Summary
The Ro 60 kDa autoantigen is a major target of the immune response in patients with systemic lupus erythematosus. In vertebrate cells, Ro binds misfolded small RNAs and likely functions in RNA quality control. In eukaryotes and bacteria, Ro also associates with small RNAs called Y RNAs. We present structures of unliganded Ro and Ro complexed with two RNAs at 1.95 and 2.2 Å resolution, respectively. Ro consists of a von Willebrand Factor A domain and a donut-shaped domain composed of HEAT repeats. In the complex, a fragment of Y RNA binds on the outer surface of the HEAT repeat ring, and single-stranded RNA binds in the toroid hole. Mutagenesis supports a binding site for misfolded RNAs that encompasses both sites, with a single-stranded end inserted into the toroid cavity. Our experiments suggest that one role of Y RNAs may be to regulate access of other RNAs to Ro.
Introduction
Although many RNA molecules must fold into complex structures in order to function, even relatively small RNAs, such as tRNAs, 5S rRNAs, and the spliceosomal U snRNAs, have a propensity to misfold into nonfunctional conformations (Herschlag, 1995; Schroeder et al., 2004). Abnormal RNAs can also be generated in other ways, including transcriptional errors, post-transcriptional editing events and incorrect processing. While multiple mechanisms by which defective or truncated mRNAs are recognized and targeted for degradation have been described (Maquat, 2004; Parker and Song, 2004), all these pathways require at least one round of translation, making them inaccessible to RNAs that are not translated. Far less is known of the quality control mechanisms by which misfolded and otherwise abnormal noncoding RNAs are recognized or of the fate (refolding vs. degradation) of misfolded structural RNAs.
A protein that likely functions in a quality control pathway for defective noncoding RNAs is the Ro 60 kDa protein (reviewed by Chen and Wolin, 2004). This RNA-binding protein was first described as a major autoantigen in patients suffering from two systemic rheumatic diseases, systemic lupus erythematosus and Sjogren’s syndrome. In patients with lupus, anti-Ro antibodies are highly associated with specific photosensitive skin lesions and with neonatal lupus, a syndrome in which mothers with anti-Ro antibodies give birth to infants with similar photosensitive skin lesions and a cardiac conduction defect, complete heart block (Patel and Werth, 2002; Buyon et al., 2004). Interestingly, mice lacking Ro develop a lupus-like syndrome consisting of glomerulonephritis, autoantibodies against chromatin and ribosomes and sunlight sensitivity, suggesting that the normal function of Ro is important for prevention of autoimmunity (Xue et al., 2003).
In all cells which have been studied, Ro is both nuclear and cytoplasmic. In the cytoplasm, Ro associates with small RNAs of unknown function called Y RNAs. Orthologues of the human Ro/Y RNA complexes have been described in other mammals, the frog Xenopus laevis, the nematode Caenorhabditis elegans and the eubacterium Deinococcus radiodurans (Hendrick et al., 1981; O’Brien et al., 1993; Farris et al., 1995; Van Horn et al., 1995; Chen et al., 2000).
Studies in vertebrate cell nuclei have revealed that Ro associates with misfolded small RNAs. In Xenopus oocytes, Ro binds a large population of variant pre-5S rRNAs that are inefficiently processed to mature 5S RNA and are eventually degraded (O’Brien and Wolin, 1994). The variant pre-5S rRNAs that Ro binds are longer at the 3’ end than mature 5S rRNAs, due to a failure of RNA polymerase III to terminate at the first transcription signal. These RNAs also contain point mutations that cause the RNAs to misfold into an alternative structure (O’Brien and Wolin, 1994; Shi et al., 1996). Recently, Ro was also found associated with misfolded variant U2 snRNAs in mouse embryonic stem cells (Chen et al., 2003). As Ro is the only protein described to interact with misfolded small RNAs in cells, Ro is proposed to function in quality control of defective noncoding RNAs (Chen et al., 2003).
In both mouse and bacterial cells, Ro is important for cell survival after ultraviolet (UV) irradiation. An orthologue of Ro in the radiation-resistant eubacterium Deinococcus radiodurans contributes to survival after UV irradiation (Chen et al., 2003). Consistent with a role in the handling of intracellular damage, both the Ro orthologue and the Y RNA are upregulated after irradiation. Further, mouse embryonic stem cells lacking Ro display increased sensitivity to UV, but not ionizing, radiation. Following UV irradiation, both Ro and the Y RNA undergo alterations in their subcellular distribution, changing from largely cytoplasmic to mostly nuclear (Chen et al., 2003). Although the mechanism by which Ro facilitates cell survival after UV irradiation is unknown, it was proposed that Ro targets UV-damaged RNAs for degradation (Chen et al., 2000; Chen et al., 2003).
Ro appears to interact with a variety of RNA structural elements. The binding site for Ro on Y RNAs is a highly conserved bulged helix (Wolin and Steitz, 1984; Pruijn et al., 1991; Green et al., 1998). As Ro binding is sequence-specific and dependent on two bulges that widen the major groove, Ro was proposed to bind in the major groove of the conserved Y RNA helix (Green et al., 1998). In contrast, while Ro recognition of misfolded pre-5S rRNAs requires the formation of an abnormal helix, neither the exact sequence of the helix nor the presence of bulges is important for binding (Shi et al., 1996). Moreover, Ro only binds misfolded pre-5S rRNAs that contain a 3′ extension, which is not present in mature 5S RNA (O’Brien and Wolin, 1994). Despite the differences in the requirements for Ro binding to Y RNAs and misfolded pre-5S rRNAs, misfolded pre-5S rRNAs compete with Y RNA for binding, suggesting the two RNAs bind to at least partly overlapping sites (Green et al., 1998).
To understand how Ro interacts with several different types of RNAs, and to determine how Ro discriminates between misfolded and properly folded RNAs, we have undertaken structural studies. Here we present both the 1.95 Å structure of unliganded Ro and the 2.2 Å structure of Ro complexed with two RNA fragments: a segment of Y RNA and a single strand of RNA. The Ro protein consists of two domains. One domain adopts a Rossmann fold similar to that of the von Willebrand factor A (vWFA) domain, found in a number of extracellular matrix proteins and proteins involved in cell adhesion, including integrins (Whittaker and Hines, 2002). In Ro, this domain contains a divalent cation binding site, which in integrins functions as a conformational switch and is involved in ligand binding. The second domain consists of a series of HEAT repeats arranged into a toroid. Remarkably, this domain contains a positively charged central hole, which binds single-stranded RNA. We report that Ro has two overlapping RNA binding sites: one on the outer surface of Ro that binds Y RNAs and a second, which includes the central cavity, that is likely responsible for binding misfolded RNAs. Since Y RNA binding would sterically block misfolded RNAs from binding, we propose that one role of Y RNAs is to regulate access of Ro to other RNAs.
Results and Discussion
The full-length Ro protein from Xenopus laevis was crystallized both in its unliganded and RNA-bound form. The structure of the unliganded protein was determined at 1.95 Å and has been refined to R= 21.3% and Rfree=25.7%. The refined model includes residues 5-537, except for the disordered loops 136–143 and 336–340, and 489 water molecules. The structure of the Ro/RNA complex was determined at 2.2 Å. The crystallographic asymmetric unit contains two protein/RNA complexes. The protein model includes residues 5–537 except for those in the loop 336–340, which is disordered. In addition to protein, each complex consists of a fragment of Y RNA and also a single stranded RNA oligonucleotide. We crystallized Ro in the presence of two strands of RNA which, when annealed, represent the minimal Ro binding site on Y RNA. It appears that the bound single-stranded RNA is one of the RNA strands that was inadvertently present in excess in the crystallization mix, and the modeled sequence is taken from the Y RNA. In one of the complexes, five of the residues in the single strand are ordered. In the second complex, there are seven ordered nucleotide residues. The two additional nucleotides are involved in crystal contacts. In addition to the protein/RNA complexes, the refined model includes 408 water molecules, two magnesium ions, and two acetate ions from the crystallization buffer. The structure has been refined to R=20.2% and Rfree=23.8% (Table 1).
Table 1.
Refinement Statistics
| Ro/RNA complex | |
| Resolution (Ǻ) | 20–2.2 |
| R/Rfree (%) | 20.2/23.8 |
| Rmsd bond lengths (Ǻ) | 0.0064 |
| Rmsd bond angles (degrees) | 1.269 |
| Rmsd improper angles (degrees) | 0.745 |
| Rmsd dihedral angles (degrees) | 21.13 |
| Rmsd on B factor: mainchain/sidechain | 1.25/2.28 |
| Ramachandran Plot (2 complexes per asymmetric unit) | |
| Residues in most favored region | 886 |
| in additionally allowed region | 78 |
| in generously allowed region | 4 |
| in disallowed region | 0 |
| No. of water molecules | 408 |
| Unliganded Ro | |
| Resolution (Ǻ) | 20.0–1.95 |
| R/Rfree (%) | 21.3/25.7 |
| Rmsd bond lengths (Ǻ) | 0.0081 |
| Rmsd bond angles (degrees) | 1.335 |
| Rmsd improper angles (degrees) | 0.854 |
| Rmsd dihedral angles (degrees) | 20.9 |
| Rmsd on B factor: mainchain/sidechain | 1.07/1.72 |
| Ramachandran Plot | |
| Residues in most favored region | 440 |
| in additionally allowed region | 33 |
| in generously allowed region | 1 |
| in disallowed region | 0 |
| No. of water molecules | 489 |
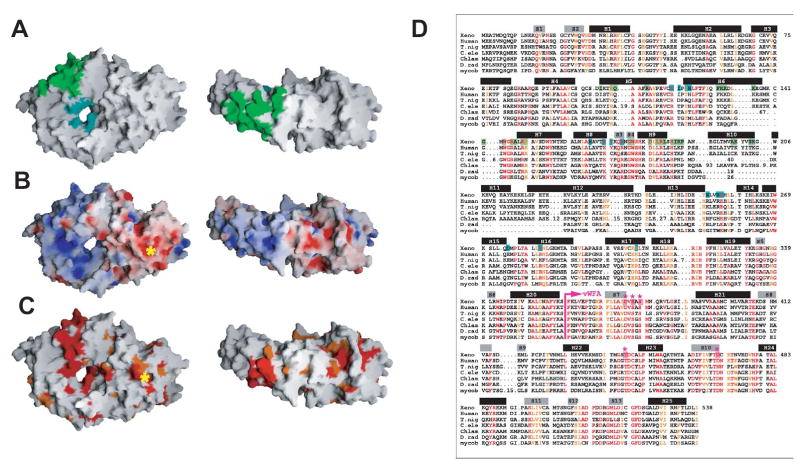
Overview of the Structure
In both the unliganded Ro structure and the structure of the ribonucleoprotein complex, the Ro protein forms an elliptical toroid, which measures 85 and 55 Å across and is 25–35 Å thick (Figures 1). The elliptical central cavity measures ~10–15 Å across. Ro consists of two domains. The domain that interacts with RNA is almost entirely helical and forms a closed loop (residues ~30–360), which is clasped shut by the second domain (residues ~5–30 and 360–537). The loop domain is reminiscent of HEAT repeat proteins such as importin-β (Cingolani et al., 1999) or the PR65/A subunit of protein phosphatase 2A (Groves et al., 1999), though the arrangement of repeats is less regular in Ro with distortion in the region encompassing helices H8 and H15. Except in this distorted region, the helices face the top and bottom surfaces of the toroid while the connecting loops line the central channel. Importin-β is similar to Ro in that it uses HEAT repeats to form a central cavity, in this case to enclose a domain of importin-α. In importin-β however, these repeats are arranged into a continuous spiraling superhelix, where helices line the central cavity and interact in a modular fashion with residues of importin-α (Cingolani et al., 1999). The central channel of Ro and a narrow band of residues along the outer surface of the loop domain are basic (Figure 2B). In the structure of the Ro/RNA complex, a 20 nt fragment of Y RNA is bound on the outer, basic surface of the Ro loop domain, where it interacts with residues that are highly conserved in all Ro proteins (see below), while the ssRNA is bound in the central positively charged cavity (Figures 1A and 2). Residues in this cavity that interact with RNA are also well conserved. To our knowledge, Ro is the first protein shown to interact with nucleic acids using HEAT repeats. The pumilio homology domain interacts with single-stranded RNA ends using the related ARM repeats (Wang et al., 2002), but its RNA binding surface differs from both RNA binding surfaces in Ro.
Figure 1. Structures of Ro and the Ro-RNA complex.
A. A molecular surface representation of Ro with bound RNA shown in pink. Ro is an elliptical toroid. A fragment of Y RNA is bound on its outer surface, and a singlestranded RNA oligonucleotide is bound in the central cavity. The Y RNA fragment is oriented such that in the full-length RNA, the 5′ and 3′ termini would be at the top of the figure (labeled), and the remainder of the RNA (see Figure 3C) would project towards the reader. The MIDAS motif, a divalent cation binding site, is shown in light blue. A 90° rotation about a horizontal axis relates views on the right and left.
B. Domain structure of Ro, with orientations as in panel A. Ro consists of a toroid of HEAT repeats and a von Willebrand Factor A (vWFA) domain. Ro is colored from blue at the N-terminus to red at the C-terminus. The MIDAS motif in the vWFA domain is marked with a grey sphere. Helices are labeled H1 through H25, and beta strands are labeled S1 through S13.
C. Superimposed backbone traces of unliganded Ro and the Ro-RNA complex. The carbon alpha trace of Ro from the ribonucleoprotein complex (pink) is superimposed on that of unliganded Ro (blue). At left, the carbon alpha traces are aligned using residues 144–278, and at right they are aligned using residues 360–537. Loop 136–143, close to the Y RNA binding surface of Ro, becomes ordered upon RNA binding, and loop 168–175, involved in single stranded RNA binding, rearranges. Asterisks mark the positions of residue 278 (blue) and loops 144–278 and 168–175.
Figure 2. Ro surfaces that interact with RNA.
A. A molecular surface representation of the Ro structure. Surfaces that interact with Y RNA are green while those that interact with single stranded RNA are teal. The views are identical to Figures 1A and 1B.
B. Ro colored by electrostatic surface potential. Surfaces that bind RNA have a positive potential, and the vWFA domain is mostly acidic. The asterisk marks the MIDAS motif.
C. Ro colored by sequence conservation according to the alignment in panel D. Surface areas where sequences are strictly conserved and similar are red and orange, respectively. Surfaces that interact with Y RNA and with single stranded RNA are conserved as are the characteristic MIDAS motif residues.
D. Sequence alignment of Ro proteins from X. laevis, humans, T. nigris, C. elegans, C. reinhardtii, D. radiodurans, and the mycobacteriophage Bxz1 gp220 protein. Residues that are identical in at least 6 of the 7 sequences are red, and residues that are similar (L=V=I, F=Y=W=H, S=T, E=D, R=K, N=Q) in at least 6 of the 7 sequences are orange. Residues in X. laevis Ro that interact with Y RNA are green, and those that interact with single stranded RNA are teal. The beginning of the vWFA domain is indicated by a violet arrow, and residues in the MIDAS motif (D-x-S-x-S…T…D) are violet and marked with violet asterisks. Alpha helices and beta strands are indicated by black and grey bars, respectively, and are numbered as in Figure 1B.
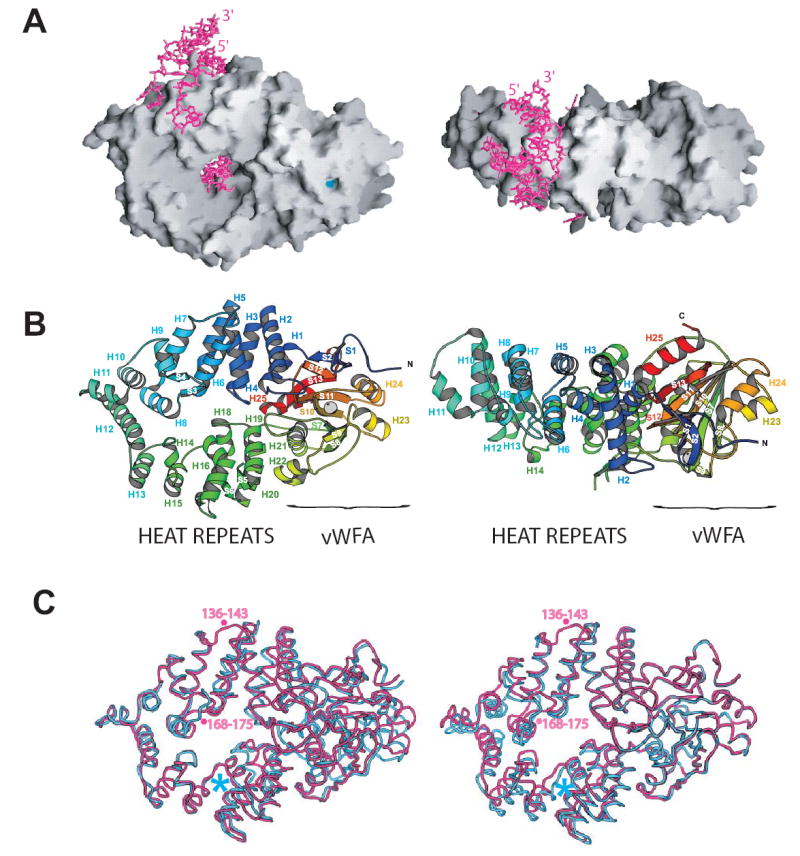
The second domain of Ro adopts a Rossmann fold similar to that of the von Willebrand Factor A (vWFA) domain found in the eponymous platelet adhesion protein and also in the integrins (Whittaker and Hines, 2002). Its core, a five-stranded parallel beta sheet encompassing strands S5, S6, S7, S8, S10, S11and S13, is sandwiched between two sets of parallel alpha helices, H23, H24 and H21, H22, H25 (Figure 1B). The plane of the sheet is normal to the long axis of Ro. Five highly conserved residues at the C-terminal edge of the beta sheet form a divalent cation binding site, termed the MIDAS motif (metal-ion dependent adhesion site) in integrins, where the motif was first described (see Shimaoka et al., 2002, for review). In the integrins, this motif (D-x-S-x-S…T…D) is a conformational switch that is critical both for transmitting changes that accompany ligand binding and for ligand binding itself. A glutamate or an aspartate from the ligand completes the MIDAS metal coordination. The metal coordination observed in Ro is similar to that in the ligand-bound integrins (Figure 3). In the structure of the Ro/RNA complex, crystallized in the presence of 100 mM magnesium acetate, a magnesium ion is directly coordinated by S378 and S380 of the MIDAS signature sequence D-X-S-X-S, T445, two water molecules, and an acetate ion (Figure 3B). The acetate may be a mimetic for a glutamate or an aspartate in an as yet unknown interaction partner of Ro. Residues D376 and D469 of Ro coordinate one of the water molecules. The second water molecule in the Ro MIDAS motif is hydrogen bonded to Y47, a conserved residue near the N-terminus of Ro. The metal coordination site is very similar in the unliganded Ro structure, except that in the absence of magnesium acetate in the crystallization solutions, water molecules have been modeled into the Mg++ and the acetate sites. The thirty most N-terminal residues of Ro pack against the sides of the vWFA domain, and the N-terminal strands S1 and S2 form a small sheet with strand S12 in the vWFA fold. One surface of the vWFA domain, which includes the MIDAS motif, is highly acidic (Figure 2B).
Figure 3. The MIDAS Motif.
A. View of the MIDAS motif with Ro in an orientation as in Figure 1B, left panel. Ro is colored as in 1B. Residues in the MIDAS motif are indicated.
B. A closer view of the MIDAS motif with Ro in a different orientation. The magnesium ion, shown in pink, is coordinated by S378, S380, T445, an acetate ion (Ac) and two water molecules (w). D376 and D469 form hydrogen bonds to one of these water molecules while Y47 at the Ro N-terminus forms a hydrogen bond to the second water molecule.
We note that other RNA-binding proteins, whose structures have not yet been determined, may have a similar architecture to the Ro protein. Both the human TEP1 and its likely homolog Tetrahymena p80 share sequence homology with the HEAT repeat domain of the Ro protein and also contain an adjacent vWFA domain (Anantharaman et al., 2002; Bateman and Kickhoefer, 2003). TEP1 is a component of large cytoplasmic ribonucleoprotein particles called vaults. Although the function of vaults is unknown, TEP1 binds small RNAs that are similar in size to Y RNAs (Kickhoefer et al., 2001). Although Ro residues critical for Y RNA binding are not conserved in TEP1, certain residues in the central cavity that interact with single-stranded RNA are conserved. Thus, TEP1/p80 may also fold into a toroid that interacts with single-stranded RNA.
Conformational changes in Ro upon ligand binding
A comparison of the structure of Ro bound to RNA with that of unliganded Ro shows conformational changes (Figure 1C). The rms deviation of the carbon alpha traces for the two structures is 1.51 Å. The vWFA domain is very similar in both structures (rmsd for this domain alone is 0.5 Å), but a conformational change takes place in the RNA-binding closed loop domain. A fragment encompassing residues ~144–278, which includes helices H6-H15, rotates by about 13 degrees. Such flexibility has also been observed in other HEAT repeat proteins (Cingolani et al., 1999) and in ARM repeat proteins (Wang et al., 2002; Huber et al., 1997). There are additional conformational changes in two loops that are involved in RNA binding directly or are close to regions that contact RNA. Residues 136–143, close to the Y RNA binding region, are disordered in the unliganded Ro structure and become ordered in the RNA-bound structure, and the loop encompassing residues 168–175 moves further into the basic cavity as Ro binds single stranded RNA. The flexibility in Ro’s RNA binding surface may allow it to accommodate its varied RNA partners.
Observed structural changes upon RNA binding are confined to the HEAT repeat ring, and we have noted no significant changes in the vWFA domain upon RNA binding. Nevertheless, given the long range conformational changes in integrin structure that accompany ligand binding at the MIDAS motif (Shimaoka et al., 2002), it is possible that the binding at the MIDAS motif of as yet unidentified effectors mediates structural rearrangements.
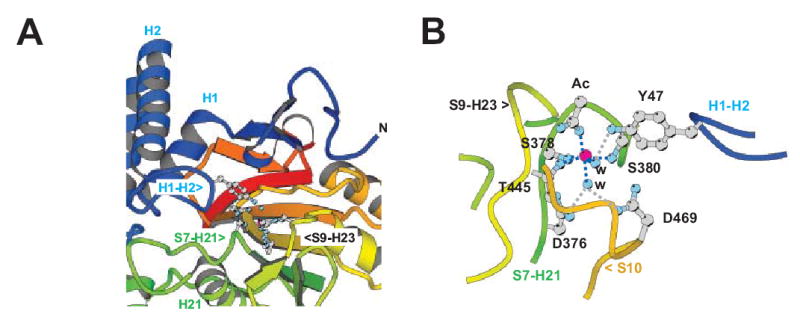
The conformation of bound Y RNA and its interactions with Ro
In the Ro/RNA structure, the Y RNA fragment forms a frayed duplex, where the unpaired ends are part of a bulge in the complete ~100 nt Y RNA (Figure 4). Helix H9 (182–187) in Ro is positioned as if to wedge apart the RNA stem. The unpaired nucleotides in the bulge, G9-A10 and U11–U12, are flipped away from the RNA stem and located to either side of this helix. The bases of A10 and G9 are stacked, as are those of U11 and U12. The side chain of L185 in helix H9 is intercalated between the bases of C8 and G9 and makes hydrophobic interactions with both. The conserved residue W177 is stacked with the G9 base. C8 in the stem is unpaired and interacts with N2 in the H187 side chain, which is at the C-terminus of helix H9. C8 and the histidine imidazole stack over the adjacent base pair C7:G13 to form one end of the stem region. The H187 imidazole appears to be protonated since N1 is perfectly positioned to form a second hydrogen bond to U11 phosphate. C8 also interacts with the L185 carbonyl via N4.
Figure 4. Ro interactions with Y RNA and single-stranded RNA.
A. Secondary structure diagrams for Xenopus laevis Y3 RNA. Two alternate conformers that may form are shown (Green et al., 1998). RNA bases that were critical for Ro binding in biochemical experiments are pink (Green et al., 1998). The Ro binding site is conserved in all known Y RNAs.
B. Fragment of Y RNA bound on the outer surface of Ro. The two RNA oligonucleotides annealed to form the Y RNA mimic are shown in pink and teal. The identity of the nucleotides and the numbering system used in the text are indicated.
C. Details of the Ro-Y RNA interaction. Helices from the Ro HEAT repeat domain are shown as cylinders and side chains that interact with RNA are indicated. Helix H9 of Ro appears wedged into the Y RNA duplex, and unpaired nucleotides, which would be part of a bulge in full length Y RNA, are arranged on either side of it. H187 (pink) interacts with C8 at the end of the duplex region.
D. Summary of Ro interactions with Y RNA. Uracil bases that are iodinated at the C5 position are indicated with asterisks. Hydrogen bonding interactions (<3.5 Å) are shown by solid lines, and possible electrostatic interactions (< 7 Å) are shown with dotted lines. Atoms in the nucleotide bases that are involved in hydrogen bonds with protein are indicated in grey. Conserved and similar residues in Ro are red and orange, as in Figure 2D. Residues involved in # stacking interactions are boxed in yellow, and residues with extensive van der Waals interactions with nucleotide bases are shown in teal. For clarity, interactions with the RNA ribose rings are not indicated. Interactions mediated by a carbonyl O or an amide N from the protein backbone rather than by a side chain are indicated by “CO” or “N”, respectively.
E. Difference (Fo-Fc) electron density for single stranded RNA bound in the central cavity of Ro. The density, from a 2.2 Å map, is shown at three times the r.m.s. variation of the map. The refined model for this portion of RNA is shown in stick presentation.
F. Structure of the single stranded RNA bound in the cavity of Ro. Bases U6-A10 (pink) are ordered in both Ro/RNA complexes in the asymmetric unit while G4 and G5 (light pink) are ordered only in one complex.
G. Ro interactions with single stranded RNA bound in the central cavity. The notations are as in panel D. Only interactions that are the same in both Ro/RNA complexes in the asymmetric unit are shown.
Ro recognizes Y RNA both by specific interactions with the nucleotide bases and by shape complementarity. The major groove of the stem region adjacent to the bulge is widened to 18 Å, allowing for direct contacts with the bases. Almost all of the hydrogen bonding interactions are to nucleotide bases along one strand, specifically to G4, G5, U6, C7, and C8, all of which are part of the helical stem region, and to G9 and A10 in the bulged region. The residues involved in hydrogen bonding to the nucleotide bases are highly conserved in all Ro proteins (Figures 2C, 2D, and 4). Critical residues include R146, which forms hydrogen bonds to both G4 and G5, R149, which forms hydrogen bonds to G5 and U6, and W144, which interacts with the C7 N4 via its carbonyl. The crucial role of H187 and its interaction with C8 was described above. D181 and K133 along with the main chain carbonyl oxygen of G176 interact with G9, a bulged residue, and K133 hydrogen bonds to the nucleotide base of A10. Interactions with the second strand are few, and only R146, which interacts with C15 by way of a water molecule, is conserved. Additionally, K200 forms a hydrogen bond with the base of U11.
Ro makes numerous contacts with the Y RNA backbone. Most of the residues in loops 142–149 and 176–187 are involved in hydrophobic or van der Waals contacts to the backbone or hydrogen bonds to the 2’OH groups. A number of residues (see Figure 4: K108, Q109, K136, G142, K150, A147, R184) form hydrogen bonds to the phosphate groups of one RNA strand. Additionally, several basic residues near the protein-RNA interface could contribute to electrostatic interactions with the charged phosphates (R113, K132, R149).
Single-stranded RNA is bound in the positively charged central cavity
A single strand of RNA, apparently one of the strands used to form the Y RNA duplex, is bound in the basic central channel. In one complex in the asymmetric unit, the electron density is consistent with nucleotides U6-A10, while in the second complex nucleotides G4 and G5 are also ordered by their participation in crystal contacts. The sequence encompassing U6-A10 is bound almost identically in both protein-RNA complexes in the asymmetric unit. The RNA is in an extended conformation that is stabilized by stacking interactions and van der Waals contacts with Ro (Figures 4E to 4G): R255 and R283 stack on either side of C7, R318 makes van der Waals contact with the C8 base, M166 and R252 sandwich the G9 base, and L313 packs against the A10 base. Side chains from residues T123, K170, Y171, R174, and the main chain nitrogen of L278 are involved in hydrogen bonds to the phosphate backbone, and several other basic residues (R255, R283, R307) are close enough to the phosphate backbone for favorable electrostatic interactions. There are also hydrogen bond interactions to the C8 and G9 bases: R120 interacts with O2 and N3 of C8 while the main chain nitrogens of D275 and R252 interact with N1 and N2 of G9.
We do not yet know the degree of specificity with which Ro binds single-stranded RNA. The ordered binding in the crystals would indicate that some level of specificity may be possible. The predominance of backbone and stacking interactions argues for a low level of specificity, however. The plasticity of the binding site also suggests that many sequences might be accepted.
Recognition of misfolded RNA
It is probable that Ro uses the cavity to interact with misfolded RNAs, rather than Y RNAs. While all Y RNAs contain a large single-stranded internal loop (Figure 4A), the loop is dispensable for Ro binding and is not protected by Ro in nuclease digestion experiments (Wolin and Steitz, 1984; Green et al., 1998). However, all misfolded pre-5S rRNAs associated with Ro contain 8–10 additional gene-encoded nucleotides at the 3’ end, due to readthrough of the first termination signal (O’Brien and Wolin, 1994). Enzymatic structure probing experiments reveal that these 3’ nucleotides are singlestranded (Shi et al., 1996; G. F. and S. L.W., manuscript in preparation). To test the importance of the 3’ extension for Ro binding, we performed electrophoretic mobility shift assays (EMSA) using a mutant pre-5S that is bound by Ro (Shi et al., 1996). Removal of the 3′ extension decreased the affinity of Ro for the RNA by more than tenfold (Figure 5A and data not shown). As the 3’ extension is important for Ro binding to misfolded pre-5S rRNA, we propose that the 3’ end binds in the inner hole.
Figure 5. Binding of wild-type and mutant Ro proteins to Y RNA and misfolded pre-5S rRNA.

A. Effect of shortening the 3’ end of pre-5S rRNA on Ro binding. 32P-labeled misfolded X. laevis oocyte pre-5S rRNA (left) or pre-5S rRNA lacking 8 nt of the 3′ trailer (right) was incubated without protein (lane 1) or with the indicated concentrations of Ro protein (lanes 2–9). The misfolded pre-5S rRNA contains several mutations that cause it to fold efficiently into the structure recognized by Ro (Shi et al., 1996). Protein-RNA complexes were separated from unbound RNA by native gel electrophoresis.
B. Binding of mutant Ro proteins to RNA. 32P-labeled Xenopus Y3 RNA (left panels) or misfolded pre-5S rRNA (right panels) was incubated either without protein (lane 1) or with increasing amounts of wild-type Ro (top row) or the indicated mutant proteins (bottom four rows). Protein concentrations are given in nM above lanes 2–9. Protein-RNA complexes were separated from unbound RNA in native gels.
To further explore the role of the central cavity in binding RNA, we mutated residues in the cavity that interact with single stranded RNA in the Ro/RNA structure (K170A/R174A, R255S/R283S, and R120S) and tested their affinities for both Y RNA and misfolded pre-5S rRNA using EMSA. As would be expected if Y RNA does not bind in the central cavity, none of the mutations had a measurable effect on the affinity of Ro for Y RNA (Figure 5B and Table 2). In contrast, the double mutant K170A/R174A had an ~ 7-fold decrease in affinity for misfolded pre-5S rRNA (Figure 5B and Table 2), confirming that residues in the cavity are important for binding this RNA. It is plausible that these residues interact with the phosphate backbone of the 5S rRNA single-stranded 3’ end much as they interact with the single strand of RNA observed in the structure.
Table 2.
Binding of mutant Ro proteins to RNA
| Kd xY3 (nM) | Kd pre-5S (nM) | |
|---|---|---|
| wild-type | 5.2 ± 2.5 | 25.0 ± 13.5 |
| Residues that interact with Y RNA | ||
| R146S R149S | 21.9 ± 1.5 | 48.5 ± 14.4 |
| H187S | 153.2 ± 13.6 | 105.2 ± 20.2 |
| R184A | 30.8 ± 5.5 | 187.0 ± 40.2 |
| K108A Q109A R184A | 198.3 ± 10.3 | 135.3 ± 12.2 |
| K136A | 4.8 ± 2.3 | 22.3 ± 7.5 |
| Residues in the central cavity | ||
| K170A R174A | 4.7 ± 2.6 | 179.5 ± 24.1 |
| R255S R283S | 3.4 ± 0.3 | 46.8 ± 0.2 |
| R120S | 5.6 ± 0.7 | 34.1 ± 4.8 |
In contrast to K170A/R174A, several mutations had little effect on pre-5S rRNA binding. R255 and R283, which stack on either side of a cytosine base in the Ro/RNA complex and may contribute to electrostatic interactions with the single stranded RNA phosphate backbone, do not appear to be important for binding misfolded pre-5S rRNA. Likewise, R120, which forms hydrogen bonds to a cytosine base in the Ro/RNA complex structure, is not important for binding pre-5S rRNA. The absence of a mutation effect on pre-5S rRNA binding at these positions may suggest, as noted above, that the interactions with single stranded RNA within the central cavity may vary for different RNA species. Further, we show below that misfolded pre-5S rRNAs interact with an extensive surface on Ro not confined only to the central cavity. Thus, the effects of mutations that alter only a small part of the interaction surface may be expected to be subtle.
In addition to a free 3’ end, Ro recognition of variant pre-5S rRNAs requires that the RNA misfold to form an alternative helix (Shi et al., 1996). As Ro binding protects this helix and adjacent helices from ribonuclease digestion (Shi et al., 1996), Ro likely also contacts helical elements in the misfolded RNA. We propose that these duplex portions of the misfolded 5S rRNA bind on the positively charged outer surface of the HEAT repeat domain, partly the same surface to which Y RNA binds. To explore the role of the Y RNA binding site in the binding of misfolded 5S rRNA, we mutated residues that interact with Y RNA in the Ro/RNA structure (H187S, R184A, K136S, R146S/R149S, and K108/Q109/R184). We measured the affinities of the Ro mutants for both Y RNA and 5S rRNA by EMSA. Some of the mutations in the Y RNA binding site affected not only Y RNA binding, as expected, but also the binding of misfolded 5S rRNA (Figure 5B and Table 2). The H187S and K108A/Q109A/R184A mutants had the most significant effects on Y RNA binding with, respectively, ~30- and ~38-fold decreases in affinity. Their effects on misfolded 5S rRNA were more modest but still significant, with ~4–5-fold decreases in affinity (Table 2). Residue R184 also appears to be important for both Y RNA and 5S rRNA binding, as the R184A mutant has an ~6–7- fold reduced binding affinity for both of these RNAs. Some residues that are important for Y RNA binding appear relatively unimportant for 5S rRNA binding. For example, mutating R146 and R149, residues involved in specific interactions with Y RNA bases, affects Y RNA binding but has little or no effect on 5S rRNA binding. Together these experiments suggest that although Y RNA and misfolded 5S rRNA may not interact with Ro in the same way, they bind to overlapping surfaces. These results are consistent with previous data revealing that misfolded pre-5S rRNA competes with Y RNAs for binding to purified Ro (Green et al., 1998).
Our mutagenesis results agree with a model in which duplex portions of the 5S rRNA bind to the positively charged outer surfaces of the Ro HEAT repeat ring, including parts of the Y RNA binding site, while the single stranded 5S rRNA terminus extends into the cavity and strengthens the binding interaction.
This model suggests that one role of Y RNAs may be to sterically block misfolded and other RNAs from binding to Ro. Possibly, then, Y RNA plays a regulatory role in small RNA quality control in that its dissociation from Ro allows these misfolded RNAs to bind. The mechanism by which Y RNAs may dissociate from Ro is currently unclear. Conformational changes associated with single-stranded RNA binding, nuclear localization, and/or association with other proteins or co-factors could all result in a weakened affinity for Y RNA, allowing its dissociation.
The implications of the central cavity for misfolded RNA binding
The central channel is wide enough to accommodate single but not doublestranded RNA, and it is likely that short RNA segments simply thread into the channel. Such threading requires the single stranded portion to have a free end. A loop, for example, could not bind. The use of a hole, or elongated cavity, to recognize ends has been previously observed in proteins that recognize peptide or nucleic acid termini. The ring-shaped lambda exonuclease, for example, can bind DNA only by sliding over the ends, biasing the enzyme to cleavage in terminal regions (Carter and Radding, 1971).
It is also possible, however, that the HEAT repeat ring can open to allow RNA to enter the channel. For example, the vWFA domain could initially unclasp the Ro N-terminus, then re-bind it to encircle the RNA. Interestingly, the MIDAS motif in the vWFA domain is close to the interface with the N-terminal region of Ro, with one N-terminal residue, Y47, forming part of the secondary coordination sphere of the MIDAS magnesium ion. Conceivably, the MIDAS motif could play a role in conformational changes that accompany ring opening and closing. Transient opening of the loop might allow longer single stranded regions to bind more rapidly and might allow even for the binding of large RNA loops.
Concluding Remarks
We have proposed that the central basic cavity in Ro binds the 3’ ends of misfolded RNAs while a positively charged, conserved region near the central cavity, which includes the Y RNA binding site, binds other portions of misfolded RNAs. One possibility is that Ro binding assists in destabilizing misfolded helices, facilitating either correct folding or degradation. Since vWFA domains involved in cell adhesion frequently mediate protein-protein interactions, we speculate that in Ro this domain serves as a platform for recruiting additional protein components of the quality control pathway, such as helicases and/or ribonucleases. It is also conceivable that Ro assists these downstream components in their function, perhaps, given its toroidal shape, as a processivity factor.
Experimental Procedures
Protein Expression and Purification
The Ro protein was overexpressed in insect cells infected with recombinant baculovirus carrying the Xenopus laevis Ro cDNA (Green et al., 1998). To prepare mutants of Ro, the cDNA encoding X. laevis Ro (O’Brien et al., 1993) was inserted between the BamHI and HindIII sites in the pFastbac1 vector (Invitrogen). Mutations (Table 2) were introduced using the QuickChange site-directed mutagenesis kit (Stratagene). Plasmids were sequenced to confirm the presence of the expected mutation. Recombinant baculovirus was obtained using the Bac-to-Bac expression system (Invitrogen) following manufacturer instructions.
For both wild type and mutant Ro, High Five cells were grown to a density of 1.5–2x106 cells/ml in Excel 405 medium (JRH), infected at 3–5 plaque forming units/cell, and harvested 72 hr postinfection. Cells were resuspended in buffer A (100 mM NaCl, 25 mM Tris pH 8.0, 50mM DTT, 3 mM MgCl2, 1 mM PMSF) and lysed by sonication. Protein purification was as described (Green et al., 1998), except for high levels of DTT (~50 mM) present throughout purification. Briefly, the lysate was applied to a heparin column (Amersham) and eluted with a linear salt gradient in buffer B (20 mM Tris pH 8.0, 10% glycerol, 50 mM DTT, 550 mM NaCl). Fractions containing Ro were pooled, concentrated, and loaded onto a Superdex200 gel filtration column (Amersham).
The protein was eluted in buffer C (100 mM NaCl, 20 mM Tris pH 8.0, 10% glycerol, 50 mM DTT) and concentrated to 10–15 mg/ml. Yields from 1L of insect cell culture varied between 5–10 mg.
After several attempts, a highly selenomethionine-substituted protein was expressed in insect cells. The High Five cells were infected as described above, but eight hr postinfection, the Excel 405 medium was exchanged for a medium deficient in methionine and yeastolate (JRH). At about 20 hr post-infection, seleno-L-methionine (Sigma) was added to a final concentration of 50 mg/ml. Cells were harvested and protein purified as above. The yield from 1L of insect cell culture was ~3 mg. Several attempts to quantitate selenium substitution by electrospray mass spectrometry were unsuccessful. A protein that was only 40% seleno-substituted, as determined by electrospray mass spectrometry, was produced when Excel 405 deficient in methionine but not yeastolate was used as a depletion medium and the depletion period was seven hr instead of twelve. Selenomethionine-substituted Ro was purified like the native protein, except that 1.25 mM Tris (2-carboxyethyl) phosphine hydrochloride (TCEP) was used in place of DTT.
Complex formation
Gel-purified RNA oligomers were purchased from Dharmacon. The oligomers were resuspended in Buffer D (1.5 mM MgCl2, 125 mM NaCl, 20 mM Tris pH 7.5) and quantitated on the basis of absorbance at 260 nm. To form the Y RNA fragment, two strands of RNA were mixed in roughly stoichiometric quantities and annealed. The RNA was heated to 95°C for two minutes and then plunged into an ice bath. Ro was added to the RNA and incubated on ice for one hour. To determine the correct ratio of protein to RNA for crystallization, we performed gel shift assays (Batey et al, 2001).
Crystallization and Data Collection
Crystallization of unliganded Ro
Crystallization of the unliganded protein was difficult as only some batches of protein crystallized and the quality of the crystals varied. Rod-shaped crystals were grown at room temperature by hanging-drop vapor diffusion against a well containing 25% PEG 4000, 200 mM sodium formate, 30 mM NaH2PO4, and 100 mM Tris HCl pH 8.5. Crystals grew to a maximum dimension of 300 x 100 x 100 μm within a week. They belong to spacegroup P212121 (a=68.8, b=91.7, and c=93.9 Å). The best crystals diffracted to 1.8 Å. Crystals were mounted in a loop directly from the hanging drop and were flash frozen in liquid nitrogen prior to data collection. All data were collected at - 160°C at beamline X25 at Brookhaven National Laboratory. Data were integrated, scaled, and merged using the program HKL2000 (Otwinowski and Minor, 1997).
Attempts to grow high quality crystals of selenomethionine-substituted Ro were unsuccessful in the absence of RNA.
Protein-RNA complex
The Ro protein was complexed with a series of RNA duplexes containing its recognition site. The RNA sequences varied in length and in overhang. The best crystals were obtained with an RNA 20-mer (see Figure 4D) using the hanging-drop vapor diffusion method. The reservoir solution contained 29% 2-methyl-2,4-pentanediol, 50 mM sodium acetate pH 3.6, and 100 mM magnesium acetate. The drops were composed of 1 μl of Ro/RNA complex at ~10 mg/ml and 0.5 μl of reservoir solution. Plate shaped crystals grew to a maximum dimension of 300 x 100 x 100 μm within a week. The crystals belong to spacegroup P21 (a= 60.6, b=153.4, c=80.5 Å and beta=92.8°). The crystals diffract to 2.2 Å. In contrast to crystals of unliganded Ro, crystals of the Ro/RNA complex grew reproducibly from different batches of protein, and the selenomethionine substituted protein crystallized readily. In preparation for heavy atom soaking experiments, crystals were transferred to a solution containing 50 mM TrisCl pH 7 and 1.25mM TCEP, in place of the sodium acetate and DTT in the crystallization solution. Crystals were loop mounted directly from this solution and flash frozen in liquid nitrogen prior to data collection.
Structure Determination
The structure of the Ro/RNA complex was determined by the multiple isomorphous replacement method and anomalous scattering. As a reference data set, we used data from a crystal in which the protein was 40% selenomethionine- substituted and in which four uridines of the RNA duplex were substituted with iodine at the C5 position (Figure 4D). Crystals grown with unsubstituted protein were not isomorphous with crystals grown from protein with ≥40% selenomethionine substitution. Nor were crystals grown with unsubstituted RNA isomorphous to those obtained with the iodinated RNA. We used the iodine anomalous signal from the reference crystal and data from three heavy atom derivatives to determine the phases. The selenium derivative was prepared from the highly substituted selenomethionine protein. To prepare a lead and a uranyl derivative, we soaked a reference crystal in either 1 mM lead acetate or 1.5mM uranyl for one day. Data sets were collected at beamlines X25 and X29 at Brookhaven National Laboratory (Table 3, Supplementary Data), and data were integrated, scaled, and merged using the program HKL2000 (Otwinowski and Minor, 1997). The program Solve (Terwilliger and Berendzen, 1999) was used to solve the iodine anomalous Patterson map and to calculate the first phases. These were used to determine the positions of the lead and uranyl sites in two of the other derivatives. Improved density modified phases calculated using the iodine, the lead, and the uranyl data were sufficient to locate thirtytwo of forty selenium sites in difference Fourier maps. Incorporation of the phase information from the selenium derivative and subsequent density modification by solvent flipping in CNS (Brunger et al., 1998) yielded very clear 3.0 Å electron density maps. We used the program O (Kleywegt and Jones, 1997) in model building. We were guided by the positions of the selenomethionine side chains in aligning the sequence of Ro to a model of the polyalanine backbone. We built the Y RNA fragment into the density guided by the iodine positions, which fix the sequence alignment for the Y RNA. A partial model comprising the two protein/Y RNA complexes in the asymmetric unit was first refined at 2.6 Å. The model was rebuilt, then the phases were extended to 2.2 Å. Several rounds of manual rebuilding and refinement followed. Although density for the single strand of RNA in the positively charged central cavity of Ro was clear in the initial maps, this RNA was modeled only in later stages of the refinement when higher resolution maps were available. The RNA was modeled into very strong, continuous Fo- Fc difference density (signal/noise> 3.0, shown in Figure 4E). The sequence of purines and pyrimidines, evident from the electron density, and the absence of iodines in this strand are consistent with our sequence assignment. We have modeled a single magnesium ion, two water molecules, and an acetate into a conserved MIDAS motif pocket in Ro. The coordination geometry and bond distances for the magnesium ion are consistent with those observed in other MIDAS motif structures (Baldwin et al., 1998).
Table 3.
Data Collection and Phasing Statistics
| Reference (I anom)* | Semet* | UO2Ac2 | PbAc2* | High res. Ro/RNA | Unliganded | |
|---|---|---|---|---|---|---|
| Wavelength (Ǻ) | 1.4 | 0.9797 | 1.1 | 0.9486 | 1.1 | 0.979 |
| Resolution (Ǻ) | 3.0 | 3.0 | 3.0 | 3.2 | 2.2 | 1.95 |
| Spacegroup | P21 | P21 | P21 | P21 | P21 | P212121 |
| Cell Constants | ||||||
| a (Ǻ) | 60.1 | 60.4 | 60.3 | 60.5 | 60.3 | 68.8 |
| b (Ǻ) | 153.6 | 153.9 | 153.9 | 153.4 | 153.6 | 91.7 |
| c (Ǻ) | 80.4 | 80.6 | 80.3 | 80.5 | 80.6 | 93.9 |
| β (in degrees) | 92.9 | 92.8 | 93.0 | 93.1 | 92.8 | |
| Completeness (%) (highest resolution bin) | 97.8 (97.1) | 99.8 (99.8) | 79.8 (79.8) | 99.9 (100) | 98.4 (86.2) | 96.5 (98.9) |
| Avg redundancy | 3.7 | 2.8 | 2.3 | 3.8 | 3.7 | 4.5 |
| Rsym (%) (highest resolution bin) | 6.7 (11.7) | 9.2 (16.6) | 6.1 (12.2) | 10.2 (19.6) | 7.9 (22.8) | 6.1 (24.5) |
| I/σ (highest resolution bin) | 23.95 (14.96) | 16.0 (7.8) | 16.8 (7.6) | 18.6 (8.9) | 17.9 (4.7) | 19.4 (3.5) |
| Phasing Statistics (Solve) | ||||||
| Sites/asymmetric unit | 8 | 36 | 2 | 2 | ||
| Phasing Power (Solve) | ||||||
| Acentric/centric | 2.51/1.19 | 0.54/0.46 | 0.44/0.40 | |||
| Anomalous | 0.57 | 0.28 | 0.25 | |||
| FOM (highest resolution bin) | 0.52 (0.51) | |||||
optimized anomalous diffraction data sets
Rsym = ∑∑j|Ij−<I>|/∑Ij
Phasing power= rms|FH|/ɛ, where the denominator is the lack of closure error
Anomalous phasing power= rms|F+-F−|/ɛ, where the denominator is lack of closure error FOM=figure of merit= ∫ P(θ)exp(iθ)/∫ P(θ)dθ, where P is the probability distribution of the phase angle θ.
Prior to refinement using CNS (Brunger et al., 1998), 10% of all reflections were set aside to monitor Rfree. Non-crystallographic symmetry restraints were maintained in the initial stages of refinement, then released. Rounds of manual rebuilding were followed by cycles of torsion angle dynamics, positional refinement, and individual Bfactor refinement. For the final model, R= 20.2% and Rfree= 23.8% over all data between 20.0 and 2.2 Å. Refinement statistics are listed in Table 1.
The structure of the unliganded Ro was determined by molecular replacement techniques as implemented in CNS (Brunger et al., 1998). As a search model, we used the structure of Ro in the refined ribonucleoprotein complex. The protein model was rebuilt with the help of omit maps. Since the crystallization solution did not contain divalent cations and because other MIDAS motif structures have been determined with water at the magnesium site (Baldwin et al., 1998), we have modeled water in place of a magnesium ion in the MIDAS motif pocket. Rounds of rebuilding were followed by cycles of refinement as described for the Ro/RNA complex. For the final model, R=21.3% and Rfree=25.7% over all data between 20.0 and 1.95 Å. Data collection and refinement statistics are listed in Table 1.
Coordinates have been deposited in the Protein Data Bank with accession numbers 1YVP and 1YVR. Figures were prepared using the programs GRASP (Nicholls, 1993), Molscript (Kraulis, 1991), and O (Kleywegt and Jones, 1997).
Electrophoretic mobility shift assays
The clones containing Xenopus Y3 RNA (xY3 RNA) and misfolded pre-5S rRNA (pre-5S stem mt 1+2 RNA; Shi et al., 1996) under control of the T7 promoter have been described (Shi et al., 1996; Green et al., 1998). After linearization with DraI, xY3 RNA and pre-5S rRNAs were synthesized in the presence of [α-32P] rUTP (400 Ci/mmol; Amersham) using T7 RNA polymerase as described (Shi et al, 1996). To generate misfolded pre-5S rRNA lacking the last 8 nucleotides, a DNA template for T7 transcription was synthesized from overlapping DNA oligonucleotides. All RNAs were gel-purified and quantitated by scintillation counting.
For gelshift assays, purified RNAs were resuspended in binding buffer (20 mM Tris-HCl, pH 7.5, 125 mM NaCl, 1 mM EDTA, 1.5 mM MgCl2, 2 mM DTT, 10% glycerol), refolded by heating to 90°C for 2 min and frozen on dry ice. After thawing slowly on ice the RNAs were diluted to 1.5 fmol/μl. Binding reactions contained 1.5 fmol xY3 RNA or misfolded pre-5S rRNA and 24 to 6144 fmol of native Ro protein or a mutant Ro protein, 0.3 _g/μL E.coli tRNA, and 0.01% NP-40 in binding buffer. The total reaction volume was 10 μl. Reactions were incubated at room temperature for 30 min and then at 4°C for 30 min, and loaded onto 5% polyacrylamide (38:2 acrylamide:bisacrylamide)/5% glycerol gels running at 2 V/cm (prerun at 16 V/cm for 10 min at 4°C). Gels were run at 4°C for 10 min at 2 V/cm, followed by 16 V/cm in 0.5X TBE (50mM Tris, 45 mM boric acid, and 1.25 mM EDTA) at 4°C for 2 hr and 30 min. The fraction of bound RNA (Θ) was quantitated using a PhosphorImager (Molecular Dynamics). Using Imagequant, the data were fitted by nonlinear least squares analysis as a function of protein concentration using the equation:
where Θ is the fraction of bound RNA and Kd is the dissociation constant. Each Kd is the average of at least three experiments.
Acknowledgments
We are grateful to D.W. Rodgers, A.M. Pyle, and E. Ullu for extensive discussions regarding this manuscript. We thank the staff at beamlines X12C, X25, and X29 at the Brookhaven National Laboratory for assistance in data collection and D. Hildesheim at the National Cell Culture Center for overexpressing some of the Ro protein used in these studies. A.F.S. was supported by a Brown-Coxe fellowship and a fellowship from the Arthritis Foundation. This work was funded by grants from the G. Harold and Leila Y. Mathers Foundation, the Pew Charitable Trust (K.M.R.) and the N.I.H. (R01-GM70521 to K.M.R. and R01-GM48410 to S.L.W.). S.L.W. is an Investigator of the Howard Hughes Medical Institute.
References
- Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30:1427–1464. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin ET, Sarver RW, Bryant GL, Curry KA, Fairbanks MB, Finzel BC, Garlick RL, Heinrikson RL, Horton NC, Kelley LL, et al. Cation binding to the integrin CD11b I domain and activation model assessment. Structure. 1998;6:923–935. doi: 10.1016/s0969-2126(98)00093-8. [DOI] [PubMed] [Google Scholar]
- Bateman A, Kickhoefer V. The TROVE module: a common element in telomerase, Ro, and vault ribonucleoproteins. BMC Bioinformatics. 2003;4:49. doi: 10.1186/1471-2105-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Sagar MB, Doudna JA. Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle. J Mol Biol. 2001;307:229–246. doi: 10.1006/jmbi.2000.4454. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Buyon JP, Rupel A, Clancy RM. Neonatal lupus syndromes. Lupus. 2004;13:705–712. doi: 10.1191/0961203304lu2008oa. [DOI] [PubMed] [Google Scholar]
- Carter DM, Radding CM. The role of exonuclease and beta protein of phage lambda in genetic recombination II. Substrate specificity and the mode of action on lambda exonuclease. J Biol Chem. 1971;246:2502–2512. [PubMed] [Google Scholar]
- Chen X, Quinn AM, Wolin SL. Ro ribonucleoproteins contribute to the resistance of Deinococcus radiodurans to ultraviolet irradiation. Genes Dev. 2000;14:777–782. [PMC free article] [PubMed] [Google Scholar]
- Chen X, Smith JD, Shi H, Yang DD, Flavell RA, Wolin SL. The Ro autoantigen binds misfolded U2 small nuclear RNAs and assists mammalian cell survival after UV irradiation. Curr Biol. 2003;13:2206–2211. doi: 10.1016/j.cub.2003.11.028. [DOI] [PubMed] [Google Scholar]
- Chen X, Wolin SL. The Ro 60 kDa autoantigen: insights into cellular function and role in autoimmunity. J Mol Med. 2004;84:232–239. doi: 10.1007/s00109-004-0529-0. [DOI] [PubMed] [Google Scholar]
- Cingolani G, Petosa C, Weis K, Muller CW. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- Farris AD, O’Brien CA, Harley JB. Y3 is the most conserved small RNA component of Ro ribonucleoprotein complexes in vertebrate species. Gene. 1995;154:193–198. doi: 10.1016/0378-1119(94)00823-b. [DOI] [PubMed] [Google Scholar]
- Green CD, Long KS, Shi H, Wolin SL. Binding of the 60-kDa Ro autoantigen to Y RNAs: evidence for recognition in the major groove of a conserved helix. RNA. 1998;4:750–765. doi: 10.1017/s1355838298971667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves MR, Hanlon N, Turowski P, Hemmings BA, Barford D. The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell. 1999;96:99–110. doi: 10.1016/s0092-8674(00)80963-0. [DOI] [PubMed] [Google Scholar]
- Hendrick JP, Wolin SL, Rinke J, Lerner MR, Steitz JA. Ro small cytoplasmic ribonucleoproteins are a subclass of La ribonucleoproteins: further characterization of the Ro and La small ribonucleoproteins from uninfected mammalian cells. Mol Cell Biol. 1981;1:1138–1149. doi: 10.1128/mcb.1.12.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- Huber AH, Nelson WJ, Weis WI. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- Kickhoefer VA, Liu Y, Kong LB, Snow BE, Stewart PL, Harrington L, Rome LH. The Telomerase/vault-associated protein TEP1 is required for vault RNA stability and its association with the vault particle. J Cell Bio. 2001;52:157–64. doi: 10.1083/jcb.152.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt G.J., Jones, T.A. (1997). Model Building and Refinement Practice. In Methods in Enzymology, CW Carter and RM Sweet, eds. (New York, NY; Academic Press), vol. 277, pp. 208–230. [DOI] [PubMed]
- Kraulis PJ. Molscript: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- Nicholls, A. (1993). GRASP: graphical representation and analysis of surface properties. (Columbia University, New York).
- O’Brien CA, Margelot K, Wolin SL. Xenopus Ro ribonucleoproteins: members of an evolutionarily conserved class of cytoplasmic ribonucleoproteins. Proc Natl Acad Sci USA. 1993;90:7250–7254. doi: 10.1073/pnas.90.15.7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien CA, Wolin SL. A possible role for the 60-kD Ro autoantigen in a discard pathway for defective 5S rRNA precursors. Genes Dev. 1994;8:2891–2903. doi: 10.1101/gad.8.23.2891. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. (1997). Processing X-ray diffraction data collected in oscillation mode. In Methods in Enzymology, CW Carter and RM Sweet, eds. (New York, NY; Academic Press), vol. 276, pp. 307–326. [DOI] [PubMed]
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Patel P, Werth V. Cutaneous lupus erythematosus: a review. Dermatol Clin. 2002;20:373–385. doi: 10.1016/s0733-8635(02)00016-5. [DOI] [PubMed] [Google Scholar]
- Pruijn GJ, Slobbe RL, van Venrooij WJ. Analysis of protein--RNA interactions within Ro ribonucleoprotein complexes. Nucleic Acids Res. 1991;19:5173–5180. doi: 10.1093/nar/19.19.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat Rev Mol Cel Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- Shi H, O’Brien CA, Van Horn DJ, Wolin SL. A misfolded form of 5S rRNA is complexed with the Ro and La autoantigens. RNA. 1996;2:769–784. [PMC free article] [PubMed] [Google Scholar]
- Shimaoka M, Takagi J, Springer TA. Conformational regulation of integrin structure and function. Annu Rev Biophys Biomol Struct. 2002;31:485–516. doi: 10.1146/annurev.biophys.31.101101.140922. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated Mad and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Horn DJ, Eisenberg D, O’Brien CA, Wolin SL. Caenorhabditis elegans embryos contain only one major species of Ro RNP. RNA. 1995;1:293–303. [PMC free article] [PubMed] [Google Scholar]
- Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- Whittaker CA, Hynes RO. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell. 2002;13:3369–3387. doi: 10.1091/mbc.E02-05-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Steitz JA. The Ro small cytoplasmic ribonucleoproteins: identification of the antigenic protein and its binding site on the Ro RNAs. Proc Natl Acad Sci USA. 1984;81:1996–2000. doi: 10.1073/pnas.81.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D, Shi H, Smith JD, Chen X, Noe DA, Cedervall T, Yang DD, Eynon E, Brash DE, Kashgarian M et al. A lupus-like syndrome develops in mice lacking the Ro 60-kDa protein, a major lupus autoantigen. Proc Natl Acad Sci USA. 2003;100:7503–7508. doi: 10.1073/pnas.0832411100. [DOI] [PMC free article] [PubMed] [Google Scholar]