Abstract
This study analyzed gene expression patterns and global genomic alterations in hepatocellular carcinomas (HCC), hepatoblastomas (HPBL), tissue adjacent to HCC and normal liver tissue derived from normal livers and hepatic resections. We found that HCC and adjacent nonneoplastic cirrhotic tissue have considerable overlap in gene expression patterns compared to normal liver. Several genes including Glypican 3, spondin-2, PEG10, EDIL3 and Osteopontin are over-expressed in HCC vs. adjacent tissue whereas Ficolin 3 is the most consistently under-expressed gene. HCC can be subdivided into three clusters based on gene expression patterns. HCC and HPBL have clearly different patterns of gene expression, with genes IGF2, Fibronectin, DLK1, TGFb1, MALAT1 and MIG6 being over-expressed in HPBL versus HCC. In addition, specific areas of the genome appear unstable in HCC, with the same regions undergoing either deletion or increased gene dosage in all HCC.
Abbreviations: AT, tissue adjacent to tumor; HCC: hepatocellular carcinoma; HPBL: hepatoblastoma of childhood; NL, normal liver; FDR, false discovery rate; DE, differentially expressed; PAM, Prediction Analysis for Microarrays; SAM, Significance Analysis for Microarrays
Hepatocellular carcinomas (HCC) and hepatoblastomas of childhood (HPBL) are two types of liver cancer with high mortality and morbidity and international prevalence. There have been several recent studies of patterns of gene expression and molecular classification of HCC.1–4 The studies demonstrated that HCC can be clustered in subgroups of gene expression patterns that have different prognostic and clinical behavior. Other recent studies also examined similarities between HCC precursor lesions (low and high grade liver nodules) and demonstrated significant similarities but also differences between HCC and precursor lesions.5
In this study, we also focused on gene expression of HCC and HPBL, but from a different perspective than previous studies. We utilized a set of tissues from normal liver (NL), HCC, HPBL and tumor adjacent (AT) tissues and determined gene expression patterns not as a ratio of tumor vs. normal, but rather as absolute separate values for each unique tissue. This allowed standard but stringent statistical analysis not feasible when gene expression is only viewed as a fold change over normal tissues. Identification of gene expression patterns of liver tumors from this perspective allows identification of the main differences between the tumor subtypes and the adjacent non-tumor (but often cirrhotic) liver; it also offers the potential of defining new therapeutic and diagnostic modalities. Our findings include some genes already shown to increase in HCC, thus validating our overall approach. Our results also revealed many other genes, not so far involved with biology of liver tumors. In addition, we carried a whole genome analysis of 27 HCC and determined chromosomal loci with genetic abnormalities common to most of the HCC.
Materials and Methods
See Supplemental information at the HEPATOLOGY website: http://interscience.wiley.com/jpages/0270-9139/suppmat/index.html.
Tissues
A total of 37 hepatocellular carcinomas and 7 hepatoblastomas were used for the study. Due to differences in the amount of tumor available, there were many cases in which adjacent tissue was not available (required for identification of tumor margins for diagnostic purposes). There were also cases in which the tumor size was very small and all required for diagnostic purposes, but tissue adjacent to the tumor was available. Overall, 32 samples of liver with cirrhosis adjacent to the tumor were used for the study. Due to these considerations, each one of these “tissues adjacent to tumor” was investigated as a separate item, and not in relation to a specific tumor to which it may relate. The statistical analysis examined the “tissues adjacent to tumor” as a population of its own. There was no adjacent tissue use for hepatoblastomas and comparisons to normal adjacent tissue reflect adjacent liver with cirrhosis. Normal livers (from donor liver tissue) were obtained from 29 cases.
Characteristics of HCC and HPBL
The diameter of HCC at the time of resection ranged from 15 to 0.9 cm (mean: 5.14 cm, median: 3.80 cm, standard error of the mean: 0.49 cm). None of the tumors was associated with infection with hepatitis B virus. Five HCC cases were associated with infection with hepatitis C (identified in Fig. 2C). All HCC were trabecular type, with varying degrees of atypia. HPBL (7 cases) were mixed fetal and embryonal (6 cases) or pure fetal (1 case).
Fig. 2.
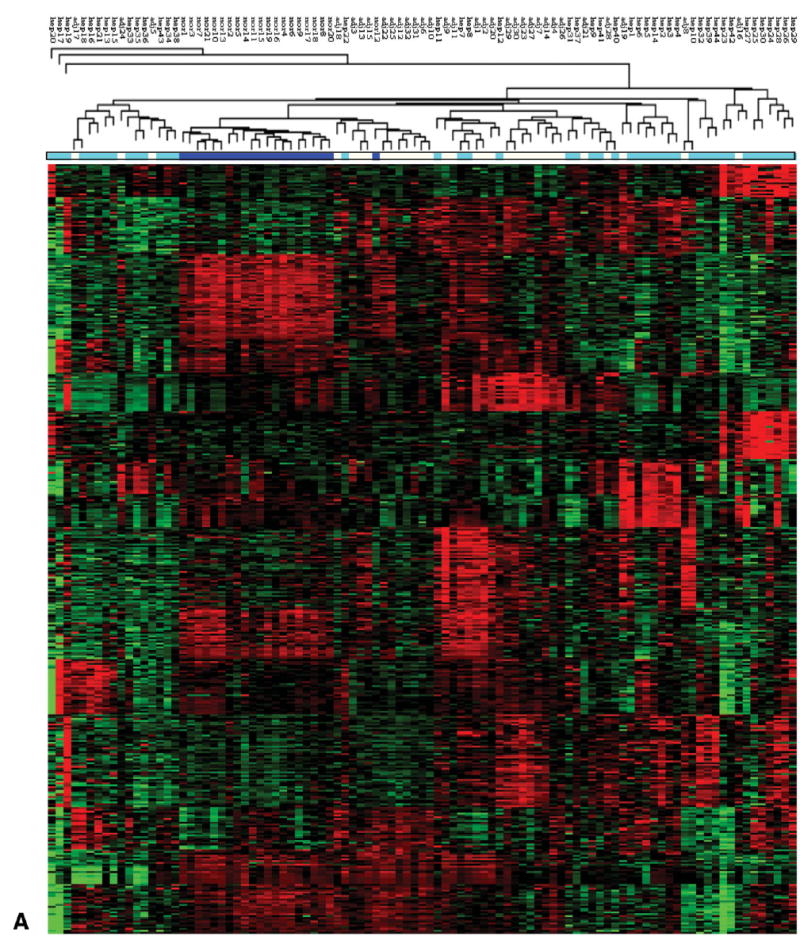

Sub classification of HCC using gene expression clustering. (A) Heat map of hierarchical clustering of all tissue samples (NL, AT, HCC and HPBL) using 419 tight clustered genes, derived by algorithms specified in Materials and Methods. The color bar under the hierarchical tree shows the sample information. Dark blue: normal samples. Light blue: tumor samples. White: tumor-adjacent samples. (B) Heat map of hierarchical clustering of all tumor samples (HCC and HPBL) using 419 tight clustered genes. Hepatoblastomas cluster between the two yellow lines. (C) Heat map of hierarchical clustering of tumor samples excluding 7 HPBL samples from children. HCC cases positive for hepatitis C are marked with red arrowheads. The two cases of fibrolamellar carcinomas are marked with black arrowheads.
Results
Statistical Comparison of Overall Gene Expression Patterns Between the Ttissue Groups
SAM software was applied to detect differentially expressed genes between NL vs. HCC, NL vs. AT and AT vs. HCC. In both NL vs HCC and NL vs AT comparisons, more than 2,000 genes (Table 1) were detected to be up-regulated and down-regulated respectively when the false discovery rate (FDR; i.e., expected false positive rate) threshold is chosen to be 0.05. The large number of differentially expressed genes suggests that normal samples are very different from tumor and adjacent samples. On the other hand, in the comparison of AT vs. HCC, a much smaller number of differentially expressed genes were detected (1127 and 140 of up- and down-regulated genes respectively). The degree of similarity (or divergence) of gene expression patterns between the groups was also assessed using PAM in order to determine the cross-validated probabilities for prediction in each pair-wise comparison of the three groups. The minimal number of genes required for good separation of class assignment probabilities for each sample was determined by PAM. The plots shown in Fig. 1 demonstrate the class assignment probabilities and prediction results. The number of genes needed to distinguish AT vs. HCC (222-gene model) is much higher than NL vs. HCC (41-gene model) and NL vs AT (73-gene model) (see Fig. 2 legend). Data in Table 1 and Fig. 1 demonstrate that AT and HCC are more similar to each other in gene expression patterns than to NL
Table 1.
Number of Differentially Expressed Genes*
| Normal vs. Tumor | Normal vs. Adjacent | Adjacent vs. Tumor | |
|---|---|---|---|
| Upregulated | 2,340 | 2,270 | 1,127 |
| Downregulated | 1,730 | 2,036 | 140 |
| Total Differentially expressed Genes | 4,070 | 4,306 | 1,267 |
(among 8398 genes that have average intensities larger than 100) obtained from SAM analysis in normal vs. tumor, normal vs. adjacent, and adjacent vs. tumor comparisons. The FDR threshold is chosen to be 0.05.
Fig. 1.
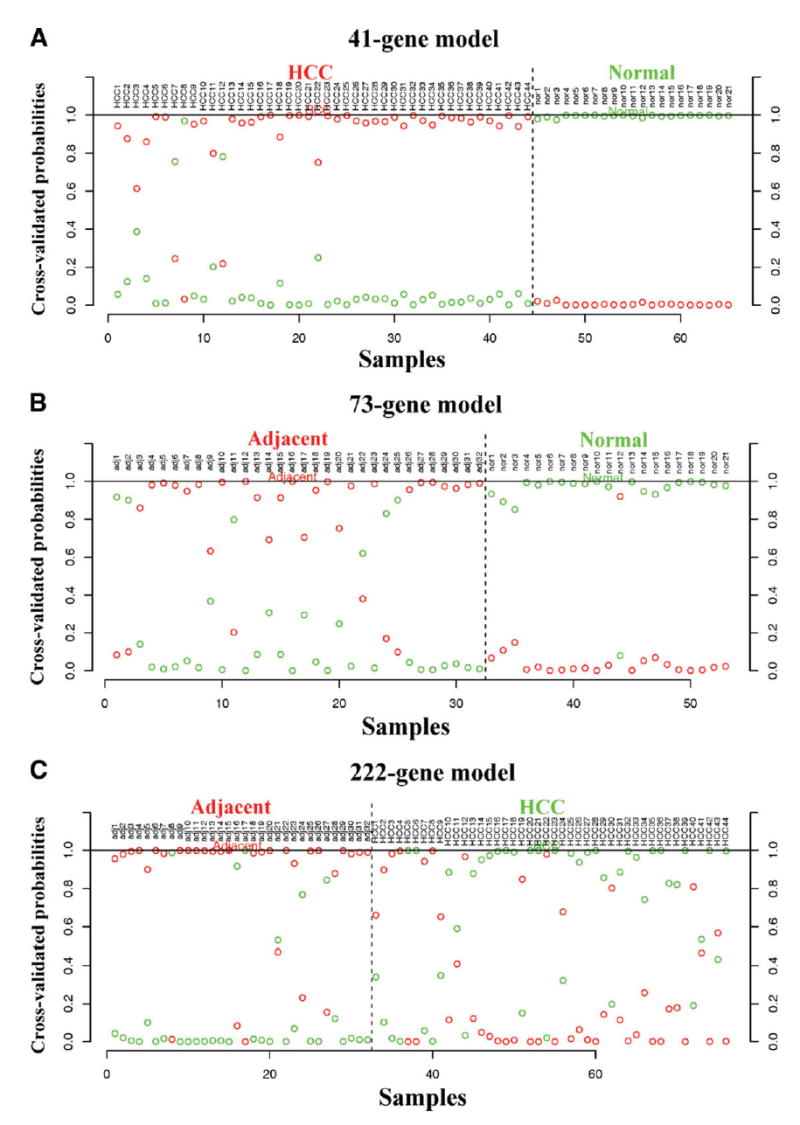
Classification model for NL, AT and HCC using gene expression profiles. For each tissue sample on x-axis, two cross-validated posterior probabilities (on y-axis) of the sample being predicted to either the red or the green group are shown. The sample is then predicted to the group with higher posterior probability (0.5). The presented classification models for each comparison are chosen with the minimum number of genes that allow good posterior probability separation. The title of each plot shows the minimal number of genes needed in each classification model that allows separation. (A) NL vs. HCC. (B) NL vs. AT. (C) AT vs. HCC. The data show that at least 222 genes are required for separation of AT vs. HCC, whereas 41 genes and 73 genes respectively are sufficient for HCC vs. normal liver and AT vs. normal liver.
Cluster Analysis of HCC and HPBL
Tight clustering analysis was first applied to obtain 15 clusters of tightly correlated expression patterns consisting of 419 genes, using specific algorithms to determine tightness of clustering, as specified in the Methods section. This analysis was totally unsupervised and served as a filtering procedure to exclude irrelevant genes not tightly correlated with any gene clusters. All 97 samples were then clustered by hierarchical clustering. The hierarchical tree, sample information and heat map are shown in Fig. 2A. All normal samples (dark blue) except for one were clustered together. The tumor (light blue) and tumor-adjacent (gray) samples are somewhat but not entirely separated, confirming the previous finding in classification analysis (Fig. 1).
We further attempted to cluster only tumor samples in order to look for potential tumor subtypes. Figure 2B shows a hierarchical clustering result of all tumor samples. All HPBL clustered together (tumors between the two yellow lines, Fig. 2B). These results indicate a distinctive gene expression pattern in HPBL vs. HCC. When HPBL were removed, the clustering of the remaining (adult HCC) samples showed two major clusters (12 and 20 samples respectively) and a minor cluster with 3 samples (Fig. 2C). We further performed classification analysis on the two major sub-clusters with PAM software. A cross validation error rate as low as ≈9% was reached.
Genomic Array Analysis of HCC Aiming to Identify Common Sites of Genome Amplification or Deletion
We performed oligonucleotide genome arrays on 27 HCC samples (and 27 matched normal controls) to measure the gene copy numbers in the cancer genome, utilizing a 4000 gene array (see Methods in Supplement). The 27 cases utilized were the subset of HCC classified in either Cluster A or Cluster B for which adjacent or normal tissue was available. The results of the genome array analysis indicate that several regions in chromosome 10, 12, 8 and 7 frequently contain gene copy number alterations (Fig. 3A). To investigate whether these abnormalities resulted in gene expression alteration, we aligned the fold change in mRNA expression with that of gene copies for each gene, and we found that many of these changes did not result in dramatic alterations in gene expression (Fig. 3B). Two genes, glypican 3 (GPC3) and TGFβ-inducible early response gene (TIEG), have high concordance between gene copy number and gene expression alteration (Fig. 3B): 5.4 fold increase in gene copy number and 19 fold increase in mRNA quantity for GPC3, and 3.3 fold and 2 fold for TIEG. Other genes with smaller degree of concordance were also noted. Concordance analysis in a sample by sample manner revealed that samples with increase of gene copy generally had higher level expression of mRNA for both GPC3 (Fig. 3C) and TIEG (data not shown). However, over-expression of GPC3 in some samples is independent of gene copy number, suggesting that other mechanisms may also be involved. Immunostains with antibodies specific for GPC3 and TIEG were performed utilizing an HCC tissue micro-array, composed of 484 samples of hepatic tissue, including 319 benign (normal or liver adjacent to HCC) liver tissues and 165 HCC. As shown in Fig. 4A, there is an increase of GPC3 and TIEG expression in HCC. To investigate whether increased expression of these two genes signaled a poorer clinical outcome (including metastasis or recurrence of HCC within 5 years), we scored the intensity of GPC3 expression in a scale of 0, 1, 2. We divided the HCC samples into expression scores of GPC3 < 0.5 vs. to those with GPC3 >1. As shown in Fig. 5, samples with GPC3 scores ≥1 have rate of metastasis or recurrence at 65%, while samples with scores ≤0.5 have a rate of 44%. Log-rank test indicated that the difference was statistically significant. These results implicate a potential role of GPC3 in determining aggressiveness of HCC and usefulness of GPC3 expression in predicting HCC outcome.
Fig. 3.
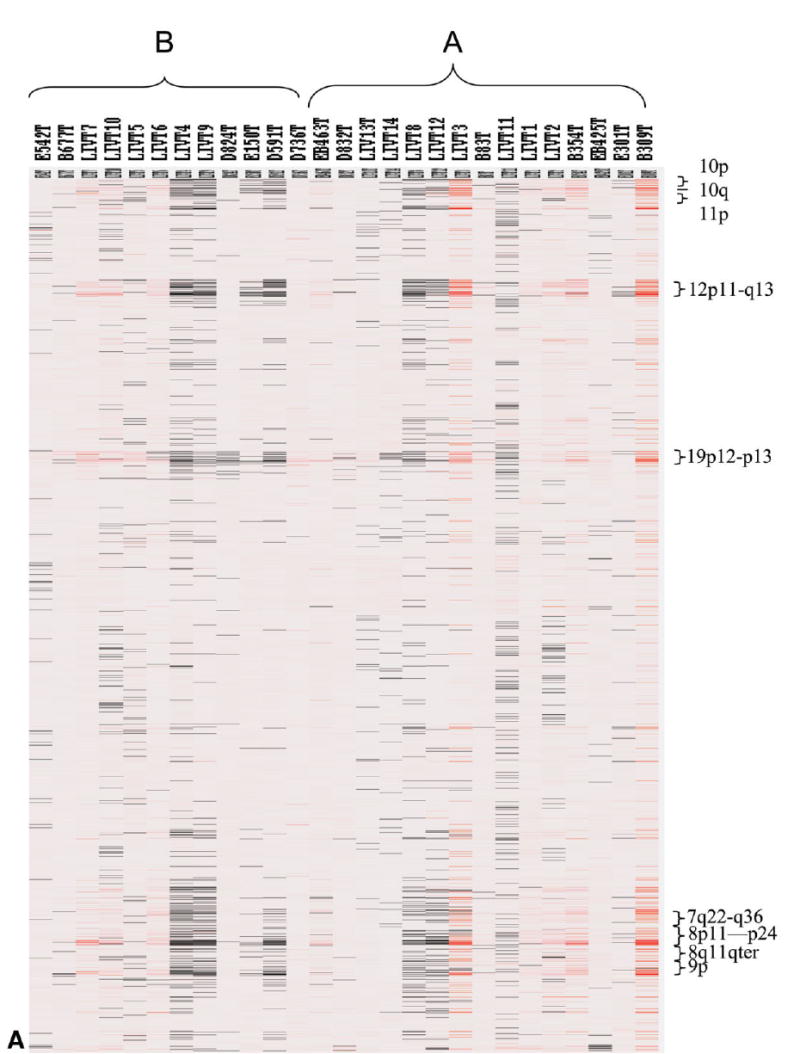

Genomic alterations in HCC. (A) Heat map display of genome copy alteration in HCC samples belonging to Clusters A and B. Red color indicates increase in gene copies, black indicates decreased copies and white indicates no changes. The chromosome areas with marked changes are shown on the right y-axis. Several loci show alterations in copy numbers (increase or decrease) across all HCC. (B) Genome-wide concordance analysis of gene copy number and expression changes. Average copy number of each locus in HCC genome (blue) was plotted along with the physical location in each of the chromosomes. The corresponding average mRNA levels of each of the genes in the HCC samples (red) were expressed as a ratio with that of NL samples and plotted as log2 in the graph. GPC3 and TIEG are indicated by arrows. (C) Concordance analysis GPC3 gene copy and mRNA expression in the 27 HCC samples. Red: Ratio of gene expression for HCC/NL; Blue: Gene copy number.
Fig. 4.
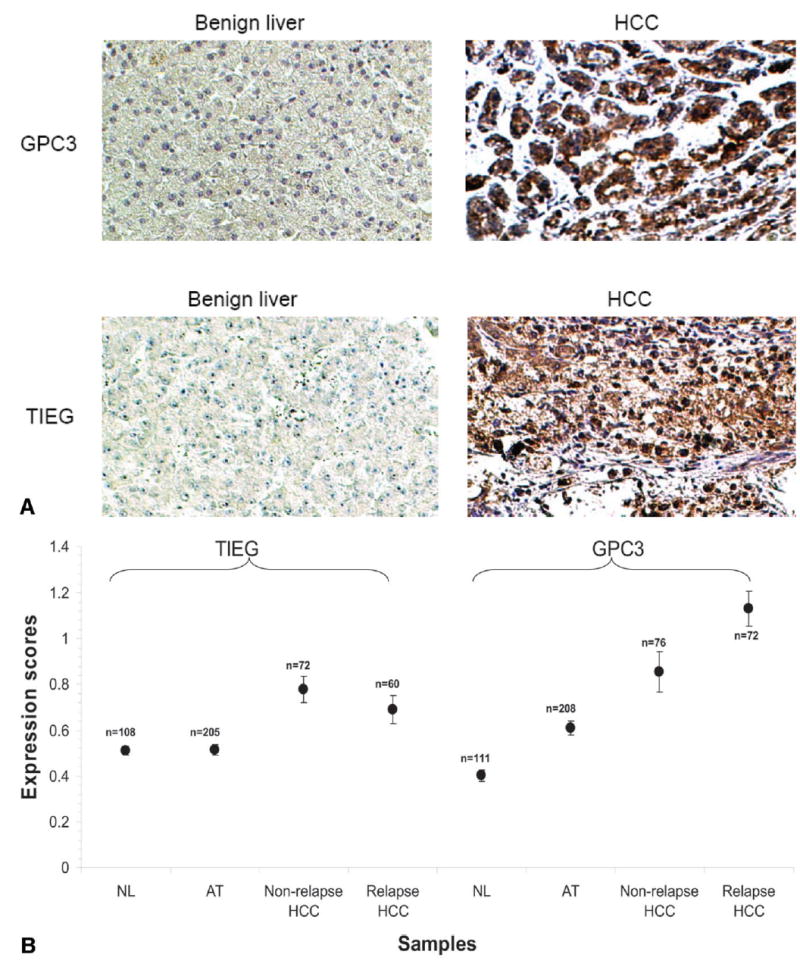
Over-expression of GPC3 and TIEG in hepatocellular carcinoma. (A) Representative photomicrographs of GPC3 and TIEG protein expression in benign (normal liver and tumor adjacent) liver tissues and HCC samples. (B) Expression scores of GPC3 and TIEG in NL, AT, HCC non-relapse and HCC relapsed samples (see Results).
Fig. 5.
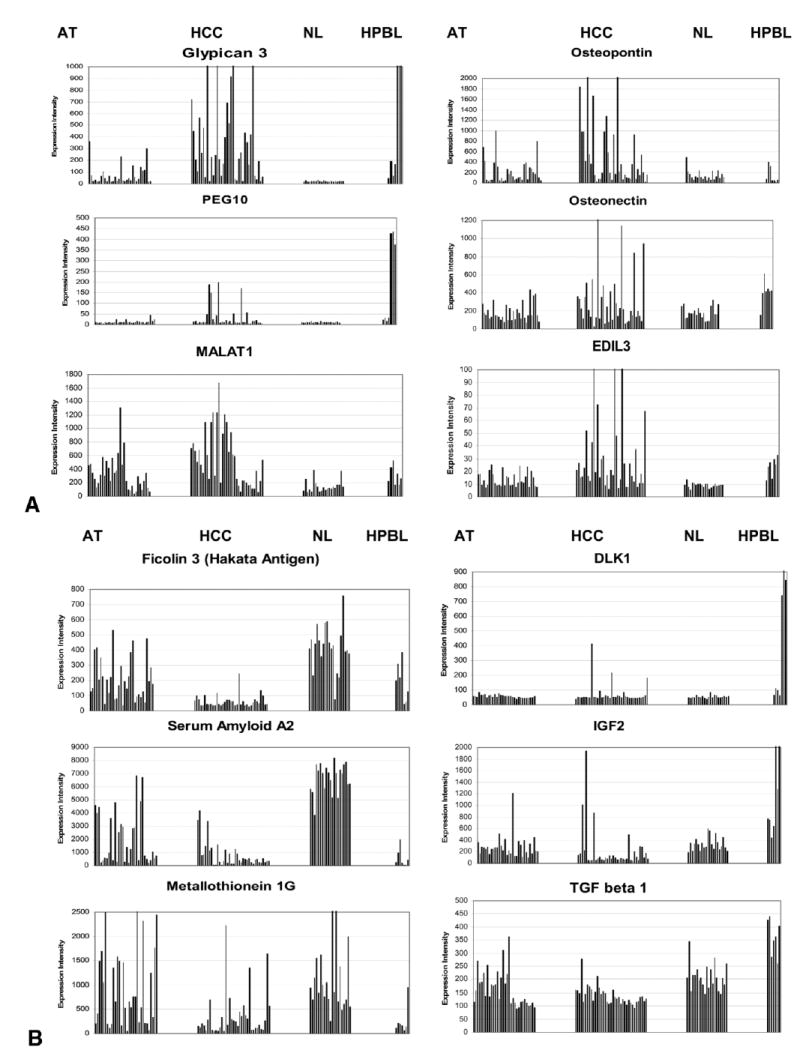
Comparative patterns of expression of different genes in AT, HCC, NL and HPBL. Genes were chosen from among the most over-expressed genes in different tissues, to illustrate the different patterns of gene expression in the various tissue categories. Each tissue category is separated from the others in the graph. The categories (AT, HCC, NL and HPBL) are aligned and labeled for each column of graphs at the top of the figure. Each bar illustrates the expression intensity (Affymetrix) in one specific tissue sample. Each of the different tissue categories included the following number of tissue samples: AT: 32, HCC: 37, NL: 21, HPBL: 7. Some of the genes (e.g. Glypican 3, PEG10, osteonectin and EDIL3) are over-expressed in both HCC and HPBL, but not in NL and AT. Others (e.g. osteopontin, MALAT1) are elevated in HCC but also, to a lesser degree, in AT. Ficolin 3 is under-expressed specifically in HCC and Serum Amyloid 2 (and, to a lesser extent, Metallothionein 1G) is under-expressed in both HCC and HPBL. DLK1 and IGF2 (and, to a lesser degree, TGF-beta 1) are uniquely over-expressed in HPBL. Graphs were derived from the individual points used for the statistical analysis of samples described in Supplemental Tables 1–8.
Patterns of Gene Expression in HCC Compared to Adjacent Tissue and Normal Liver
The results in Figs. 1-2 were derived by applying statistical methodology on the entire set of tissues in order to determine general patterns of similarities and differences in gene expression between the groups. We wanted however to also determine the specific genes which are maximally different (over- or under-expressed) between any two of the groups. To that effect, we analyzed the rank order of over- or under- expression of all genes in comparisons between the different histologic groups (NL, HCC, HPBL and AT). In this part of the study we excluded any genes in which the differences between the categories were not statistically significant. The appropriate P value of statistical significance for each comparison was adjusted by the Benjamini-Hochberg procedure for multiple comparison.6 Some genes with apparent biological importance which were significant at a P value greater than the Benjamini-Hochberg standard but nonetheless less than .05 are also specified as such and discussed separately. The results are summarized below and shown in detail in Supplemental Tables 1-8.
The overall methodology and the results for some of the differentially expressed genes emerging from these comparisons are illustrated in Fig. 5A-B. When each tissue sample is expressed as a separate bar, it is evident that some of the genes are differentially expressed in either HCC alone (e.g., Glypican, ficolin, osteopontin) or in both HCC and adjacent cirrhotic tissue (e.g., serum amyloid A2, MALAT1, metallothionein 1G). Glypican is over-expressed in most HPBL as in the HCC. Other genes (e.g., PEG10) are dramatically over-expressed in some HCC and a subset of HPBL. Some genes (e.g., DLK1 and IGF2) are over-expressed in a subset of HPBL. This approach goes beyond mere statistical significance to demonstrate heterogeneity of expression of genes within each category.
Summary of Genes Differentially Expressed in the Comparison Groups
For details, see Supplemental Tables 1–8.
A. HCC vs. NL and AT
Over-expressed genes. The list of the top 120 over-expressed genes in HCC vs. NL (Supplemental Table 1), includes many proteins related to matrix, or matrix signaling, including Glypican 3 (19.15X), COL1A2 (4.80X), Galectin 3 (4.01X), SPARC-like 1 (hevin, 3.69X), Lumican (3.30X), Osteopontin (3.27X), Collagen Type IV (2.97X), Versican (2.56X), Collagen type III (2.48X), Thrombospondin (2.14X), Collagen Type V (2.10X), and Osteonectin (2.07). Several of these genes are also overexpressed in HCC vs. AT (Suppl. Table 3). Notable is the persistent over-expression (HCC vs. AT)) in Glypican 3 and Collagen Type I. Osteonectin, osteopontin and alpha fetoprotein are also over-expressed (but not satisfying Benjamini-Hochberg restriction). Another matrix protein, Spondin 2, was found elevated (2.11X) in HCC specifically vs. AT but not vs. NL.
Under-expressed Genes (Supplemental Table 2)
There are decreases in gene expression patterns unique to the neoplastic change. These include many members of the Metallothionein family (HCC/AT: MT 1X: 40%, 1B: 49%, 1E: 49%, 1A: 49%, 1F: 56%), and Serum amyloid protein A1. Ficolin (also known as Hakata antigen), is down-regulated only in HCC, whereas it is highly expressed in both cirrhotic nodules and normal livers.
Comparison of Gene Expression Patterns Between HCC and Hepatoblastomas (HPBL)
(Supplemental Tables 5 and 6). Hepatoblastomas did cluster distinctly from HCC, as shown in Fig. 2B.
Genes increased in HPBL vs. HCC: Mitogen-inducible gene 6 (Mig6: 3.88X), TGFβ1 (2,58X). The genes DLK1 (7.64X) and IGF2 (6.71X) were also over-expressed, but at criteria less stringent than the Benjamini-Hochberg (P values at 0.01157 and 0.04352 respectively). The reason for this is shown in Fig. 5. DLK1, IGF2 and PEG10 are over-expressed very prominently, but only in a subset of HPBL (same for all three genes) (Fig. 4).
Genes decreased in HPBL vs. HCC: Genes in this category include Interferon alpha inducible protein 27 (23%), Galectin 4 (24%), ubiquitin 2 (42%) and alpha-1 adrenergic receptor (35%).
Comparison of Gene Expression Patterns Between the Two Main Groups of HCC
Detail data on thedifferences in gene expression patterns between Clusters A and B are shown in Supplemental Tables 7–8.
Genes Over-expressed in Cluster A
(The number in parenthesis show the ratio of expression intensity of Cluster A/Cluster B.). Increase are several members of the cytochrome P450 family of genes (CYP 7A1: 6.82X, CYP 3A4: 3.69X, CYP 3A7: 2.47X, CYP 3A5: 2.14X). Also increased are several members of the alcohol dehydrogenase family (1B: 2.47X, 5A1: 2.11X, 1A: 2.09X, 3A2: 2.08X, and 1C: 2.03X). PEG10 is expressed primarily in Cluster A (3.69X). Also notable is the increased expression of Insulin Receptor substrate 1 (IRS1: 2.82X) and Erb-B3, (2.35X). Many of these genes did not meet the stringent criteria of Benjamini-Hochberg (P value< .00117), but all were over-expressed at a P value< .002.
Genes Over-expressed in Cluster B
The combined over-expression of the hypoxia inducible factor 1 alpha (HIF1α: 2.14X) and the gene adrenomedullin (2.18X), well known to be dependent on HIF1α,7 suggests that hypoxia is a more dominant factor in Cluster B than Cluster A. Both of these genes met the Benjamini-Hochberg criteria (P value< .00117). Also increased in Cluster B (but only at a P< .004) were the genes Chemokine (C-C) ligand 7 (5.64X), Regulator of G-protein signaling 2 (4.04X), SOCS3 (4.00X), C-reactive protein (4.00X), TIMP-1 (3.58X), IGFBP 5 (2.54X), IGF-BP 7 (2.25X), and collagen types IV (2.29X) and VI (1.99X).
Discussion and Biologic Significance of the Findings General Aspects
Our results impact upon several issues related to biology of liver cancer, HCC or HPBL. Our aim was to identify existing patterns of gene expression in HCC and HPBL that constitute a “signature” gene expression. While our findings may not necessarily link to tumor prognosis, they nonetheless have successfully identified unique expression patterns that can become the basis for future diagnostic and therapeutic targeting. Our approach is further validated by the fact that some of the genes “discovered” in this study as highly associated with HCC, have already been found in other independent studies to be exclusively associated with liver cancer. Other genes, however, discovered in this study, had not been detected in previous literature and should be studied further. We also studied the RNA of each tissue sample separately (and not as a mixed sample of HCC and normal). This makes it possible to carry out the type of statistical analysis shown in Fig. 5 and Supplemental Tables.
As with previous studies, we also did an unsupervised clustering of all tissues, including HCC, HPBL, adjacent tissue and normal liver. As shown in Figs. 1-2, the clustering analysis shows that an algorithm can be constructed that allows separation of the categories. We believe, however, that sole focus on this approach may lead to the wrong conclusion of large differences in gene expression between HCC and adjacent tissues. The number of genes which are uniquely over- or under-expressed in HCC versus adjacent tissue (P value Benjamini-Hochberg standard) is exceedingly small (Supplemental Tables 3 and 4).
Biologic Significance of Differentially Expressed Genes Between Tumor-Adjacent Tissue and HCC
Growth-related genes over-expressed in both HCC and AT should reflect the hyper-proliferative status of both HCC and cirrhotic nodules. Others, however, do not appear to have obvious connection to cell growth. The coordinate regulation of these genes in both HCC and AT shows a common biologic set of alterations between HCC and cirrhosis. Nonetheless, there were genes distinctly altered only in HCC. These deserve special attention, as they may become the basis for tumor detection or future therapies. These include:
Glypican 3. This protein (GPC3) is the most over-expressed in HCC and HPBL, and not in AT (or NL). Several recent studies have shown that Glypican 3 is over-expressed uniquely in HCC.8–11 GPC3 is a member of a family of six proteins which are anchored to the plasma membrane by GPI linkage and bind heparin sulfate pro-teoglycans. GPC3 regulates availability of Wnt proteins.12 We also show in this study that there is an apparent increase in gene dosage for GPC3, suggesting that this gene is amplified in some HCC (Fig. 3). In a very recent review13 it was shown that Glypicans act as alternate co-activators of growth factor receptors when syndecan is not available or rapidly consumed.
Paternally expressed 10 (PEG10) is a paternally expressed imprinted gene, member of a group of imprinted genes residing on chromosome locus 7q21.14 It is over-expressed mainly in HCC belonging to Cluster A group and in HPBL. A recent study showed that PEG10 is over-expressed in HCC and regenerating mouse liver. Transfection of PEG10 to HCC cell lines increased their tumorigenicity.15 PEG10 exerts its effects by binding to an apoptosis related protein SIAH1.16
Alpha fetoprotein (2.0X) is another gene in this category. There is extensive literature on AFP over-expression in about 50% of HCC.17
EGF-like repeats and discoidin I-like domains 3 (EDIL3) is a member of a family of extracellular matrix proteins with multiple EGF-like repeats. There is no previous literature relating EDIL3 to HCC or any other neoplasia. EDIL3 is a minor splicing variant of Del1, for which there is extensive literature on its role in promotion of angiogenesis via interaction with integrin a(v)b3.18 It induced by angiogenesis related factors, including VEGF1,19 a factor known to be produced by hepatocellular carcinomas.20
Osteopontin. Several previous studies link osteopontin with worse prognosis of HCC.21,22
Ficolin 3 (Hakata antigen) is decreased selectively in HCC in comparison both to adjacent tissue (0.4X) and donor liver (0.2X). The pattern of its distribution is shown in Fig. 5B. There is no previous literature associating ficolin family members with cancer. Ficolin 3 is expressed in hepatocytes and biliary cells and is excreted in the bile.23,24 Ficolins are either secreted or attached to plasma membrane. They act as a site of aggregation of MASP proteases and are associated with the lectin pathway of complement activation.25
Both DLK1 and PEG10 are paternally imprinted and this suggests that there may be a broader pattern of imprinting irregularities in HCC, as is seen with many other types of tumors.26
Biologic Significance of Differentially Expressed Genes Between Sub-groups of HCC
Recent studies showed that HCC may be subdivided into subgroups based on patterns of gene expression.1–3 Our results show that that there are three clusters of HCC, though cluster C contained only 3 HCC cases. It is not clear from our studies whether the clusters reported in previous studies correspond to the main clusters seen in Fig. 2C. In our study, HIF1a (and the dependent adrenomedullin7) was over-expressed in Cluster B (Supplemental Table 8). HIF1a was also predominantly expressed in one of the two clusters of the previous study. Several cytochrome P450 family members were over-expressed in Cluster A, suggesting a higher level of differentiation. Genes PEG10 (anti-apoptotic), IRS1 (a “shuttle kinase” for the insulin receptor) and Erb-B3 (a growth promoting gene increased in multiple tumors27,28) are also expressed in Cluster A. HIF1-alpha and the dependent gene adrenomedullin were more expressed in Cluster B. The other genes discussed above (Glypican 3) appear common to all HCC and equally expressed between the clusters. The results in Fig. 3A compare genomic alterations between the two main clusters of HCC. There are areas of genomic instability that are commonly seen across the board in most HCC. In the same genomic area, however, some tumors exhibit deletions whereas others exhibit apparent increase in gene dosage. There are no unique genomic differences between HCC Clusters A and B, although in the common areas of genetic instability, HCC in cluster A tend to have increase in gene dosage vs. decrease. Also, there was no identifiable difference in histology or size between the tumors in the different clusters. We also did not find any correlation between the size of the tumor at the time of resection and its classification into any of the clusters shown in Fig. 2C (data not shown). This suggests that the major gene expression determinants are defined soon after tumor initiation. Our tumors, reflecting the patient groups in our institution, did not contain patients infected with Hepatitis B virus. Of interest is the apparent aggregation of most of the cases expressing hepatitis C virus (Fig. 2C). Though one of the cases is outside the cluster of the other six, the apparent aggregation of six of seven cases in cluster B suggests that expression of HCV is a major determinant for gene expression patterns in HCC. The two fibrolamellar carcinomas included in the group do not sort in the same cluster.
Biologic Significance of Genes Differentially Expressed Between HPBL and HCC
Increased in HPBL vs. HCC (Supplemental Table 5).
Insulin-like growth factor 2 (IGF2). This gene is the most over-expressed in HPBL in relation to HCC (6.71X). There is extensive literature on the association of IGF2, hepatoblastomas and other tumors as part of the Beckwith-Wiedemann syndrome.29,30
Mitogen inducible gene 6 (MIG6) (3.88X). This protein appears to inhibit the effects of both EGF and HGF receptors, and as such the increase seen in HPBL is unexpected.31,32
Delta-like 1 homolog (DLK1) is also an imprinted paternally expressed gene,33,34 in a different locus than PEG10. DLK1 protein inactivates GAS1, a gene associated with growth arrest and interacts with Notch-1 signaling.35,36 Recent literature shows increased expression of DLK1 in embryonic hepatocytes and transiently amplifying ductular cells (oval cells).37 There is extensive previous literature documenting emergence of fetal patterns of gene expression in HCC, and similarities between HCC and oval cells.4,38 DLK1 was greatly elevated in 3 of 7 HPBL, suggesting a possibility of HPBL subtypes based on DLK1 expression.
Transforming Growth factor beta 1 (TGFb1) (increased 2.58X). This well known cytokine increases in many tumors, including HCC.39 It is not increased, however, in HCC vs. NL or AT, thus its increase in HPBL vs. HCC is of true functional significance. There is no other previous association of TGFb1 with HPBL. TGFb1 is associated with connective tissue synthesis, often a prominent feature of HPBL histology.40
PEG10 is over-expressed in HPBL and in Cluster A of HCC.
The studies overall reveal substantial similarities between HCC and adjacent cirrhotic tissue. Nonetheless, specific gene alterations are uniquely characteristic of the HCC and distinct patterns exist between HCC and HPBL. The across-the-board genomic areas of instability in HCC, with the same areas showing increase or decease in gene dosage in different tumors, are also surprising and need to be explored. Of importance, our methodology revealed results with some genes that had been independently “discovered” in previous independent literature. This was not the case with previous studies of global genomic analysis and it does validate our overall approach to define the gene expression and genomic alteration signatures of liver cancer.
Supplementary Material
Footnotes
Potential conflict of interest: Nothing to report.
Supported by NIH Grants CA30241 and CA35373 (P.I. GKM), CA098249 (P.I. JHL) and the Rangos Fund for Enhancement of Pathology Research.
References
- 1.Lee JS, Thorgeirsson SS. Genetic profiling of human hepatocellular carcinoma. Semin Liver Dis. 2005;25:125–132. doi: 10.1055/s-2005-871192. [DOI] [PubMed] [Google Scholar]
- 2.Lee JS, Thorgeirsson SS. Genome-scale profiling of gene expression in hepatocellular carcinoma: classification, survival prediction, and identification of therapeutic targets. Gastroenterology. 2004;127:S51–S55. doi: 10.1053/j.gastro.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 3.Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, Durnez A, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. HEPATOLOGY. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 4.Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, Mikaelyan A, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 5.Nam SW, Park JY, Ramasamy A, Shevade S, Islam A, Long PM, Park CK, et al. Molecular changes from dysplastic nodule to hepatocellular carcinoma through gene expression profiling. HEPATOLOGY. 2005;42:809–818. doi: 10.1002/hep.20878. [DOI] [PubMed] [Google Scholar]
- 6.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 7.Garayoa M, Martinez A, Lee S, Pio R, An WG, Neckers L, Trepel J, et al. Hypoxia-inducible factor-1 (HIF-1) up-regulates adrenomedullin expression in human tumor cell lines during oxygen deprivation: a possible promotion mechanism of carcinogenesis. Mol Endocrinol. 2000;14:848–862. doi: 10.1210/mend.14.6.0473. [DOI] [PubMed] [Google Scholar]
- 8.Capurro M, Wanless IR, Sherman M, Deboer G, Shi W, Miyoshi E, Filmus J. Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology. 2003;125:89–97. doi: 10.1016/s0016-5085(03)00689-9. [DOI] [PubMed] [Google Scholar]
- 9.Sung YK, Hwang SY, Park MK, Farooq M, Han IS, Bae HI, Kim JC, et al. Glypican-3 is overexpressed in human hepatocellular carcinoma. Cancer Sci. 2003;94:259–262. doi: 10.1111/j.1349-7006.2003.tb01430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakatsura T, Yoshitake Y, Senju S, Monji M, Komori H, Motomura Y, Hosaka S, et al. Glypican-3, overexpressed specifically in human hepato-cellular carcinoma, is a novel tumor marker. Biochem Biophys Res Commun. 2003;306:16–25. doi: 10.1016/s0006-291x(03)00908-2. [DOI] [PubMed] [Google Scholar]
- 11.Zhu ZW, Friess H, Wang L, Abou-Shady M, Zimmermann A, Lander AD, Korc M, et al. Enhanced glypican-3 expression differentiates the majority of hepatocellular carcinomas from benign hepatic disorders. Gut. 2001;48:558–564. doi: 10.1136/gut.48.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capurro MI, Xiang YY, Lobe C, Filmus J. Glypican-3 promotes the growth of hepatocellular carcinoma by stimulating canonical Wnt signaling. Cancer Res. 2005;65:6245–6254. doi: 10.1158/0008-5472.CAN-04-4244. [DOI] [PubMed] [Google Scholar]
- 13.Selleck SB. Shedding light on the distinct functions of proteoglycans. Sci STKE 2006. 2006:pe17. doi: 10.1126/stke.3292006pe17. [DOI] [PubMed] [Google Scholar]
- 14.Ono R, Kobayashi S, Wagatsuma H, Aisaka K, Kohda T, Kaneko-Ishino T, Ishino F. A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics. 2001;73:232–237. doi: 10.1006/geno.2001.6494. [DOI] [PubMed] [Google Scholar]
- 15.Tsou AP, Chuang YC, Su JY, Yang CW, Liao YL, Liu WK, Chiu JH, et al. Overexpression of a novel imprinted gene, PEG10, in human hepatocellular carcinoma and in regenerating mouse livers. J Biomed Sci. 2003;10:625–635. doi: 10.1159/000073528. [DOI] [PubMed] [Google Scholar]
- 16.Okabe H, Satoh S, Furukawa Y, Kato T, Hasegawa S, Nakajima Y, Yamaoka Y, et al. Involvement of PEG10 in human hepatocellular carcinogenesis through interaction with SIAH1. Cancer Res. 2003;63:3043–3048. [PubMed] [Google Scholar]
- 17.Marrero JA. Screening tests for hepatocellular carcinoma. Clin Liver Dis. 2005;9:235–251. doi: 10.1016/j.cld.2004.12.006. vi. [DOI] [PubMed] [Google Scholar]
- 18.Rezaee M, Penta K, Quertermous T. Del1 mediates VSMC adhesion, migration, and proliferation through interaction with integrin alpha-(v)beta(3) Am J Physiol Heart Circ Physiol. 2002;282:H1924–H1932. doi: 10.1152/ajpheart.00921.2001. [DOI] [PubMed] [Google Scholar]
- 19.Aoki M, Kanamori M, Ohmori K, Takaishi M, Huh NH, Nogami S, Kimura T. Expression of developmentally regulated endothelial cell locus 1 was induced by tumor-derived factors including VEGF. Biochem Biophys Res Commun. 2005;333:990–995. doi: 10.1016/j.bbrc.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Hirohashi K, Yamamoto T, Uenishi T, Ogawa M, Sakabe K, Takemura S, Shuto T, et al. CD44 and VEGF expression in extrahepatic metastasis of human hepatocellular carcinoma. Hepatogastroenterology. 2004;51:1121–1123. [PubMed] [Google Scholar]
- 21.Iso Y, Sawada T, Okada T, Kubota K. Loss of E-cadherin mRNA and gain of osteopontin mRNA are useful markers for detecting early recurrence of HCV-related hepatocellular carcinoma. J Surg Oncol. 2005;92:304–311. doi: 10.1002/jso.20388. [DOI] [PubMed] [Google Scholar]
- 22.Gotoh M, Sakamoto M, Kanetaka K, Chuuma M, Hirohashi S. Overexpression of osteopontin in hepatocellular carcinoma. Pathol Int. 2002;52:19–24. doi: 10.1046/j.1440-1827.2002.01316.x. [DOI] [PubMed] [Google Scholar]
- 23.Akaiwa M, Yae Y, Sugimoto R, Suzuki SO, Iwaki T, Izuhara K, Hamasaki N. Hakata antigen, a new member of the ficolin/opsonin p35 family, is a novel human lectin secreted into bronchus/alveolus and bile. J Histochem Cytochem. 1999;47:777–786. doi: 10.1177/002215549904700607. [DOI] [PubMed] [Google Scholar]
- 24.Sugimoto R, Yae Y, Akaiwa M, Kitajima S, Shibata Y, Sato H, Hirata J, et al. Cloning and characterization of the Hakata antigen, a member of the ficolin/opsonin p35 lectin family. J Biol Chem. 1998;273:20721–20727. doi: 10.1074/jbc.273.33.20721. [DOI] [PubMed] [Google Scholar]
- 25.Sorensen R, Thiel S, Jensenius JC. Mannan-binding-lectin-associated serine proteases, characteristics and disease associations. Springer Semin Immunopathol. 2005;27:299–319. doi: 10.1007/s00281-005-0006-z. [DOI] [PubMed] [Google Scholar]
- 26.Bjornsson HT, Cui H, Gius D, Fallin MD, Feinberg AP. The new field of epigenomics: implications for cancer and other common disease research. Cold Spring Harb Symp Quant Biol. 2004;69:447–456. doi: 10.1101/sqb.2004.69.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 28.Carraway KL, 3rd, Sliwkowski MX, Akita R, Platko JV, Guy PM, Nuijens A, Diamonti AJ, et al. The erbB3 gene product is a receptor for heregulin. J Biol Chem. 1994;269:14303–14306. [PubMed] [Google Scholar]
- 29.Li X, Adam G, Cui H, Sandstedt B, Ohlsson R, Ekstrom TJ. Expression, promoter usage and parental imprinting status of insulin-like growth factor II (IGF2) in human hepatoblastoma: uncoupling of IGF2 and H19 imprinting. Oncogene. 1995;11:221–229. [PubMed] [Google Scholar]
- 30.Prawitt D, Enklaar T, Gartner-Rupprecht B, Spangenberg C, Lausch E, Reutzel D, Fees S, et al. Microdeletion and IGF2 loss of imprinting in a cascade causing Beckwith-Wiedemann syndrome with Wilms’ tumor. Nat Genet. 2005;37:785–786. doi: 10.1038/ng0805-785. author reply 786–787. [DOI] [PubMed] [Google Scholar]
- 31.Xu D, Makkinje A, Kyriakis JM. Gene 33 is an endogenous inhibitor of epidermal growth factor (EGF) receptor signaling and mediates dexamethasone-induced suppression of EGF function. J Biol Chem. 2005;280:2924–2933. doi: 10.1074/jbc.M408907200. [DOI] [PubMed] [Google Scholar]
- 32.Pante G, Thompson J, Lamballe F, Iwata T, Ferby I, Barr FA, Davies AM, et al. Mitogen-inducible gene 6 is an endogenous inhibitor of HGF/Met-induced cell migration and neurite growth. J Cell Biol. 2005;171:337–348. doi: 10.1083/jcb.200502013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi S, Wagatsuma H, Ono R, Ichikawa H, Yamazaki M, Tashiro H, Aisaka K, et al. Mouse Peg9/Dlk1 and human PEG9/DLK1 are paternally expressed imprinted genes closely located to the maternally expressed imprinted genes: mouse Meg3/Gtl2 and human MEG3. Genes Cells. 2000;5:1029–1037. doi: 10.1046/j.1365-2443.2000.00390.x. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt JV, Matteson PG, Jones BK, Guan XJ, Tilghman SM. The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev. 2000;14:1997–2002. [PMC free article] [PubMed] [Google Scholar]
- 35.Baladron V, Ruiz-Hidalgo MJ, Nueda ML, Diaz-Guerra MJ, Garcia-Ramirez JJ, Bonvini E, Gubina E, et al. dlk acts as a negative regulator of Notch1 activation through interactions with specific EGF-like repeats. Exp Cell Res. 2005;303:343–359. doi: 10.1016/j.yexcr.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Baladron V, Ruiz-Hidalgo MJ, Bonvini E, Gubina E, Notario V, Laborda J. The EGF-like homeotic protein dlk affects cell growth and interacts with growth-modulating molecules in the yeast two-hybrid system. Biochem Biophys Res Commun. 2002;291:193–204. doi: 10.1006/bbrc.2002.6431. [DOI] [PubMed] [Google Scholar]
- 37.Jensen CH, Jauho EI, Santoni-Rugiu E, Holmskov U, Teisner B, Tygstrup N, Bisgaard HC. Transit-amplifying ductular (oval) cells and their hepatocytic progeny are characterized by a novel and distinctive expression of delta-like protein/preadipocyte factor 1/fetal antigen 1. Am J Pathol. 2004;164:1347–1359. doi: 10.1016/S0002-9440(10)63221-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sell S. Cellular origin of hepatocellular carcinomas. Semin Cell Dev Biol. 2002;13:419–424. doi: 10.1016/s1084952102001295. [DOI] [PubMed] [Google Scholar]
- 39.Matsuzaki K, Date M, Furukawa F, Tahashi Y, Matsushita M, Sugano Y, Yamashiki N, et al. Regulatory mechanisms for transforming growth factor beta as an autocrine inhibitor in human hepatocellular carcinoma: implications for roles of smads in its growth. HEPATOLOGY. 2000;32:218–227. doi: 10.1053/jhep.2000.9145. [DOI] [PubMed] [Google Scholar]
- 40.Roberts AB, McCune BK, Sporn MB. TGF-beta: regulation of extracellular matrix. Kidney Int. 1992;41:557–559. doi: 10.1038/ki.1992.81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


