Abstract
A major question in plant physiology is how the large amount of sucrose made in leaves is transported to the rest of the plant. Although physiological, biochemical, and anatomical investigations have been performed in this field, to date there have been very few genetic studies. Using a reverse genetic screen, we have identified mutant Arabidopsis plants containing transferred DNA insertions in the gene encoding a phloem-specific sucrose transporter, SUC2. SUC2 is thought to function in loading sugar from the apoplast into the conducting sieve tubes. In the homozygous state, these mutations resulted in stunted growth, retarded development, and sterility. The source leaves of mutant plants contained a great excess of starch, and radiolabeled sugar failed to be transported efficiently to roots and inflorescences. These data provide genetic proof that apoplastic phloem loading is critical for growth, development, and reproduction in Arabidopsis and that SUC2 is at least partially responsible for this step.
During the transition from water to land, plants evolved tissues specialized for the production, utilization, and storage of fixed carbon. In this regard, plant organs are generally divided into two categories, sources and sinks. Sources are organs such as mature leaves that export photoassimilates, and sinks are organs such as roots and flowers that import photoassimilates. To coordinate this division of labor, a tissue evolved that is highly specialized for the long-distance transport of sugars. This tissue, collectively called the phloem, allows for the movement of the photoassimilates from source to sink.
The mechanism by which sugars are loaded into the phloem has long been a subject of great interest to plant biologists (see reviews in refs. 1–3). In mature leaves, sugars are produced in the photosynthetic mesophyll cells and then moved into the conducting cells of the phloem known as the sieve elements. Microscopic observations have determined that some plant species have numerous unique intercellular conduits, called plasmodesmata, which join the sieve elements or their closely associated companion cells and the other cells in source leaves. Plasmodesmata are plant-specific structures that act as bridges across the cell wall and allow solutes to move between adjoining protoplasts without being transported across a membrane. The collective protoplast of these cells connected by plasmodesmata is called the symplast. Plants in which sugars follow an entirely symplastic pathway from the mesophyll cells to the sieve elements without being transported across the plasma membrane are called symplastic loaders.
In an alternative mode of phloem loading, sugars must pass through the apoplast, defined as the extracellular cell wall space, on the path from mesophyll to sieve element. These apoplastic loaders require sugar transporters in the plasma membranes of their sieve elements and/or companion cells to move sugars into these symplastically isolated cells. These transporters must move the sugars against a gradient to concentrate them within the sieve elements, thus providing the conditions for the osmotically driven pressure flow mechanism of phloem transport. Sucrose transporters use the proton motive force created by H+-ATPases to move sucrose across the plasma membrane against its concentration gradient.
Molecular studies on the nature of phloem loading have mostly involved solanaceous species, such as tobacco, potato, and tomato, and the model higher plant Arabidopsis thaliana. For genetic studies, Arabidopsis provides the most exhaustive and facile means of determining the essential components of phloem loading and transport. Some recent studies have uncovered physiological differences between the solanaceous species and Arabidopsis. For example, whereas immunolocalization studies on H+-ATPases and sucrose transporters have localized these proteins to the plasma membranes of companion cells of Arabidopsis, in potato and tobacco, the same proteins have been localized to the plasma membranes of sieve elements (4–7). von Schaewen et al. (8) tested the hypothesis that sucrose transporters are important in phloem loading in tobacco and Arabidopsis by apoplastically expressing a transgenic invertase in these plants, theoretically rendering sucrose uptake from the apoplast ineffective. The tobacco plants were quite stunted in growth, but the Arabidopsis plants were not (8). These results seem to indicate that tobacco is an apoplastic loader but Arabidopsis is not, although negative and indirect results are difficult to interpret. A traditional genetic strategy for testing this hypothesis by disrupting endogenous genes has not been reported, and this is the approach we have chosen.
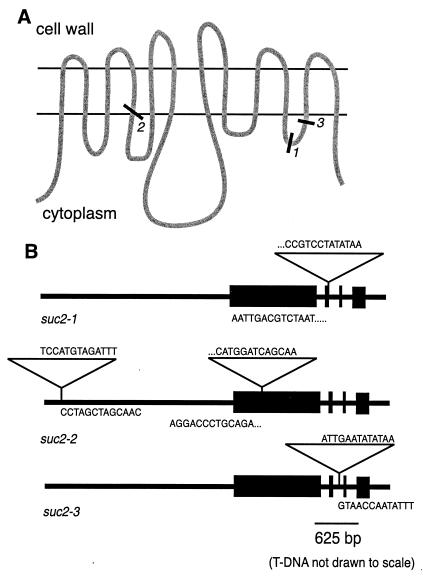
SUC2, the phloem-specific sucrose transporter in Arabidopsis (5, 6), belongs to a large family of transporters including hexose transporters from plants, mammals, algae and fungi, and amino acid transporters from plants (see reviews in refs. 1 and 9–13). These proteins have 12 membrane-spanning domains with a large cytoplasmic loop between domains 6 and 7 (Fig. 1A). Transmembrane domains 7–12 are very similar to 1–6, indicating that the gene may have undergone an internal duplication event. There are currently six sucrose transporter genes identified in the Arabidopsis genome (14).
Figure 1.
The mutant alleles. (A) Representation of SUC2 in the plasma membrane showing apoplastic, membrane-spanning, and cytoplasmic domains and the T-DNA insertion sites of the three mutant alleles. (B) Locations of the T-DNA insertions within the sequence of the SUC2 gene. Boxes indicate exons, and triangles indicate T-DNA. Gene-insertion junction sequences are shown.
In this paper, we report the use of a reverse genetic approach to test the in planta function of sucrose transporters in Arabidopsis. This was accomplished by isolating Arabidopsis mutants with disruptive insertions in SUC2. The resultant phenotype provides strong evidence in support of apoplastic loading as the primary method for initiating long distance transport in Arabidopsis.
Materials and Methods
Reverse Genetic Screen.
Mutant Arabidopsis lines containing random transferred DNA (T-DNA) insertions were screened as described by Krysan et al. (15). Briefly, PCR primers were designed to amplify the wild-type gene. These primers were used in combination with a T-DNA-specific primer to detect insertions within the SUC2 gene. The primers used in this screen were SUC25′, ACAGTTCGGTTGGGCTTTACAGTTATCTC; SUC23′, TTAGGTAAAATACAAACCAACCCAATGTG; and T-DNA left border, CATTTTATAATAACGCTGCGGACATCTAC. The template consisted of pooled DNA from 60,480 random T-DNA insertion lines generated at the University of Wisconsin. PCRs were performed with increasingly deconvoluted DNA template pools until individual plants containing the insertion were identified. Southern hybridizations and sequencing were done at several steps to ensure the validity of the insertion. Plants were genotyped through one PCR containing both gene-specific primers and the T-DNA-specific primer under conditions that, with template from a plant heterozygous for the insertion, gave two equally intense bands on an ethidium bromide stained gel, one indicating the presence of the wild-type allele and one the mutant allele.
Growth Conditions.
Plants were grown under many different conditions as described under Results. Mutant plants were always grown side by side with wild-type and heterozygous siblings for controlled comparison. Seeds were germinated on Murashige and Skoog media, sometimes with 1% (wt/vol) supplemental sucrose. After germination, some plants were transferred to different plates or to a 3:1 mixture of Jiffy-Mix and Perlite and then watered every 2 days with Miracle Gro-supplemented water. All plants were grown in 24-h light at 22–25°C. Mutant plants growing on sucrose-supplemented media were transferred to fresh media when necessary.
Microscopy.
Leaves were sampled in 0.05 M sodium cacodylate buffer, pH 7.2, and then fixed in buffered 5% glutaraldehyde for 60 s in a tissue histology microwave (T. Pella, Redwood, CA) (16). Secondary fixation was in buffered 2% osmium tetroxide for 1 min in the microwave. Tissue was then dehydrated through a graded acetone series, 1 min per concentration, in the microwave, and infiltrated and embedded in Spurr's resin over several days at room temperature. Sections were stained with toluidine blue for light microscopy.
14C Feeding.
A source rosette leaf from each plant to be fed was crimped with forceps, and a 2-μl drop of [U-14C]sucrose (Amersham Pharmacia) was placed on the leaf. The drop was left for 1 h and then rinsed off. The plants were left for 5 h before being dissected and exposed to phosphor screens (Molecular Dynamics). Initial experiments showed that these screens were more sensitive than x-ray film. After 24–48 h, the screens were read. The dissected plant parts were then left shaking in scintillation mixture for 24 h before being read in a liquid scintillation counter. For normalized quantitative analysis, the measured radioactivity detected within a plant part was compared with the total radioactivity detected within that plant. In total, four wild-type, two heterozygous, and three homozygous plants were treated.
Results
Isolation and Characterization of Mutant Alleles.
Using a reverse genetic screen (15, 17), we identified three mutant alleles of the SUC2 gene. PCR primers designed to amplify SUC2 were used in combination with primers specific to T-DNA left and right borders. Pooled template DNA was isolated from 60,480 T-DNA-transformed lines generated at the University of Wisconsin and available to researchers at the Arabidopsis Knockout Facility (www.biotech.wisc.edu/Arabidopsis/default.htm).
Individual heterozygous plants containing the three mutant alleles were identified. Allele suc2–1 contains a single insertion in the second exon (Fig. 1B), and suc2–2 contains two insertions—one in the first exon and one approximately 2.4 kb upstream of the start codon within the presumed promoter region (18). suc2–3 contains one insertion within the second intron.
As described earlier (17), T-DNA insertion is a highly effective mutagen for creating null alleles. Because of its propensity to insert as concatemers of 10 kb, most T-DNA insertions result in loss of transcript, and functional protein is absent in the homozygous mutant plants. If mRNA is transcribed, the T-DNA sequence also contains stop codons, resulting in early transcription termination (17). Fig. 1A shows approximate points within the protein where truncation would occur should translation of the shorter transcript be successful. These potentially truncated SUC2 proteins would most likely be nonfunctional, as studies on the HUP1 hexose transporter of Chlorella have shown the functional importance of both the 11th transmembrane domain and the C-terminal region (19, 20).
Out of 72 offspring of a selfed suc2–3 heterozygote, 19 plants were wild-type, 40 were heterozygous, and 13 were homozygous. In applying χ2 analysis with the null hypothesis of Mendelian segregation for a single locus, P > 0.05, which meant that the null hypothesis was not rejected. The mutant allele shows regular segregation.
General Phenotype.
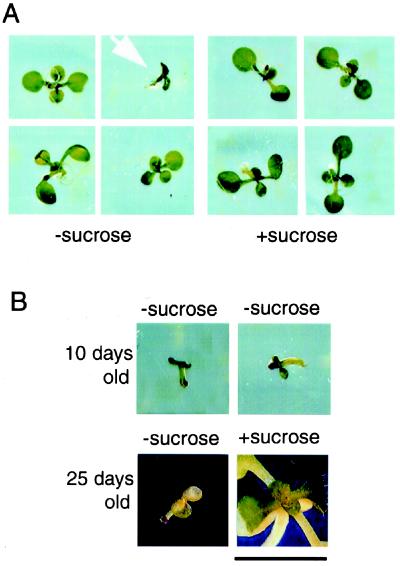
When germinated on media with supplemental sucrose, the suc2 homozygotes appear not to be different from wild-type or heterozygous seedlings. But phenotypic differences are visible early in development if segregating seeds are germinated on media without supplemental sucrose. Fig. 2A shows 10-day-old seedlings germinated without (left) and with (right) 1% sucrose. All seedlings germinated with sucrose, regardless of genotype, have fully expanded cotyledons, well developed primary roots, and expanding rosette leaves. However, when germinated without sucrose, suc2 mutant seedlings are smaller than wild-type seedlings. They have yellowing, translucent cotyledons, very short primary roots, and no rosette leaves. The mutant seedlings will not develop beyond this stage without sucrose.
Figure 2.
The arrested growth and sucrose rescue of suc2 mutant seedlings germinated without supplemental sucrose. (A) Segregating progeny of a suc2–1 heterozygote germinated without or with supplemental sucrose. With no sucrose, the 10-day-old homozygous mutant seedling (arrow) is much smaller and slower in development than its heterozygous and wild-type siblings, having only cotyledons and very short roots. In contrast, all seedlings grown with supplemental sucrose have fully expanded cotyledons and expanding rosette leaves, regardless of genotype. (B) Two suc2–1 homozygotes germinated without sucrose and then transferred to fresh plates either with or without sucrose. At 10 days, both seedlings show stunted growth, have short roots, and have not produced rosette leaves, in contrast to heterozygous and wild-type siblings (see A). Two days later, both seedlings were transferred to fresh media, either without or with sucrose. The seedling transferred to fresh media without sucrose did not produce rosette leaves and eventually yellowed and died. The seedling transferred to sucrose, however, was rescued. It started producing rosette leaves and a root. (Bar = 1 cm.)
The developmentally blocked suc2 mutant seedlings can be partially rescued by the addition of sucrose. Fig. 2B shows two suc2 mutant seedlings germinated without sucrose and grown for 10 days. They are both small and translucent, lacking rosette leaves and well developed primary roots. Two days later, both populations were transferred to fresh media, either without or with 1% supplemental sucrose. At 25 days after planting, the seedling without added sucrose was still small and even more translucent and yellow, and it produced no rosette leaves (Fig. 2B). By contrast, the seedling transferred to sucrose developed a green rosette like a wild-type plant. These results demonstrate that the mutant phenotype is masked on sucrose. There is also a small percentage of wild-type plants that appear stunted and do not produce rosette leaves when germinated on media lacking sucrose. These plants, however, fail to be rescued through exogenous sucrose (data not shown).
All segregating offspring of a SUC2 heterozygote germinated with supplemental sucrose look identical in the first few weeks. However, after being transferred to soil with no exogenous sucrose available, suc2 homozygous mutant plants become dwarfed and slower in development than their heterozygous and wild-type siblings. Fig. 3A shows a suc2 homozygous mutant plant next to its heterozygous sibling. The homozygote is smaller, has fewer rosette leaves, and is not yet at the reproductive stage exhibited by the heterozygote. Fig. 3 B and C show mutant plants at a closer view. All rosette leaves are very small and shaped more like juvenile than adult leaves (21). Also, the leaves are darker green than those of heterozygotes, and the purple areas indicate sectors of anthocyanin accumulation. Although mutant plants occasionally flower (Fig. 3C), they never produce viable seed. The homozygotes are also much slower in overall development, bolting 1–3 weeks later than wild-type or heterozygous siblings and living 3–4 weeks longer.
Figure 3.
Further phenotypic aspects of suc2 mutants. (A) suc2–3 heterozygous and homozygous siblings. These 50-day-old plants were given supplemental sucrose for 16 days before being transferred to soil. The heterozygote is producing seed; the homozygote is dwarfed and nonreproductive. (B and C) A closer view of two homozygotes from this population. The very small rosette leaves are starting to turn purple because of anthocyanin accumulation. The plant in C has started to flower, but no mutant plants have ever produced viable seeds. (Bar = 1 cm.)
The general fitness of suc2 mutant plants and their capacity to produce flowers is directly correlated with the amount of time supplemental sucrose is supplied. Table 1 shows that plants grown longer on sucrose are relatively larger and produce more inflorescences and flowers. None of these plants is as large as a wild-type plant, and none has ever produced viable seed. When grown with continuous supplemental sucrose, the mutants exhibit a very disorderly and chaotic morphology, producing green tissue that is partially leaf-like and partially callus-like. The lifespan of these plants is also greatly extended; they can thrive on fresh sucrose media for over 6 months (Table 1).
Table 1.
Characteristics of suc2 homozygotes
| Genotype | Days on sucrose | Size | Bolts/flowers | Viable seed |
|---|---|---|---|---|
| SUC2 | 0 | +++++ | +++++ | + |
| suc2-1 | 0 | − | − | − |
| suc2-1 | 8 | + | − | − |
| suc2-1 | 8 | ++ | − | − |
| suc2-1 | 8 | ++ | − | − |
| suc2-1 | 16 | +++ | + | − |
| suc2-1 | 16 | +++ | ++ | − |
| suc2-1 | 315 | ++++ | +++ | − |
Anatomy and Physiology of the Mutants.
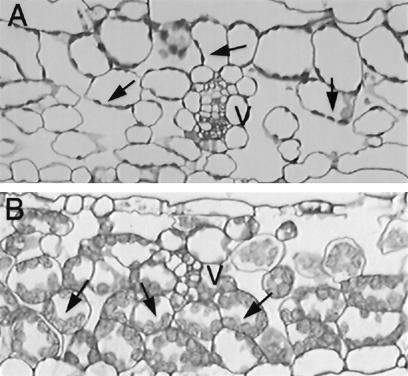
Mutant plants also differ from wild-type plants anatomically. Cross sections through equivalent source rosette leaves (determined by relative position within the rosette) of wild-type and mutant siblings growing in soil show obvious differences. The chloroplasts in the mesophyll cells of the wild-type leaf are small (Fig. 4A). Those of the mutant leaf are much larger and contain large starch granules (Fig. 4B). This build up of starch is consistent with the hypothesis that sucrose is not being transported out of the leaf.
Figure 4.
Internal phenotype of suc2 mutants. Cross sections of rosette leaves of wild-type (A) and suc2–1 homozygote (B) siblings grown under identical conditions. V indicates vascular bundles. The mesophyll cell chloroplasts of the wild-type leaf (arrows) are small and contain no obvious stored starch. Those of the mutant, however, are large and contain visible starch, indicating that sugars are probably not being efficiently transported out of these leaves.
To test directly how well sucrose is transported out of source leaves in suc2 mutant plants, we fed source leaves of wild-type and mutant plants with 14C-labeled sucrose and analyzed how the sucrose moved within the plant.
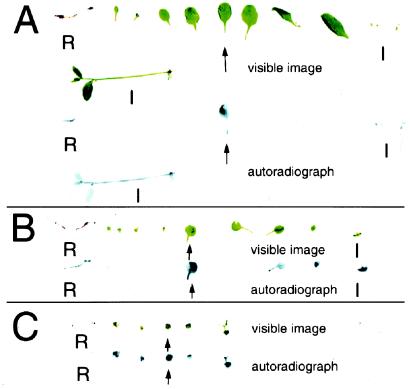
Fig. 5 shows the results of these experiments for two wild-type and one sibling mutant plant, all grown under identical conditions. The visible images of dissected plants, laid out as they were exposed to a phosphor screen for the detection of 14C, are shown directly above the resulting autoradiographs. The fed leaves are indicated with arrows.
Figure 5.
Results of movement of 14C-sucrose in wild-type and suc2 mutant plants. Shown are dissected parts and the corresponding autoradiographs of plants fed 14C-sucrose in drops applied to a crimped rosette leaf (arrow). The organs are laid out in the following order: roots (R), cotyledons and rosette leaves in developmental order, and inflorescences (I), both primary and axillary. All plants are siblings grown under identical conditions. (A) In this wild-type plant, most of the labeled sucrose moved to the roots and axillary and primary inflorescences. No label is found in rosette leaves other than the fed one. (B) In this smaller wild-type plant, included to rule out differences between mutant and wild type being based solely on size, again most of the labeled sucrose moved to roots and inflorescences, with a small amount moving to younger rosette leaves (possibly still sinks). (C) In this suc2–1 mutant plant, most of the labeled sugar moved to other rosette leaves, both younger and older than the fed leaf. No label moved longer distances to the primary sink organs like roots or inflorescences.
In the case of the wild-type plants, most of the label had moved out of the fed leaf and into the roots and the primary and axillary inflorescences. Only a few young sink leaves of the second wild-type plant had imported the labeled sucrose. However, the distribution of the label is drastically different in the mutant plant. The fed leaf retained much more of the label. The labeled sucrose that was exported is found primarily throughout the rosette, both in younger, sink leaves and older, presumably source leaves. There is relatively little label in the roots and inflorescences, even as compared with the second wild-type plant, which is more equivalent in size and stage in life cycle.
After liquid scintillation counting of individual plant parts, the distribution of labeled sucrose was quantitatively analyzed. Because of inconsistencies in leaf size and degree of crimping, the leaves fed 14C-sucrose did not absorb the same quantity of radioactivity. Therefore, all quantitative comparisons are normalized as a ratio of the radioactivity of a given plant part to the total amount of radioactivity present within that individual plant.
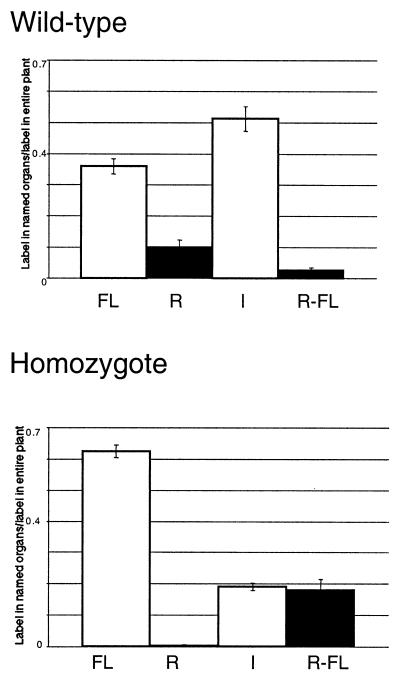
Fig. 6 shows quantitative comparisons between wild-type and suc2 mutant plants. The label found in these plants is divided among four compartments: the fed leaf, roots, inflorescences, and non-fed rosette leaves. In wild-type plants, the majority of the label is in the inflorescences. The next most heavily labeled category is the fed leaf, followed by the roots. Only a very small percentage of the label is found elsewhere in the rosette. In the homozygotes, most of the label is still in the fed leaves with about an equal amount in the rest of the rosette and inflorescences. An extremely small percentage of the label moved to the roots.
Figure 6.
Quantitative comparison of labeled sucrose movement within wild-type and mutant plants. Bar graphs with standard deviation indicate distribution of radioactive label in fed wild-type and homozygous mutant plants 5 h after feeding. y axis values are the ratio of radioactivity detected by liquid scintillation counting in indicated organs to the total radioactivity detected for that plant. The organ groupings are as follows: FL, fed rosette leaf; R, roots; I, inflorescences; and R-FL, rosette without fed leaf, i.e., all other rosette leaves.
Discussion
The loading of sucrose into the sieve elements of source leaves is an important and basic physiological process in plants. It has been predicted that sucrose transporters are essential for this process in species that are apoplastic phloem loaders. We have sought to test these models using a reverse genetic screen.
This paper characterizes the result of a mutation in a specific plant sucrose transporter gene. SUC2 has been shown through both reporter gene and immunolocalization studies to be expressed within the phloem tissues, specifically in the companion cells (5, 6). Previous studies in other species have investigated the in planta function of phloem-specific sucrose transporters through the apoplastic expression of transgenic invertase (8, 22–24) or by expression of an antisense sucrose transporter mRNA (25–29). General phenotypic effects in the resultant transgenic tobacco, potato, or tomato plants were stunted growth, impaired root development, accumulation of carbohydrates within source leaves, reduced ability to export sugars out of these leaves, and crinkled or curled leaves, sometimes with necrotic areas. A maize mutant, sed1, also showed some of these phenotypes (30), although the gene responsible has not yet been identified. These examples give ideas of what export deficient mutants might look like, but there are always possible pleiotropic effects when working with an artificially derived system (transgenic invertase) or antisense suppression of one member of a large gene family.
The overall phenotype caused by the disruption of the SUC2 gene, i.e., stunted growth, might have been overlooked in a traditional forward genetic screen. Although it is an expected phenotype based on the hypothesis that SUC2 is important in phloem transport, it is also a very general and ubiquitous phenotype that could have many different causes. The reverse genetic screen allowed us to recognize this phenotype specifically as a result of decreased phloem transport.
All of the general phenotypic characteristics discussed here were observed in mutant plants of all three independent alleles. This is strong evidence that the phenotype results from disruption of the SUC2 gene rather than a mutation nearby or a general secondary result of T-DNA transformation. In plants outcrossed to wild-type plants for multiple generations, the SUC2 insertions and the phenotypes segregated together (data not shown). Although we have not tested whether SUC2 protein (truncated, functional, or otherwise) is present in the mutant plants, the common phenotypes of three independent alleles with different insertion sites in the same gene argues against the presence of a biologically active protein.
Mutant seedlings germinated without supplemental sucrose are unable to develop beyond the cotyledon expansion stage, which may be when a seedling would begin to require sugars other than those stored during embryogenesis. According to this heterotrophic transition model, starving cells in the mutant plants are able to take up exogenously supplied sucrose through the roots, using it to enable further growth and development. This explains why mutant seedlings halted in development by lack of supplemental sucrose are able to be rescued if moved to media with sucrose (Fig. 2B) and also why mutant plants left on supplemental sucrose for longer periods of time, and therefore able to take up more sucrose from the media, are more likely to produce larger rosettes and inflorescences (Table 1). This is comparable with the concept of “luxury uptake” in mineral nutrition, where it has been long known that plants take up and store nutrients above and beyond their current needs for future utilization (31).
Microscopic observation of cells of mutant and wild-type leaves also supports the hypothesis that sugars are not being exported from the leaves. The observed buildup of stored starch is not normal for a mature exporting leaf. A leaf unable to export sucrose would at first start photosynthesizing, but the inability to export most of the sucrose produced would result in an abundance of sugars, leading to the production of protective anthocyanins and the shutdown of the photosynthetic machinery (32). It is also possible that some or all of the phenotypes observed with the suc2 mutants are caused by reduced levels of sucrose as a hormonal signal rather than as a nutrient (33).
Finally, the 14C-sucrose feeding experiments also support the idea of an inefficient and abnormal system of sucrose movement in suc2 mutant plants. Although the mutants are able to move some labeled sucrose out of their leaves, a much higher percentage remains behind. Most of the transported sucrose remains within the rosette, both in younger and older leaves. This situation is in contrast to wild-type plants, where most of the labeled sucrose is moved to the inflorescences and roots.
All of this evidence points to an essential role for SUC2 in phloem loading and transport in Arabidopsis. The mutant plant is not able to complete its normal life cycle without the SUC2 protein. This suggests that Arabidopsis is indeed an apoplastic loader. By eliminating one isoform of a sizable gene family, we have shown the relative importance of the SUC2 gene product. This indicates that the sucrose transporters of Arabidopsis are not fully functionally redundant.
Although phloem loading is compromised in suc2 mutant plants, these plants are still able to transport sucrose to some extent. Homozygous mutant plants would not be able to live long if they were not able to transport sugars at all. 14C-sucrose fed to a leaf of a mutant plant was able to move to other organs but not in the same pattern as in wild-type plants, and also not with the same efficiency (Figs. 5 and 6).
There are several mechanisms by which suc2 mutant plants may be able to export sucrose from their leaves and move it throughout the plant, albeit at a much reduced efficiency. First, the SUC2 protein could still be expressed, but in a truncated form that does not function as efficiently. As discussed earlier, this is not very probable because the same phenotype was found with three different alleles that would truncate the protein at vastly different points, and all would lack the C terminus, which has been suggested to be essential for activity (19, 20). Second, it could be that SUC2 is only one of several sucrose transporters involved in phloem loading. Additional genes encoding sucrose transporters have been identified in Arabidopsis, namely SUC1 and the SUTs. A third possibility is that SUC2, which is expressed in both sink and source organs, may be more important in the retrieval of sucrose along the sieve tubes. According to this “leakage” model, sucrose would be loaded into sieve tubes in the mutants but would leak out during long distance transport into roots and inflorescences.
A fourth model is that other sucrose transporter genes are not normally expressed in phloem in source organs, but the absence of SUC2 results in their up-regulation in these areas. Evidence against this possibility arises from studies involving Vicia faba leaves fed sucrose via the xylem transpiration stream. In these leaves, the excess sugar led to a reduction in sucrose symporter activity (34). A fifth possibility is that in the absence of a sucrose transporter, Arabidopsis is able to load monomeric hexoses into its sieve tubes rather than the disaccharide sucrose. There are 26 hexose transporters in Arabidopsis (14).
This study describes direct genetic evidence for the in planta importance of a single sucrose transporter isoform in Arabidopsis. The phenotype produced when the SUC2 gene is disrupted proves that Arabidopsis is an apoplastic loader, which requires a sucrose transporter for efficient phloem transport.
Acknowledgments
We thank Rick Amasino and his laboratory, Sandra Austin-Phillips, Heather Burch, and Julie Galagan for generating the mutant lines. We acknowledge support from the Howard Hughes Medical Institute (J.G.). This work was supported by Grants DBI 9872638 from the National Science Foundation, DE-F602–88ER 13938 from the Department of Energy, and BIR-9220331 from National Science Foundation/Department of Energy/United States Department of Agriculture Collaborative Research in Plant Biology Program.
Abbreviation
- T-DNA
transferred DNA
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250473797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250473797
References
- 1.Rentsch D, Boorer K J, Frommer W B. J Membr Biol. 1998;162:177–190. doi: 10.1007/s002329900355. [DOI] [PubMed] [Google Scholar]
- 2.Komor E, Orlich G, Weig A, Koeckenberger W. J Exp Bot. 1996;47:1155–1164. doi: 10.1093/jxb/47.Special_Issue.1155. [DOI] [PubMed] [Google Scholar]
- 3.van Bel A J E. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:253–281. [Google Scholar]
- 4.Kuehn C, Franceschi V R, Schulz A, Lemoine R, Frommer W B. Science. 1997;275:1298–1300. doi: 10.1126/science.275.5304.1298. [DOI] [PubMed] [Google Scholar]
- 5.Stadler R, Sauer N. Bot Acta. 1996;109:299–306. [Google Scholar]
- 6.Truernit E, Sauer N. Planta. 1995;196:564–570. doi: 10.1007/BF00203657. [DOI] [PubMed] [Google Scholar]
- 7.DeWitt N D, Sussman M R. Plant Cell. 1995;7:2053–2067. doi: 10.1105/tpc.7.12.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Schaewen A, Stitt M, Schmidt R, Sonnewald U, Willmitzer L. EMBO J. 1990;9:3033–3044. doi: 10.1002/j.1460-2075.1990.tb07499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bush D R. Curr Opin Plant Biol. 1999;2:187–191. doi: 10.1016/S1369-5266(99)80034-X. [DOI] [PubMed] [Google Scholar]
- 10.Bush D R, Chiou T-J, Chen L. J Exp Bot. 1996;47:1205–1210. doi: 10.1093/jxb/47.Special_Issue.1205. [DOI] [PubMed] [Google Scholar]
- 11.Bush D R. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:513–542. [Google Scholar]
- 12.Sauer N, Tanner W. Bot Acta. 1993;106:277–286. [Google Scholar]
- 13.Ward J M, Kuehn C, Tegeder M, Frommer W B. Int Rev Cytol. 1998;178:41–71. doi: 10.1016/s0074-7696(08)62135-x. [DOI] [PubMed] [Google Scholar]
- 14.Lalonde S, Boles E, Hellmann H, Barker L, Patrick J W, Frommer W B. Plant Cell. 1999;11:707–726. doi: 10.1105/tpc.11.4.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krysan P J, Young J C, Tax F, Sussman M R. Proc Natl Acad Sci USA. 1996;93:8145–8150. doi: 10.1073/pnas.93.15.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giberson R T, Demaree R S J. Microsc Res Tech. 1995;32:246–254. doi: 10.1002/jemt.1070320307. [DOI] [PubMed] [Google Scholar]
- 17.Krysan P J, Young J C, Sussman M R. Plant Cell. 1999;11:2283–2290. doi: 10.1105/tpc.11.12.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imlau A, Truernit E, Sauer N. Plant Cell. 1999;11:309–322. doi: 10.1105/tpc.11.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caspari T, Stadler R, Sauer N, Tanner W. J Biol Chem. 1994;269:3498–3502. [PubMed] [Google Scholar]
- 20.Will A, Grassl R, Erdmenger J, Caspari T, Tanner W. J Biol Chem. 1998;273:11456–11462. doi: 10.1074/jbc.273.19.11456. [DOI] [PubMed] [Google Scholar]
- 21.Bowman J. Arabidopsis: An Atlas of Morphology and Development. New York: Springer; 1994. [Google Scholar]
- 22.Ding B, Haudenshield J S, Willmitzer L, Lucas W J. Plant J. 1993;4:179–189. doi: 10.1046/j.1365-313x.1993.04010179.x. [DOI] [PubMed] [Google Scholar]
- 23.Heineke D, Sonnewald U, Buessis D, Guenter G, Leidreiter K, Wilke I, Raschke K, Willmitzer L, Heldt H W. Plant Physiol. 1992;100:301–308. doi: 10.1104/pp.100.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dickinson C D, Altabella T, Chrispeels M J. Plant Physiol. 1991;95:420–425. doi: 10.1104/pp.95.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buerkle L, Hibberd J M, Quick W P, Kuehn C, Hirner B, Frommer W B. Plant Physiol. 1998;118:59–68. doi: 10.1104/pp.118.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reismeier J W, Willmitzer L, Frommer W B. EMBO J. 1994;13:1–7. doi: 10.1002/j.1460-2075.1994.tb06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemoine R, Kuehn C, Thiele N, Delrot S, Frommer W B. Plant Cell Env. 1996;19:1124–1131. [Google Scholar]
- 28.Kuehn C, Quick W P, Schulz A, Reismeier J W, Sonnewald U, Frommer W B. Plant Cell Environ. 1996;19:1115–1123. [Google Scholar]
- 29.Schulz A, Kuehn C, Reismeier J W, Frommer W B. Planta. 1998;206:533–543. [Google Scholar]
- 30.Russin W A, Evert R F, Vanderveer P J, Sharkey T D, Briggs S P. Plant Cell. 1996;8:645–658. doi: 10.1105/tpc.8.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marschner H. Mineral Nutrition of Higher Plants. New York: Academic; 1986. [Google Scholar]
- 32.Jang J-C, Sheen J. Trend Plant Sci. 1997;2:208–214. [Google Scholar]
- 33.Sheen J, Zhou L, Jang J-C. Curr Opin Plant Biol. 1999;2:410–418. doi: 10.1016/s1369-5266(99)00014-x. [DOI] [PubMed] [Google Scholar]
- 34.Chiou T-J, Bush D R. Proc Natl Acad Sci USA. 1998;95:4784–4788. doi: 10.1073/pnas.95.8.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]