted line). (Right) Coexpression with syntaxin (filled symbols) induced a hyperpolarizing shift in the inactivation properties of wild-type channels and each of the II/III loop mutants relative to their behavior in control (open symbols). The shift was estimated as the displacement along the voltage axis that would be needed to align a data point in the presence of syntaxin (left vertical arrow) with the voltage-dependent curve describing inactivation in the absence of syntaxin (right vertical arrow). Estimates of the shift were as follows: WT, ΔV1/2 > 28 mV; Nd2, ΔV1/2 =16 mV; Nd5, ΔV1/2 =12 mV; Nd6, ΔV1/2 =10 mV; Nd7, ΔV1/2 =12 mV. The shift was statistically significant (P < 0.01) in all cases. Where not shown, error bars (SEM) were smaller than the symbols.
To evaluate the importance of the II/III loop in channel modulation by syntaxin 1A, the II/III loop deletion mutants were individually expressed in oocytes in the presence and absence of syntaxin 1A and subjected to the standard descending staircase protocol. For constructs Nd5, Nd6, and Nd7, the voltages used in the protocol were modified from −60/−80/−120 mV to −40/−60/−120 mV to compensate for the shift in the position of their steady-state inactivation curves (Fig. 4A). Current amplitudes obtained at the more depolarized holding potentials were normalized by those obtained at −120 mV and were presented as current ratios (Fig. 4B). In each case, the effect of syntaxin 1A was expressed quantitatively as a displacement of the inactivation midpoint along the voltage axis (midpoint shift designated as ΔV1/2). The ΔV1/2 for wild-type channels was at least 28 mV, in reasonably good agreement with previous findings (19). In comparison, the voltage shift was 16 mV in mutant Nd2, in which the major portion of the synprint site was deleted. Further reduction of the voltage shift relative to the parent N-type channel was observed with the other II/III loop deletion mutants (Nd5, Nd6, Nd7). However, in all cases the inhibitory influence of syntaxin remained statistically significant (P < 0.05). Thus, the modulatory effect of syntaxin is facilitated by the presence of the II/III loop, but this motif cannot be solely responsible for the interaction.
The findings in Fig. 4 have at least two possible interpretations with regard to the modulatory effect of syntaxin on N-type channel gating. One possibility is that the modulation is normally transmitted via key portions of the II/III loop, but in its absence, other regions of the channel may partially substitute for this domain. The other possibility is that the II/III loop acts as an important structural anchor for syntaxin, but that some other region of the channel provides additional specificity to mediate the modulatory effect. To explore these possibilities, we examined the binding of syntaxin to the II/III loop. If the modulatory effect were transmitted via key portions of the II/III loop, one might expect the binding to be altered by point mutations in syntaxin that greatly reduce its modulatory effect, as in the Ala240→Val, Val244→Ala mutant (s1A-YK6, Fig. 3). To evaluate binding affinity, we used a quantitative displacement scintillation proximity assay (Materials and Methods). A portion of the α1B II/III loop (amino acids 730–879) that included the synprint site (15) was expressed as a GST-fusion protein, immobilized to a solid support, and allowed to interact with [35S]methionine-labeled cytosolic portion of syntaxin 1A (s1A-M267X, amino acids 1–266). Displacement of the radiolabeled syntaxin 1A fragments was used to assess the binding of cytosolic portions of WT syntaxin 1A and the s1AYK6 mutant, derived by bacterial expression.
In agreement with Sheng et al. (15), we found that the cytosolic region of wild-type syntaxin 1A (s1A-M267X) specifically interacted with the immobilized II/III loop construct, acting at micromolar concentrations to displace radiolabeled s1A-M267X (Fig. 5). However, the corresponding cytosolic region of the s1A-YK6 point mutant (s1A-YK6-M267X) was equally effective in the displacement assay, indicating that both wild-type and mutant syntaxin 1A fragments displayed similar affinity for the II/III loop. This suggests that the functional differences between wild-type syntaxin and the s1A-YK6 point mutant cannot be accounted for by the strength of syntaxin binding to the II/III loop. Additional involvement of other channel regions must be considered.
Figure 5.
Wild-type syntaxin 1A and s1AYK6 point mutant display similar affinity for the II/III loop of the N-type Ca2+ channel in a scintillation proximity displacement assay. 35S-labeled s1A-M267X binding to an immobilized recombinant GST fusion protein incorporating the II/III loop of α1B. Binding assays were performed in the presence of increasing concentrations of soluble competing proteins: wild-type syntaxin 1A fragment (s1A-M267X, square), a corresponding fragment of mutant s1A-YK6 (triangle), and a control protein (GST, circle). All three proteins were derived from bacterial expression and purified. In all scintillation proximity binding assays, nonspecific binding (determined by binding of 35S-s1A-M267X to immobilized GST) was about 23% of total binding (determined by maximal binding of 35S-s1A-M267X to immobilized GST-II/III loop fragment). The data points show mean ± SEM values for percent binding, expressed as a fraction of maximal binding (n = 3).
Discussion
Our experiments provide insights into the structural basis of syntaxin modulation of Ca2+ channel gating. Alterations in either syntaxin 1A or α1B were consistent in suggesting that the modulation involves a molecular interface between syntaxin and N-type Ca2+ channels that functions alongside the well-established binding interaction between syntaxin and the synprint site on the II/III loop of α1B (1, 2, 11–13, 15, 16). The synprint interaction would preserve colocalization of the fusion machinery and Ca2+ channels regardless of vesicular turnover, while allowing the modulatory interaction to be switched on and off at appropriate stages of the vesicular duty cycle.
Structural Determinants on Syntaxin 1A and Relationship Between Vesicular Turnover and Ca2+ Channel Modulation.
The regulation of N-type channel gating by syntaxin primarily involves the carboxyl-terminal portion of syntaxin (H3 + TMR), in line with biochemical analysis of binding interactions (15) and similar to determinants of the modulatory interaction of syntaxin with cystic fibrosis transmembrane regulator (30). The TMR of syntaxin is needed for the functional effect, most likely as a membrane anchor, but it is unlikely to provide specificity (Fig. 2). The H3 domain contains a key effector site between Ile-233 and Ala-247, critical for the functional effect on N-type channels (Fig. 3) but not for the biochemical interaction with the II-III loop (Fig. 5). The involvement of Ala-240 and Val-244 in Ca2+ channel modulation is intriguing because the accessibility of these residues would be expected to vary with the state of the release machinery (40). In a binary or ternary SNARE complex, Ala-240 and Val-244 of syntaxin would lie buried within the hydrophobic core of the four-helix bundle comprised of syntaxin, VAMP/synaptobrevin, and SNAP-25, shielded from solvent and inaccessible with regard to outside interactions (4). At synaptic release sites, the same hydrophobic interactions would hold the modulatory effect of syntaxin in check until the SNARE complex underwent dissociation. Freed from restraint, syntaxin would be available to stabilize Ca2+ channel inactivation until it was reincorporated in a new prefusion SNARE complex, in preparation for another round of exocytosis. This hypothesis would account for the finding that syntaxin's ability to inhibit presynaptic Ca2+ channels depends on Ca2+-dependent vesicular turnover (24). It also provides a structural basis for understanding how syntaxin inhibition might be modulated by other synaptic proteins such as synaptotagmin and SNAP-25 (21, 27) or cysteine string protein (29), which do not prevent syntaxin binding to the synprint site.
Structural Determinants on N-Type Channels (α1B) and Links to Other Forms of Ca2+ Channel Modulation.
We found that the synprint site on the α1B II/III loop binds to the H3 region of syntaxin 1A, in confirmation of the original biochemical experiments of Sheng et al. (15, 16) (Fig. 5). Deletions within the II/III loop region of α1B that completely eliminated the synprint site weakened the modulation of channels by syntaxin, but by no means abolished it (Fig. 4B). Evidently, the II/III loop serves an important anchoring or positioning function that may facilitate modulation but is not absolutely essential for the modulatory action. Binding experiments with GST-fusion proteins containing the II/III loop provided a complementary perspective (Fig. 5). The affinity of syntaxin for these constructs was unaffected by the point mutations in H3 region that abolished syntaxin's modulatory action, suggesting that syntaxin's modulatory action on channel gating may involve a binding interaction distinct from the synprint site. The location of the effector site on α1B for syntaxin modulation remains to be determined, but our data suggest consideration of motifs other than the II/III loop itself. The I/II loop is one candidate, because it lies close to determinants of voltage-dependent inactivation (41) and participates in modulation of gating by other intracellular effectors (42, 43). This would foster crosstalk between syntaxin interactions and G protein effects (23, 44). An alternative is the carboxyl-terminal region, also implicated in Ca2+ channel inactivation and modulation (41, 45, 46).
We hypothesize that a separation of anchoring and modulatory functions of syntaxin may offer functional advantages. A stable interaction between syntaxin and the synprint site in the II-III loop safeguards the structural integrity of the presynaptic fusion complex (15, 16). Additional interactions of a more flexible nature may be critical for regulating Ca2+ entry according to the state of the release machinery (24).
Acknowledgments
We are grateful to Yun Kee for valuable advice and to J. B. Bergsman and E. T. Kavalali for insightful discussion and other help. This work was supported by the Silvio Conte–National Institute of Mental Health Center for Neuroscience Research, the Mathers Charitable Trust, and the McKnight Foundation.
Abbreviation
- TMR
terminal transmembrane region
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220389697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220389697
References
- 1.Bennett M K, Calakos N, Scheller R H. Science. 1992;257:255–259. doi: 10.1126/science.1321498. [DOI] [PubMed] [Google Scholar]
- 2.Yoshida A, Oho C, Omori A, Kuwahara R, Ito T, Takahashi M. J Biol Chem. 1992;267:24925–24928. [PubMed] [Google Scholar]
- 3.Sollner T, Bennett M K, Whiteheart S W, Scheller R H, Rothman J E. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- 4.Sutton R B, Fasshauer D, Jahn R, Brunger A T. Nature (London) 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 5.Kee Y, Lin R C, Hsu S C, Scheller R H. Neuron. 1995;14:991–998. doi: 10.1016/0896-6273(95)90337-2. [DOI] [PubMed] [Google Scholar]
- 6.Wu M N, Fergestad T, Lloyd T E, He Y, Broadie K, Bellen H J. Neuron. 1999;23:593–605. doi: 10.1016/s0896-6273(00)80811-9. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi T, Momiyama A. Nature (London) 1993;366:156–158. doi: 10.1038/366156a0. [DOI] [PubMed] [Google Scholar]
- 8.Luebke J I, Dunlap K, Turner T J. Neuron. 1993;11:895–902. doi: 10.1016/0896-6273(93)90119-c. [DOI] [PubMed] [Google Scholar]
- 9.Wheeler D B, Tsien R W, Randall A. Science. 1994;266:828–831. doi: 10.1126/science.266.5186.830. [DOI] [PubMed] [Google Scholar]
- 10.Castillo P, Weisskopf M G, Nicoll R A. Neuron. 1994;12:261–269. doi: 10.1016/0896-6273(94)90269-0. [DOI] [PubMed] [Google Scholar]
- 11.el Far O, Charvin N, Leveque C, Martin-Moutot N, Takahashi M, Seagar M J. FEBS Lett. 1995;361:101–105. doi: 10.1016/0014-5793(95)00156-4. [DOI] [PubMed] [Google Scholar]
- 12.Leveque C, el Far O, Martin-Moutot N, Sato K, Kato R, Takahashi M, Seagar M J. J Biol Chem. 1994;269:6306–6312. [PubMed] [Google Scholar]
- 13.Martin-Moutot N, Charvin N, Leveque C, Sato K, Nishiki T, Kozaki S, Takahashi M, Seagar M. J Biol Chem. 1996;271:6567–6570. doi: 10.1074/jbc.271.12.6567. [DOI] [PubMed] [Google Scholar]
- 14.Seagar M, Takahashi M. J Bioenerg Biomembr. 1998;30:347–356. doi: 10.1023/a:1021937605818. [DOI] [PubMed] [Google Scholar]
- 15.Sheng Z H, Rettig J, Takahashi M, Catterall W A. Neuron. 1994;13:1303–1313. doi: 10.1016/0896-6273(94)90417-0. [DOI] [PubMed] [Google Scholar]
- 16.Sheng Z H, Westenbroek R E, Catterall W A. J Bioenerg Biomembr. 1998;30:335–345. doi: 10.1023/a:1021985521748. [DOI] [PubMed] [Google Scholar]
- 17.Mochida S, Sheng Z H, Baker C, Kobayashi H, Catterall W A. Neuron. 1996;17:781–788. doi: 10.1016/s0896-6273(00)80209-3. [DOI] [PubMed] [Google Scholar]
- 18.Llinas R, Steinberg I Z, Walton K. Biophys J. 1981;33:323–351. doi: 10.1016/S0006-3495(81)84899-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bezprozvanny I, Scheller R H, Tsien R W. Nature (London) 1995;378:623–626. doi: 10.1038/378623a0. [DOI] [PubMed] [Google Scholar]
- 20.Smirnova T, Fossier P, Stinnakre J, Mallet J, Baux G. Neuroscience. 1995;68:125–133. doi: 10.1016/0306-4522(95)00134-5. [DOI] [PubMed] [Google Scholar]
- 21.Wiser O, Bennett M K, Atlas D. EMBO J. 1996;15:4100–4110. [PMC free article] [PubMed] [Google Scholar]
- 22.Sutton K G, McRory J E, Guthrie H, Murphy T H, Snutch T P. Nature (London) 1999;401:800–804. doi: 10.1038/44586. [DOI] [PubMed] [Google Scholar]
- 23.Jarvis S E, Magga J M, Beedle A M, Braun J E, Zamponi G W. J Biol Chem. 2000;275:6388–6394. doi: 10.1074/jbc.275.9.6388. [DOI] [PubMed] [Google Scholar]
- 24.Bergsman J B, Tsien R W. J Neurosci. 2000;20:4368–4378. doi: 10.1523/JNEUROSCI.20-12-04368.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Degtiar V E, Scheller R H, Tsien R W. J Neurosci. 2000;20:4355–4367. doi: 10.1523/JNEUROSCI.20-12-04355.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tobi D, Wiser O, Trus M, Atlas D. Recept Channels. 1998;6:89–98. [PubMed] [Google Scholar]
- 27.Wiser O, Tobi D, Trus M, Atlas D. FEBS Lett. 1997;404:203–207. doi: 10.1016/s0014-5793(97)00130-0. [DOI] [PubMed] [Google Scholar]
- 28.Leveque C, Pupier S, Marqueze B, Geslin L, Kataoka M, Takahashi M, De Waard M, Seagar M. J Biol Chem. 1998;273:13488–13492. doi: 10.1074/jbc.273.22.13488. [DOI] [PubMed] [Google Scholar]
- 29.Nie Z, Ranjan R, Wenniger J J, Hong S N, Bronk P, Zinsmaier K E. J Neurosci. 1999;19:10270–10279. doi: 10.1523/JNEUROSCI.19-23-10270.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naren A P, Nelson D J, Xie W, Jovov B, Pevsner J, Bennett M K, Benos D J, Quick M W, Kirk K L. Nature (London) 1997;390:302–305. doi: 10.1038/36882. [DOI] [PubMed] [Google Scholar]
- 31.Qi J, Peters K W, Liu C, Wang J M, Edinger R S, Johnson J P, Watkins S C, Frizzell R A. J Biol Chem. 1999;274:30345–30348. doi: 10.1074/jbc.274.43.30345. [DOI] [PubMed] [Google Scholar]
- 32.Zhong H, Yokoyama C T, Scheuer T, Catterall W A. Nat Neurosci. 1999;2:939–941. doi: 10.1038/14721. [DOI] [PubMed] [Google Scholar]
- 33.Chapman E R, An S, Barton N, Jahn R. J Biol Chem. 1994;269:27427–27432. [PubMed] [Google Scholar]
- 34.Fernandez I, Ubach J, Dulubova I, Zhang X, Sudhof T C, Rizo J. Cell. 1998;94:841–849. doi: 10.1016/s0092-8674(00)81742-0. [DOI] [PubMed] [Google Scholar]
- 35.Liman E R, Tytgat J, Hess P. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- 36.Inoue A, Obata K, Akagawa K. J Biol Chem. 1992;267:10613–10619. [PubMed] [Google Scholar]
- 37.Ellinor P T, Zhang J-F, Horne W A, Tsien R W. Nature (London) 1994;372:272–275. doi: 10.1038/372272a0. [DOI] [PubMed] [Google Scholar]
- 38.Bennett M K, Garcia-Arraras J E, Elfernik L A, Peterson K, Fleming A M, Hazuka C D, Scheller R H. Cell. 1993;74:863–873. doi: 10.1016/0092-8674(93)90466-4. [DOI] [PubMed] [Google Scholar]
- 39.Wiser O, Trus M, Hernandez A, Renstrom E, Barg S, Rorsman P, Atlas D. Proc Natl Acad Sci USA. 1999;96:248–253. doi: 10.1073/pnas.96.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fiebig K M, Rice L M, Pollock E, Brunger A T. Nat Struct Biol. 1999;6:117–123. doi: 10.1038/5803. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J-F, Ellinor P T, Aldrich R W, Tsien R W. Nature (London) 1994;372:97–100. doi: 10.1038/372097a0. [DOI] [PubMed] [Google Scholar]
- 42.De Waard M, Liu H, Walker D, Scott V E, Gurnett C A, Campbell K P. Nature (London) 1997;385:446–450. doi: 10.1038/385446a0. [DOI] [PubMed] [Google Scholar]
- 43.Zamponi G W, Bourinet E, Nelson D, Nargeot J, Snutch T P. Nature (London) 1997;385:442–446. doi: 10.1038/385442a0. [DOI] [PubMed] [Google Scholar]
- 44.Stanley E F, Mirotznik R R. Nature (London) 1997;385:340–343. doi: 10.1038/385340a0. [DOI] [PubMed] [Google Scholar]
- 45.Qin N, Platano D, Olcese R, Stefani E, Birnbaumer L. Proc Natl Acad Sci USA. 1997;94:8866–8871. doi: 10.1073/pnas.94.16.8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker D, Bichet D, Campbell K P, De Waard M. J Biol Chem. 1998;273:2361–2367. doi: 10.1074/jbc.273.4.2361. [DOI] [PubMed] [Google Scholar]