Abstract
X linked dilated cardiomyopathy is a familial disease that is allelic to Duchenne and Becker muscular dystrophies and caused by mutations in the dystrophin gene. In several families with X linked dilated cardiomyopathy, the pattern of expression of dystrophin mutations in cardiac muscle differs from that in skeletal muscle. A number of these mutations affect transcription and splicing of the dystrophin gene in a tissue specific manner; others may affect regions of dystrophin that are presumed to have a more important role in cardiac than in skeletal muscle. These mutations are important because they highlight the fundamental differences in processing of the dystrophin gene between skeletal and cardiac tissues, as well as differences in the functional domains more relevant for one tissue or the other. This review focuses on the major mechanisms that have been proposed to explain this disorder.
Keywords: dystrophin, X linked dilated cardiomyopathy, splicing, protein domains
Dilated cardiomyopathy (DCM) is a leading cause of cardiovascular morbidity and mortality. The DCMs form a heterogeneous group of disorders with different patterns of inheritance, including autosomal dominant (∼23%), X linked (∼5%), autosomal recessive, and mitochondrial transmission.1 About 30% of all DCMs are now considered to be inherited and the remaining 70% are sporadic.1 It is becoming increasingly clear, however, that a number of “sporadic” cases of DCM may be accompanied by de novo mutations in genes clearly associated with dominant disorders, complicating the issue of genetic counselling and prevalence of genetic disorders among patients with DCM. The best example is that of laminopathies, a significant contributor to the DCMs with conduction system disease, in which at least 50% of cases are the results of de novo mutations.2
Five genes have so far been associated with X linked forms of cardiomyopathy and skeletal myopathy: tafazzin (also known as G4.5) gene,3 involved in Barth’s syndrome4; the emerin gene,5 responsible for the X linked variant of Emery-Dreifuss muscular dystrophy6; lysosome associated membrane protein 2 deficiency,7 involved in Danon’s disease8; the XK membrane transport protein,9 responsible for McLeod’s syndrome10; and the dystrophin gene,11 responsible for Duchenne (DMD) and Becker (BMD) muscular dystrophies and for X linked dilated cardiomyopathy (XLDC).12 Table 1 summarises the main features of these conditions.
Table 1.
X linked conditions responsible for cardiomyopathy and skeletal myopathy
| Disease | Gene | Protein function | Age at onset | CK | Skeletal muscle involvement | Cardiac muscle involvement |
| Barth’s syndrome (X linked recessive) | G4.4 (tafazzin) | Remodelling of cardiolipin and phosphatidylglycerol | Infancy | Normal | Proximal | Ventricular hypertrophy and dilatation |
| EDMD (X linked recessive) | Emerin | Nuclear envelope protein | Childhood | Increased | Humeroperoneal | AV conduction defects low rate of DCM |
| Danon’s disease (or XVCM-MR) (X linked dominant) | LAMP2 | Lysosome associated membrane protein | Early teens | Increased | Proximal limb girdle and axial | Biventricular concentric hypertrophy or left ventricular dilatation |
| McLeod’s syndrome (X linked recessive) | XK | Putative membrane transport protein of unknown function | Adult life | High | Minimal weakness and atrophy | DCM |
| DMD (X linked recessive) | Dystrophin | Cytoskeleton protein | 2–5 years | High | Proximal muscles | DCM |
| BMD (X linked recessive) | Dystrophin | Cytoskeleton protein | Variable | High | Weakness | DCM |
AV, atrioventricular; BMD, Becker muscular dystrophy; CK, creatine kinase; DCM, dilated cardiomyopathy; DMD, Duchenne muscular dystrophy; LAMP2, lysosome associated membrane protein 2; XVCM-MR, X linked vacuolar myopathy with cardiomyopathy and mental retardation.
Dystrophinopathy is the only condition that has been associated with exclusive cardiac involvement (XLDC). This form is secondary to mutations in the dystrophin gene, usually associated with DMD and BMD.11 DMD is the most common form of muscular dystrophy in childhood, occurring in one in 3500 male births.13 DMD is progressive, leading to loss of ambulation by 13 years. Although DMD can be inherited as an X linked condition, about one third of patients have de novo mutations. DMD is caused by a major reduction or absence in dystrophin expression at the sarcolemma.14
Cardiac involvement in DMD develops insidiously during the first decade of life, at a time when skeletal muscle weakness is already significant.14 DCM is the most common type at all ages, occasionally following a mild hypertrophic phase. The clinical pathological aspects of cardiac involvement in DMD were recently reviewed by Finsterer and colleagues.15
Despite the high incidence of cardiac involvement, most patients with DMD remain asymptomatic.16 Respiratory failure caused by diaphragm muscle weakness is the most common cause of death in DMD, which generally occurs when patients reach their 20s.
BMD is a milder allelic form of DMD with an incidence of one in 14 000 live male births, but it is almost as common as DMD among the general population due to patients’ longer survival, usually well into adulthood.17 The clinical severity varies and, while patients at the severe end of the spectrum may be able to walk in their late teens, the majority remain ambulant for life.18
Several studies have indicated that patients with BMD also have a high incidence of clinical cardiac involvement despite their milder skeletal muscle disease.19–21 In fact, some patients with BMD have initially presented with cardiomyopathy that became severe enough to require cardiac transplantation.22 The most common cause of death in BMD is heart failure. In a series of 68 patients with BMD studied from 1976 to 1993 whose diagnosis was confirmed by dystrophin testing, all had preclinical or clinical cardiac involvement by the age of 30.15,19
Female carriers of DMD and BMD, an X linked recessive disorder, have a surprisingly high incidence of cardiac involvement that progresses with age and manifests primarily as cardiomyopathy.15 Cardiac muscle biopsies of DMD and BMD carriers have a mosaic pattern of absent (DMD) or reduced (BMD) dystrophin immunoreactivity mixed with cardiomyocytes with normal dystrophin expression.23
All of the conditions described above are associated with involvement of both skeletal and cardiac muscles. An intriguing example of cardiomyopathy, without any clinical sign of skeletal muscle weakness, is XLDC, an allelic condition to DMD and BMD.
This article reviews the dystrophin gene mutations so far reported in patients with XLDC and summarises the proposed pathogenic mechanisms underlying the condition.
XLDC
XLDC was first described by Berko and Swift24 in 1987 in one large family with 11 affected young males and five manifesting carrier women, all of whom were mothers of affected sons. The condition was linked to the dystrophin gene in 1993 by Towbin12 and at the same time mutations in the dystrophin gene were identified by Muntoni and colleagues.25 No significant skeletal symptoms are present in this form. The clinical severity of the cardiac involvement in XLDC can be quite variable, ranging from an early onset and fatal cardiomyopathy to a milder form compatible with a better prognosis. The presentation, age at onset, and clinical course of the cardiomyopathy are variable. In the more common and severe form, patients typically develop congestive heart failure in their late teens or early 20s. There is rapid progression and a considerable risk of dying from ventricular failure within a year from the initial presentation.12,24,25 Female carriers of XLDC, as in DMD and BMD, may also develop cardiomyopathy later on in life but the severity is usually milder, presumably due to mosaic expression of dystrophin in the heart. Increased serum concentrations of creatine kinase are often detected in patients and carriers.24
Histological analysis of the skeletal muscle of patients with XLDC has shown a mild myopathy and variation in fibre size, with an increase in internal nuclei26 and interstitial connective tissue.27 In contrast, a severe dystrophic cardiomyopathy, characterised by notable fibrosis and variability in fibre size, is usually present in the cardiac muscle biopsies of patients with XLDC. A number of immunocytochemistry studies have shown that, although dystrophin expression is only mildly reduced in the patient’s skeletal muscle, it is absent in the cardiac muscle.25–29
The first mutations identified in XLDC were clustered towards the 5′ end of the dystrophin gene and are the majority. However, mutations in other regions further downstream were subsequently identified. To date 16 different mutations in the dystrophin gene have been reported in patients with XLDC but the precise pathogenesis of this unique phenotype has remained unclear. This is partly due to the complexity of the dystrophin gene regulation within cardiac and skeletal muscles. In recent years, it has become clear that the frame shift hypothesis,30 based on the genomic structure of the gene after a deletion, cannot explain the cases of XLDC with mutations in the 5′ end of the gene. This theory, however, well accounts for the correlation between genotype and phenotype in most other cases of dystrophinopathy: in DMD the mutation typically induces the loss of the open reading frame but this is maintained in patients with BMD. Although the frame theory does not appear to provide an explanation for XLDC cases with mutations at the 5′ end, it accounts for a number of milder cases with in-frame deletions of the hot spot region.
On the basis of the region of the mutations, Tasaki and colleagues31 have recently classified patients with XLDC into three groups: those with mutations in the 5′ end of the gene (group I)25,28,32–34 or in the central hot spot region centred around exons 48–49 (group II),31,35–37 and those with mutations in regions other than those of group I and group II (group III).26,27,33,38,39 Since a number of mutations belonging to different “categories” affect splicing of the dystrophin gene (that is, the process in which the various exons are joined together and the introns removed from the mature messenger RNA), we propose a classification based on the likely mechanism of disease, which recognises two groups (table 2): group A includes mutations affecting transcription or splicing of the dystrophin gene resulting in a more severe cardiac involvement; group B includes mutations in which specific protein domains of dystrophin are affected.
Table 2.
Literature review of dystrophin defect in XLDC patients
| Dystrophin gene mutation | CK | SK-MP | CA-MP | Reference |
| Group A | ||||
| Muscle exon/intron 1 junction deletion | High | + | – | 25 |
| Muscle exon/intron 1 junction deletion | High | ? | ? | 32 |
| Muscle exon 1 3′ splice site point mutation | Normal | + | – | 28 |
| Muscle exon 1 3′ splice site point mutation | High | ? | ? | 33 |
| Splicing point mutation in intron 1 | High | ? | ? | 33 |
| Insertion of L1 element in muscle exon 1 | High | + | ? | 34 |
| Insertion of L1 element in muscle exon 1 | High | + | ? | 34 |
| Exons 2–7 duplication | High | + | – | 27 |
| Exon 9 point mutation | High | + | – | 38 |
| Intron 11 deletion | High | + | – | 26 |
| Group B | ||||
| Exon 29 point mutation | High | + | + | 39 |
| Exon 35 missense mutation | High | ? | ? | 33 |
| Exons 45–48 deletion | Normal | ? | ? | 37 |
| Exons 45–55 deletion | High | ? | ? | 31 |
| Exon 48 deletion | High | ? | ? | 36 |
| Exon 48 deletion | Normal | ? | + | 37 |
| Exons 48–49 deletion | High | ? | ? | 35 |
| Exons 48–51 deletion | High | ? | + | 37 |
| Exons 48–53 deletion | Normal | ? | + | 37 |
| Exons 49–51 deletion | Normal | + | + | 35 |
+, present; −, absent; ?, not known; CA-MP, dystrophin protein in cardiac muscle; L1, long interspersed nuclear element; SK-MP, dystrophin protein in skeletal muscle.
THE DYSTROPHIN GENE
The dystrophin gene is the largest human gene known, spanning more than 3 Mb (that is, ∼0.1% of the total human genome) or ∼1.5% of the entire X chromosome. There are 79 exons and they account for ∼0.6% of the gene, the residual part corresponding to intervening non-coding sequences (introns) and regulatory sequences.40,41
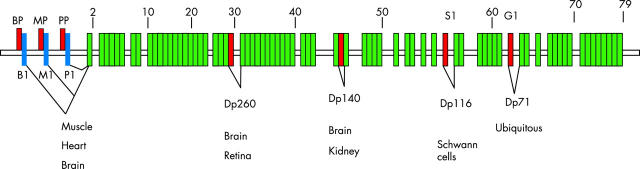
Multiple transcripts are generated from the dystrophin gene, resulting in the formation of several isoforms. These derive from a combination of different tissue specific promoters and alternative splicing of the common dystrophin exons. There are at least seven different promoters. Three of these, specific for brain, muscle and Purkinje cerebellar cells, are arranged in series within the 5′ region of the gene and initiate transcription of full length messenger RNA (14 kb) each transcribing a novel exon 1 sequence.42 Figure 1 shows the promoter arrangement.
Figure 1.
Physical map of the dystrophin gene. Green boxes indicate exons, blue boxes indicate exons specific for each promoter (B1, first brain exon; M1, first muscle exon; P1, first Purkinje exon), and red boxes indicate promoters (BP, brain promoter; G1, general type promoter; MP, muscle promoter; PP, Purkinje promoter; S1, Schwann cell promoter). Not drawn to scale.
XLDC and group A mutations
Most mutations causing XLDC are at the 5′ end of the dystrophin gene. A deletion of the muscle promoter and part of the adjacent intron 1 has been described in a large Italian family25 and more recently in a Dutch family with XLDC.32 A point mutation abolishing the consensus 3′ splice site of muscle exon 1 (that is, a mutation that would be expected to affect the splicing) has also been reported in two unrelated families.28,33 In another two unrelated Japanese families an insertion of a 5′-truncated form of human long interspersed nuclear element (LINE 1 or L1) transposable element in the 5′-untranslated region of the muscle exon 1 also resulted in XLDC.34 These three mutations display an identical transcription pattern in the skeletal muscle28,34,43 and, whenever studied, also in the cardiac muscle.28,43 Transcription studies have shown that these mutations have in common the loss of the ability to transcribe the full length muscle isoform expression in the heart and the skeletal muscle. Lack of the muscle isoform is, however, compensated by the up regulation of the brain and Purkinje isoforms in the skeletal muscle but not in the cardiac muscle.28,34,43 Mutations affecting the promoters are otherwise exceptionally rare in DMD and BMD. Deletion of muscle and brain promoters has been reported in a family with DMD44 and the deletion of muscle, brain, and Purkinje promoters in another DMD patient without cardiomyopathy.45 However, this patient with DMD was too young to be expected to have DCM. These data suggest that the brain and Purkinje promoters can compensate for the absence of dystrophin in skeletal but not cardiac muscle. Recently Feng and colleagues33 reported a novel mutation in intron 1 of the muscle transcript (IVS1 + 6T > C) in one XLDC family.33 Transcription in the skeletal and cardiac muscle has not been analysed in this case but it is possible that this mutation also affects the transcription of the muscle isoform.
In a family described by Bies and colleagues27 the mutation consisted of duplication involving exons 2–7. The mutation resulted in the skipping of exon 2, 4 or both. Expression of muscle and Purkinje isoforms was detected in both skeletal and cardiac muscle, both tissues lacking the brain isoform. Although the same mutant dystrophin transcript was present in both tissues, dystrophin was immunolocalised at the sarcolemma of skeletal muscle only and was absent in the heart. In the patient’s skeletal muscle the dystrophin concentration was reduced to 40% compared with that of control.27
A family with XLDC has been reported by Ortiz-Lopez33 in which a missense mutation in exon 9 of the dystrophin gene resulted in XLDC. In this family normal brain, muscle, and Purkinje isoform expression was detected in both cardiac and skeletal muscle tissues. However, there were no studies on the effect of this mutation on gene transcription. The observation that dystrophin protein was absent in the heart but reduced in skeletal muscle suggests that the mutation may result in tissue specific splicing of the dystrophin gene in this family.
Our group has also reported an unusual mutation involving deletion of dystrophin intron 11.26 In this family, a deletion of intron 11 has brought two naturally occurring cryptic splice sites (hidden splice sites) that are usually ignored by the splicing machinery close together, thus activating them. This results in the incorporation of part of dystrophin intron 11 as a novel exon into dystrophin mRNA transcript, abolishing the normal transcript in the cardiac muscle only. As this interrupts the reading frame, it results in the absence of dystrophin in the cardiac muscle. However, the novel exon coexists with normally spliced exon 11 to exon 12 transcript in the skeletal muscle allowing a significant residual expression of the protein (30% compared with normal control).26 Transcriptional analysis of the skeletal and cardiac muscle of this family showed an essentially normal pattern of isoform expression in the heart and skeletal muscle.26 We have found that normally spliced transcript is the only dystrophin transcript detected in patient lymphocytes. This finding not only indicates a tissue specific splicing pattern for the dystrophin gene in this family but also highlights the unpredictable nature of lymphocytes in identifying splicing mutations in patients with dystrophinopathies.
XLDC and group B mutations
Franz and colleagues reported on a family with XLDC46 in which a nonsense mutation in exon 29 caused skipping of this exon in both cardiac and skeletal muscle.39 Skipping is the mechanism by which an exon that would normally be incorporated in the mature transcript is missed out. In this patient, dystrophin is severely reduced in both skeletal and cardiac muscle.39 Recently, Feng and colleagues33 reported a missense mutation of exon 35 causing a substitution of a highly conserved asparagine by lysine in a patient with XLDC.33
Mutant dystrophin transcripts have not been well characterised in patients with mutations involving a central hot spot region but all the deletions so far identified in these patients are in frame and typically associated with milder BMD. Two patients affected by XLDC have been reported to carry in-frame deletions of exons 49–51 in one and 48–49 in the other. In the former patient almost normal concentrations of truncated protein were detected in the heart.35 Clusters of deletions of exons 45–48, exon 48, exons 48–51, and exon 48–53 have also been reported.36,37 In these patients reduced but preserved dystrophin expression was detected in the cardiac muscle.37 Recently, in-frame deletion of exons 45–55 has been described in a patient with XLDC.31
In contrast to group A patients, in whom dystrophin is totally absent within the heart, in group B patients the dystrophin concentration is reduced and no major difference in promoter usage or splicing between skeletal and cardiac muscle has been reported. Thus, in group B the mutations cause a qualitative rather than a quantitative defect in perturbing the expression of dystrophin. This suggests that a mechanism(s) other than aberrant splicing is involved in this group of patients with XLDC, possibly affecting the structure of dystrophin.
PROPOSED PATHOGENIC MECHANISMS
The finding of different mutations, dystrophin mRNA processing, and protein expression in these cases suggests that multiple pathogenic mechanisms account for all XLDC.
Mutations affecting the splicing of the dystrophin gene (group A)
Various regions of the dystrophin gene seem to be involved in this group of mutations.
Interestingly, up to 43% of dystrophin gene mutations that cause XLDC specifically affect expression of the muscle isoform. Transcription studies in all XLDC patients with mutations at the 5′ end of the dystrophin gene have detected up regulation of the brain and Purkinje isoforms of dystrophin in skeletal muscle but not in cardiac muscle.28,34,43,47 This up regulation compensates for the lack of the muscle isoform. This observation further underlines the important role of both the brain and Purkinje isoforms in maintaining stability of the cytoskeletal organisation in skeletal muscle, thus offering protection from progressive degeneration of the muscles. Up regulation of the brain and Purkinje isoforms has also been reported in the skeletal muscle of a patients with BMD with typical skeletal muscle involvement and a deletion affecting the first muscle exon 1 and the first muscle intron 1.48 Nakamura and colleagues48 proposed that the brain and Purkinje promoters may respond to reduced dystrophin expression in these patients. In addition, up regulation of the two promoters has also been detected in four patients with DMD with deletion not involving the 5′ end of the dystrophin gene.49 These observations suggest that the observed up regulation is not limited to patients with XLDC or mutations at the 5′ end of the gene. However, up regulation of the brain and Purkinje promoters would only provide an advantage to patients in whom there is no mutation in downstream exons but the defect is limited to the muscle promoter or first muscle exon.
Activation of the brain and Purkinje promoters may depend on the presence of DNA regulatory elements that may become functionally relevant under specific circumstances. In this respect, conserved DNA motifs present in the regulatory regions of several muscle specific genes have been described in the brain and Purkinje promoters.50,51 Sequence analysis of the dystrophin brain and Purkinje regions identified domains that were highly homologous to CArG boxes and CArG boxes plus E boxes, respectively; these motifs are binding sites for the myogenic determining gene (Myod, myf5, myogenin, MRF4) and serum response factor families of transcription factors.52–55 Recently, the role of a dystrophin muscle enhancer 1 (DME1) in up regulation of the brain and Purkinje isoforms in muscle cell has been shown.32 This enhancer is preserved in two of the patients with mutation involving muscle exon/intron junction deletion.25,32 More recently, Bastianutto and colleagues56 identified DME2, which contained cardiac specific transcriptional elements. This enhancer apparently has a role in dystrophin gene regulation at later stages of cardiac muscle development. The presence of two other possible enhancer elements, DME3 and DME4, has also been predicted.56 Figure 2 shows the arrangement of DMEs.
Figure 2.
A model of dystrophin gene with muscle enhancer elements. DME, dystrophin muscle enhancer.
In the family described by Bies and colleagues27 in which exons 2–7 were duplicated, the same mutant dystrophin transcript was spliced in both cardiac and skeletal muscle tissues but probably with different efficiency.27 The mutation may have caused a cardiac sensitive change in dystrophin function only. Since the dystrophin actin binding domain is essential for dystrophin function, the disruption of this interaction may lead to an exclusive impairment of cardiac muscle function. However, part of the difference was apparently also due to an alteration of dystrophin abundance in the heart, resulting from differences in transcription between cardiac and skeletal muscle. Thus, we hypothesise that a combination of disruption within the dystrophin cardiac domain and tissue specific splicing of dystrophin has a significant role in pathogenesis of XLDC in this family.
The mutation reported by Ferlini and colleagues26 involving an intronic 11 deletion is clearly a splicing mutation, as incorporation of the novel pathogenic exon is regulated in a tissue specific manner. One of the interesting features of this mutation is that all the exons are intact and thus an entirely intronic deletion is involved in XLDC.26 This observation suggests the presence of cardiac specific regulatory sequences within the dystrophin intron. Intronic enhancers have been shown to be involved in regulation of alternatively spliced exons in cell specific, differentiation stage, and tissue specific manners. These sequences presumably work by recruiting splicing factors to the 5′ splice site.57,58 Many examples of inhibitory cis-acting elements (genes that are located on one chromosome and act on or cooperate with neighbouring genes on the same chromosome) have also been described, showing that splice sites can be blocked by secondary structure or by specific or general factors binding to regulatory intronic elements near the repressed sites.57,58
Although some of the mutations reported in this group definitively affect splicing and transcription, in others there was not sufficient tissue to perform parallel studies in cardiac and skeletal muscle. An example of these cases is found in the family described by Ortiz-Lopez and colleagues38 with a missense mutation of exon 9. The authors suggest that the mutation affecting the conserved first hinge region of the protein is likely to result in loss of membrane integrity and eventual loss of function. This was probably due to the continual stress placed on the beating pump,38 therefore favouring the hypothesis of an important functional role for the mutated domain. However, no detailed studies on the effect of this mutation on gene transcription in this region could be carried out. It is important to note that the A to G substitution occurred at nucleotide +4 of exon 9, located within one exonic consensus splicing sequence. Since the 5′ and 3′ exonic extremities are involved in splicing accuracy and efficacy, it is possible that this mutation affects splicing mechanisms. In addition, the rare physiological occurrence of exon 9 skipping suggests that transcription studies in this patient may be very important for ruling out a splicing pathology.47
Mutations affecting the dystrophin domain are particularly important for its cardiac function and interaction (group B)
The great majority of patients belonging to this group carry mutations in the spectrin-like domain of dystrophin. There is no clear explanation as to why cardiac muscle is preferentially affected in them. Three possibilities can be hypothesised: firstly, trans-acting factors (protein products produced by genes on one chromosome and cooperating with or acting on genes elsewhere) in the cardiac muscle of patients with XLDC may cause misregulation of dystrophin splicing, ultimately affecting dystrophin expression in the heart; secondly, intronic cis-acting cardiac specific sequence(s) may be lost; and thirdly, some specific domains that have a specific or prevalent cardiac function may be lost.
Although the first hypothesis appears to be very unlikely, we will analyse here the available evidence in favour of or against it. In keeping with the idea that alternative splicing may have a role in these patients, Sironi and colleagues59 have recently found at least seven alternative splicing events that are involved in the dystrophin gene encompassing exons 44–58 in healthy skeletal muscle. They also noticed that deletions involving the same exons can determine diverse splicing behaviours in different patients or even different tissues of the same person.59 A study of alternative splicing events in large number of patients with BMD, carried out by the same group, suggests that altered splicing patterns in these patients do not directly derive from the gene defect but may derive from trans- rather than cis-acting factors.60 Furthermore, recent observations in up regulation of a trans-acting factor in DMD skeletal muscle has led to the proposal of the role of such factors in splicing regulation.61 Unfortunately, transcription has not been systematically studied in patients with XLDC with group B mutations. However, we analysed the transcription of dystrophin in the cardiac muscle of a patient with a deletion in this region and found a normal pattern of splicing.35,47 Therefore, neither the available transcription data nor the available protein expression data support the hypothesis that differences in splicing between cardiac and skeletal muscle have a significant role in these patients.
Regarding the second hypothesis, that the loss of intronic regions with potential regulatory functions may have a role in patients with these deletions, since the incidence of cardiac involvement is high in patients with BMD with deletions involving exon 49 but not those with deletions of only exon 48,62,63 it has been proposed that intron 48 may contain sequences important to the function of dystrophin in cardiac muscle.35 Owing to the very large size of the intron, this hypothesis has been difficult to prove.
In keeping with the “epitope” theory (the third hypothesis), the central hot spot region of the gene contains hinge regions (fig 3) that may be important for a specific function or interaction of dystrophin in the heart.64 Recently it has been shown that the dystrophin domain encoded by exons 45–48 is required for localisation of neuronal nitric oxide synthase to the sarcolemma. The authors suggested that this may relate to conformational changes induced by mutations in this region of dystrophin or to a yet unidentified protein interaction in this region.65 While neuronal nitric oxide synthase is not expressed in cardiac muscle, this study provided evidence that this region of dystrophin is involved in the interaction with dystrophin associated proteins. It can therefore be hypothesised that additional proteins relevant for heart function may interact with this domain of dystrophin.
Figure 3.
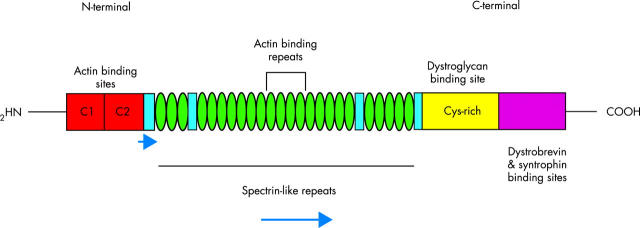
Domains of the dystrophin protein. The N-terminus contains the primary actin binding sites, whereas the C-terminus contains the β dystroglycan, dystrobrevin, and syntrophin binding sites. The N- and C-terminal domains are connected by 24 spectrin-like repeats, some of which have been shown to bind actin. The four “hinge” regions are shown as blue boxes. The blue horizontal arrows indicate regions of the gene that have been suggested to have a role in function and interaction of dystrophin in the heart.
In the family reported on by Franz and colleagues,46 a nonsense mutation in exon 29 results in exon skipping and a significant reduction of dystrophin in both cardiac and skeletal muscle.39 The detection of aberrant splicing in both tissues makes it difficult to understand the different mechanisms involved in protein expression of both these tissues. On the basis of the dystrophin interaction with actin and the sarcolemmal dystrophin associated glycoprotein complex, Franz and colleagues have hypothesised that conformational change in the mid-region of the dystrophin may profoundly affect this interaction and cause membrane instability. The rod-like domain of dystrophin is formed by 24 tandemly repeated segments, each consisting of three α helices connected by turns. Exon 29 encodes most of two of the helices and one of the turns. The deletion of exon 29 therefore results in a loss of this turn, which induces a major change in the mid-rod region.39
Recently, Feng and colleagues33 reported a missense mutation of exon 35 causing substitution of a highly conserved asparagine by lysine in a patient with XLDC. The region involved in this mutation is highly conserved in dystrophins, including chicken and dog fish, and in utrophin, which is very similar in overall structure to dystrophin.33 On the basis of the observed conservation of the mutated amino acid, the second hypothesis may be suggested. However, since there has been no report of a transcriptional study for this patient it is difficult to rule out other mechanisms.
In addition to a primary role of dystrophin in XLDC, in recent years a potential link between this protein and environmental conditions has also been identified. Dystrophin is proteolytically cleaved by coxsackieviral protease 2A, functionally impaired, and morphologically disrupted in cultured cardiomyocytes as well as in the intact mouse heart infected with coxsackievirus B3, and this has been proposed to contribute to the pathogenesis of inflammatory DCM.66,67 In addition it has been reported that the hearts of dystrophin deficient mice are more susceptible to enterovirus induced cardiomyopathy.68
There is also increasing evidence for secondary changes in dystrophin in heat failure. Recently, Vatta and colleagues69 have shown that decreased contractile function is associated with selective disruption of the amino terminus of dystrophin in patients with end stage cardiomyopathy (idiopathic dilated or ischaemic). The authors hypothesised that an unidentified protein may interact with and stabilise the amino terminus of dystrophin. It is this interaction that is disrupted in end stage heart failure. This disruption was reversible in some patients after left ventricular assistance device support. These observations support the hypothesis that changes in cytoskeletal proteins and in particular dystrophin may provide a final common pathway for contractile dysfunction in heart failure and that these changes may well be reversible by reduction of mechanical stress.69
SUMMARY
We have summarised the mutations identified so far in patients with XLDC. A number of these mutations affect transcription, splicing, or both in the dystrophin gene in a tissue specific manner. Others are more likely to be involved in destabilising regions of dystrophin and have a more important role in cardiac than in skeletal muscle. Overall, these mutations are important, as they highlight the fundamental differences in the processing of the dystrophin gene between skeletal and cardiac tissues, as well as the functional domains more critical for one tissue or the other. Recent evidence also suggests that dystrophin may have a significant role in acquired infective or ischaemic cardiomyopathies. Further studies are necessary for understanding the underlying pathogenesis of DCM.
Acknowledgments
This work was supported by the British Heart Foundation. The authors thank Afshin Cohen for reading the manuscript.
Abbreviations
BMD, Becker muscular dystrophy
DCM, dilated cardiomyopathy
DME, dystrophin muscle enhancer
DMD, Duchenne muscular dystrophy
XLDC, X linked dilated cardiomyopathy
REFERENCES
- 1.Bowles NE, Bowles KR, Towbin JA. The “final common pathway” hypothesis and inherited cardiovascular disease: the role of cytoskeletal proteins in dilated cardiomyopathy. Herz 2000;25:168–75. [DOI] [PubMed] [Google Scholar]
- 2.Bonne G , Mercuri E, Muchir A, et al. Clinical and molecular genetic spectrum of autosomal dominant Emery-Dreifuss muscular dystrophy due to mutations of the lamin A/C gene. Ann Neurol 2000;48:170–80. [PubMed] [Google Scholar]
- 3.Bione S , D’Adamo P, Maestrini E, et al. A novel X-linked gene, G4.5. is responsible for Barth syndrome. Nat Genet 1996;12:385–9. [DOI] [PubMed] [Google Scholar]
- 4.Barth PG, Scholte HR, Berden JA, et al. An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J Neurol Sci 1983;62:327–55. [DOI] [PubMed] [Google Scholar]
- 5.Bione S , Maestrini E, Rivella S, et al. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat Genet 1994;8:323–7. [DOI] [PubMed] [Google Scholar]
- 6.Emery AE. Emery-Dreifuss syndrome. J Med Genet 1989;26:637–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishino I , Fu J, Tanji K, et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature 2000;406:906–10. [DOI] [PubMed] [Google Scholar]
- 8.Danon MJ, Oh SJ, DiMauro S, et al. Lysosomal glycogen storage disease with normal acid maltase. Neurology 1981;31:51–7. [DOI] [PubMed] [Google Scholar]
- 9.Ho M , Chelly J, Carter N, et al. Isolation of the gene for McLeod syndrome that encodes a novel membrane transport protein. Cell 1994;77:869–80. [DOI] [PubMed] [Google Scholar]
- 10.Allen FH, Krabbe SMR, Corcoran PA. A new phenotype (McLeod) in the Kell blood-group system. Vox Sang 1961;6:555–60. [DOI] [PubMed] [Google Scholar]
- 11.Hoffman EP, Brown RH Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell 1987;51:919–28. [DOI] [PubMed] [Google Scholar]
- 12.Towbin JA, Hejtmancik JF, Brink P, et al. X-linked dilated cardiomyopathy: molecular genetic evidence of linkage to the Duchenne muscular dystrophy (dystrophin) gene at the Xp21 locus. Circulation 1993;87:1854–65. [DOI] [PubMed] [Google Scholar]
- 13.Mostacciuolo ML, Miorin M, Pegoraro E, et al. Reappraisal of the incidence rate of Duchenne and Becker muscular dystrophies on the basis of molecular diagnosis. Neuroepidemiology 1993;12:326–30. [DOI] [PubMed] [Google Scholar]
- 14.Engel AG, Yamamoto M, Fischbeck KH. Dystrophinopathies. In: Engel AG, Franzini-Armstrong C, eds. Myology. New York: McGraw Hill, 1994:1133–87.
- 15.Finsterer J , Stollberger C. The heart in human dystrophinopathies. Cardiology 2003;99:1–19. [DOI] [PubMed] [Google Scholar]
- 16.Nigro G , Comi LI, Politano L, et al. The incidence and evolution of cardiomyopathy in Duchenne muscular dystrophy. Int J Cardiol 1990;26:271–7. [DOI] [PubMed] [Google Scholar]
- 17.Bushby KM, Thambyayah M, Gardner-Medwin D. Prevalence and incidence of Becker muscular dystrophy. Lancet 1991;337:1022–4. [DOI] [PubMed] [Google Scholar]
- 18.Beggs AH, Hoffman EP, Snyder JR, et al. Exploring the molecular basis for variability among patients with Becker muscular dystrophy: dystrophin gene and protein studies. Am J Hum Genet 1991;49:54–67. [PMC free article] [PubMed] [Google Scholar]
- 19.Nigro G , Comi LI, Politano L, et al. Evaluation of the cardiomyopathy in Becker muscular dystrophy. Muscle Nerve 1995;18:283–91. [DOI] [PubMed] [Google Scholar]
- 20.Melacini P , Fanin M, Danieli GA, et al. Myocardial involvement is very frequent among patients affected with subclinical Becker’s muscular dystrophy. Circulation 1996;94:3168–75. [DOI] [PubMed] [Google Scholar]
- 21.Saito M , Kawai H, Akaike M, et al. Cardiac dysfunction with Becker muscular dystrophy. Am Heart J 1996;132:642–7. [DOI] [PubMed] [Google Scholar]
- 22.Quinlivan RM, Dubowitz V. Cardiac transplantation in Becker muscular dystrophy. Neuromuscul Disord 1992;2:165–7. [DOI] [PubMed] [Google Scholar]
- 23.Politano L , Nigro V, Nigro G, et al. Development of cardiomyopathy in female carriers of Duchenne and Becker muscular dystrophies. JAMA 1996;275:1335–8. [PubMed] [Google Scholar]
- 24.Berko BA, Swift M. X-linked dilated cardiomyopathy. N Engl J Med 1987;316:1186–91. [DOI] [PubMed] [Google Scholar]
- 25.Muntoni F , Cau M, Ganau A, et al. Brief report: deletion of the dystrophin muscle-promoter region associated with X-linked dilated cardiomyopathy. N Engl J Med 1993;329:921–5. [DOI] [PubMed] [Google Scholar]
- 26.Ferlini A , Galie N, Merlini L, et al. A novel Alu-like element rearranged in the dystrophin gene causes a splicing mutation in a family with X-linked dilated cardiomyopathy. Am J Hum Genet 1998;63:436–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bies RD, Maeda M, Roberds SL, et al. A 5′ dystrophin duplication mutation causes membrane deficiency of alpha-dystroglycan in a family with X-linked cardiomyopathy. J Mol Cell Cardiol 1997;29:3175–88. [DOI] [PubMed] [Google Scholar]
- 28.Milasin J , Muntoni F, Severini GM, et al. A point mutation in the 5′ splice site of the dystrophin gene first intron responsible for X-linked dilated cardiomyopathy. Hum Mol Genet 1996;5:73–9. [DOI] [PubMed] [Google Scholar]
- 29.Muntoni F , Wilson L, Marrosu G, et al. A mutation in the dystrophin gene selectively affecting dystrophin expression in the heart. J Clin Invest 1995;96:693–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monaco AP, Bertelson CJ, Liechti-Gallati S, et al. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics 1988;2:90–5. [DOI] [PubMed] [Google Scholar]
- 31.Tasaki N , Yoshida K, Haruta SI, et al. X-linked dilated cardiomyopathy with a large hot-spot deletion in the dystrophin gene. Intern Med 2001;40:1215–21. [DOI] [PubMed] [Google Scholar]
- 32.Bastianutto C , Bestard JA, Lahnakoski K, et al. Dystrophin muscle enhancer 1 is implicated in the activation of non-muscle isoforms in the skeletal muscle of patients with X-linked dilated cardiomyopathy. Hum Mol Genet 2001;10:2627–35. [DOI] [PubMed] [Google Scholar]
- 33.Feng J , Yan JY, Buzin CH, et al. Comprehensive mutation scanning of the dystrophin gene in patients with nonsyndromic X-linked dilated cardiomyopathy. J Am Coll Cardiol 2002;40:1120–4. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida K , Nakamura A, Yazaki M, et al. Insertional mutation by transposable element, L1, in the DMD gene results in X-linked dilated cardiomyopathy. Hum Mol Genet 1998;7:1129–32. [DOI] [PubMed] [Google Scholar]
- 35.Muntoni F , Di Lenarda A, Porcu M, et al. Dystrophin gene abnormalities in two patients with idiopathic dilated cardiomyopathy. Heart 1997;78:608–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melis MA, Cau M, Deidda F. Mutation of dystrophin gene in two families with X-linked dilated cardiomyopathy [abstract]. Neuromuscul Disord 1998;8 (suppl 3–4) :P244. [Google Scholar]
- 37.Arbustini E , Diegoli M, Morbini P, et al. Prevalence and characteristics of dystrophin defects in adult male patients with dilated cardiomyopathy. J Am Coll Cardiol 2000;35:1760–8. [DOI] [PubMed] [Google Scholar]
- 38.Ortiz-Lopez R , Li H, Su J, et al. Evidence for a dystrophin missense mutation as a cause of X-linked dilated cardiomyopathy. Circulation 1997;95:2434–40. [DOI] [PubMed] [Google Scholar]
- 39.Franz WM, Muller M, Muller OJ, et al. Association of nonsense mutation of dystrophin gene with disruption of sarcoglycan complex in X-linked dilated cardiomyopathy. Lancet 2000;355:1781–75. [DOI] [PubMed] [Google Scholar]
- 40.Roberts RG, Coffey AJ, Bobrow M, et al. Exon structure of the human dystrophin gene. Genomics 1993;16:536–8. [DOI] [PubMed] [Google Scholar]
- 41.Den Dunnen JT, Grootscholten PM, Dauwerse JG, et al. Reconstruction of the 2.4 Mb human DMD gene by homologous YAC recombination. Hum Mol Genet 1992;1:19–28. [DOI] [PubMed] [Google Scholar]
- 42.Ahn AH, Kunkel LM. The structural and functional diversity of dystrophin. Nat Genet 1993;3:283–91. [DOI] [PubMed] [Google Scholar]
- 43.Muntoni F , Melis MA, Ganau A, et al. Transcription of the dystrophin gene in normal tissues and in skeletal muscle of a family with X-linked dilated cardiomyopathy. Am J Hum Genet 1995;56:151–7. [PMC free article] [PubMed] [Google Scholar]
- 44.Den Dunnen JT, Casula L, Makover A, et al. Mapping of dystrophin brain promoter: a deletion of this region is compatible with normal intellect. Neuromuscul Disord 1991;1:327–31. [DOI] [PubMed] [Google Scholar]
- 45.Frisso G , Sampaolo S, Pastore L, et al. Novel deletion at the M and P promoters of the human dystrophin gene associated with a Duchenne muscular dystrophy. Neuromuscul Disord 2002;12:494–7. [DOI] [PubMed] [Google Scholar]
- 46.Franz WM, Cremer M, Herrmann R, et al. X-linked dilated cardiomyopathy: novel mutation of the dystrophin gene. Ann N Y Acad Sci 1995;752:470–91. [DOI] [PubMed] [Google Scholar]
- 47.Ferlini A , Sewry C, Melis MA, et al. X-linked dilated cardiomyopathy and the dystrophin gene. Neuromuscul Disord 1999;9:339–46. [DOI] [PubMed] [Google Scholar]
- 48.Nakamura A , Ikeda S, Yazaki M, et al. Up-regulation of the brain and Purkinje-cell forms of dystrophin transcripts, in Becker muscular dystrophy. Am J Hum Genet 1997;60:1555–8. [PMC free article] [PubMed] [Google Scholar]
- 49.Sironi M , Bardoni A, Felisari G, et al. Transcriptional activation of the non-muscle, full-length dystrophin isoforms in Duchenne muscular dystrophy skeletal muscle. J Neurol Sci 2001;186:51–7. [DOI] [PubMed] [Google Scholar]
- 50.Walsh K . Cross-binding of factors to functionally different promoter elements in c-fos and skeletal actin genes. Mol Cell Biol 1989;9:2191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Makover A , Zuk D, Breakstone J, et al. Brain-type and muscle-type promoters of the dystrophin gene differ greatly in structure. Neuromuscul Disord 1991;1:39–45. [DOI] [PubMed] [Google Scholar]
- 52.Muscat GE, Gustafson TA, Kedes L. A common factor regulates skeletal and cardiac alpha-actin gene transcription in muscle. Mol Cell Biol 1988;8:4120–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gustafson TA, Kedes L. Identification of multiple proteins that interact with functional regions of the human cardiac alpha-actin promoter. Mol Cell Biol 1989;9:3269–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lassar AB, Buskin JN, Lockshon D, et al. MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase enhancer. Cell 1989;58:823–31. [DOI] [PubMed] [Google Scholar]
- 55.Blackwell TK, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science 1990;250:1104–10. [DOI] [PubMed] [Google Scholar]
- 56.Bastianutto C , De Visser M, Muntoni F, et al. A novel muscle-specific enhancer identified within the deletion overlap region of two XLDC patients lacking muscle exon 1 of the human dystrophin gene. Genomics 2002;80:614–20. [DOI] [PubMed] [Google Scholar]
- 57.Adams MD, Rudner DZ, Rio DC. Biochemistry and regulation of pre-mRNA splicing. Curr Opin Cell Biol 1996;8:331–9. [DOI] [PubMed] [Google Scholar]
- 58.Chabot B . Directing alternative splicing: cast and scenarios. Trends Genet 1996;12:472–8. [DOI] [PubMed] [Google Scholar]
- 59.Sironi M , Cagliani R, Pozzoli U, et al. The dystrophin gene is alternatively spliced throughout its coding sequence. FEBS Lett 2002;517:163–6. [DOI] [PubMed] [Google Scholar]
- 60.Sironi M , Cagliani R, Comi GP, et al.Trans-acting factors may cause dystrophin splicing misregulation in BMD skeletal muscles. FEBS Lett 2003;537:30–4. [DOI] [PubMed] [Google Scholar]
- 61.Bakay M , Zhao P, Chen J, et al. A web-accessible complete transcriptome of normal human and DMD muscle. Neuromuscul Disord 2002;12 (suppl 1) :125–41. [DOI] [PubMed] [Google Scholar]
- 62.Melacini P , Fanin M, Danieli GA, et al. Cardiac involvement in Becker muscular dystrophy. J Am Coll Cardiol 1993;22:1927–34. [DOI] [PubMed] [Google Scholar]
- 63.Nigro G , Politano L, Nigro V, et al. Mutation of dystrophin gene and cardiomyopathy. Neuromuscul Disord 1994;4:371–9. [DOI] [PubMed] [Google Scholar]
- 64.Winder SJ, Gibson TJ, Kendrick-Jones J. Dystrophin and utrophin: the missing links! FEBS Lett 1995;369:27–33. [DOI] [PubMed] [Google Scholar]
- 65.Wells KE, Torelli S, Lu Q, et al. Relocalization of neuronal nitric oxide synthase (nNOS) as a marker for complete restoration of the dystrophin associated protein complex in skeletal muscle. Neuromuscul Disord 2003;13:21–31. [DOI] [PubMed] [Google Scholar]
- 66.Badorff C , Lee GH, Lamphear BJ, et al. Enteroviral protease 2A cleaves dystrophin: evidence of cytoskeletal disruption in an acquired cardiomyopathy. Nat Med 1999;5:320–6. [DOI] [PubMed] [Google Scholar]
- 67.Badorff C , Berkely N, Mehrotra S, et al. Enteroviral protease 2A directly cleaves dystrophin and is inhibited by a dystrophin-based substrate analogue. J Biol Chem 2000;275:11191–7. [DOI] [PubMed] [Google Scholar]
- 68.Xiong D , Lee GH, Badorff C, et al. Dystrophin deficiency markedly increases enterovirus-induced cardiomyopathy: a genetic predisposition to viral heart disease. Nat Med 2002;8:872–7. [DOI] [PubMed] [Google Scholar]
- 69.Vatta M , Stetson SJ, Perez-Verdia A, et al. Molecular remodeling of dystrophin in patients with end-stage cardiomyopathies and reversal in patients on assistance-device therapy. Lancet 2002;359:936–41. [DOI] [PubMed] [Google Scholar]