Abstract
The Notch/Jagged signaling pathway is important for cellular differentiation and proliferation. Its dysfunction is associated with human pathologies in several tissues including liver. Point mutations in Jagged-1 gene are the cause for Alagille syndrome, associated with paucity of intrahepatic bile ducts. To determine the putative role of the trans-membrane receptor Notch and its ligand Jagged-1 in liver regeneration, we investigated the expression of Notch and Jagged-1 in rat liver following 2/3 partial hepatectomy. Immunohistochemical staining of normal rat liver showed that Notch was expressed in hepatocytes, bile duct cells and endothelial cells, whereas Jagged-1 was expressed in bile duct cells and hepatocytes. Both Notch-1 and Jagged-1 proteins were upregulated in hepatocytes after partial hepatectomy up to day 4. After partial hepatectomy, nuclear translocation of the intracellular cytoplasmic domain of Notch (NICD) increased and peaked within 15 minutes, indicating the activation of Notch. Expression of the Notch-dependent target gene (HES-1) expression increased within 30 – 60 minutes. Addition of recombinant Jagged-1 protein to primary cultures of hepatocytes stimulated hepatocyte DNA synthesis. Furthermore, injection of silencing RNA for Notch and Jagged-1 to livers 2 days before partial hepatectomy significantly suppressed proliferation of hepatocytes at days 2 to 4 of the regenerative response. In conclusion, Notch/Jagged signaling pathway is activated during liver regeneration and is potentially contributing to signals affecting cell growth and differentiation.
Abbreviations: HGF, hepatocyte growth factor; TES, tris-EDTA-sucrose; DAB, diaminobenzoic acid; AEC, aminoethylcarbazole; PBC, primary biliary cirrhosis; bHLH, basic helix-loop-helix; PSC, primary sclerosing cholangitis; NICD, Notch intracellular cytoplasmic domain; HES, hairy enhancer of split; Ct, cycle threshold; siRNA, silencing RNA; HGM, hepatocyte growth medium; PCR, polymerase chain reaction
Liver regeneration after partial hepatectomy is carried out by proliferation of mature hepatocytes, biliary epithelial cells, Kupffer cells and stellate cells.1 Multiple signaling pathways contribute to essential early and late events in this process, including activation of signaling pathways triggered by HGF, EGF, TNFa, IL6 and others.2 The role of the Notch/Jagged signaling system in liver regeneration has not been studied. This system has effects on cell growth and differentiation in many tissues.3–5 In addition, mutations in Jagged-1 are associated with paucity of intrahepatic bile ducts in humans, suggesting an important role for the Notch/Jagged signaling pathway in liver tissue growth and assembly during embryonic development.6 Recent studies have investigated the expression of Notch-1 and Jagged-1 in normal rat and human liver but also in diseased human livers and revealed an induction of Jagged-1 in cases of primary biliary cirrhosis (PBC) and primary sclerosing cholangitis (PSC).6– 8 The expression of Jagged-1 in fetal liver6 and also mutations of Jagged-1, resulting in liver disease known as Alagille syndrome,9 reveal a developmental requirement for the interaction between Notch and Jagged during liver organogenesis. Reactivation of Notch signaling in adult organs may be essential in order to form new tissue during regenerative events. In view of the existing literature, we pursued the study of changes in Notch signaling during liver regeneration.
Notch genes encode for a family of transmembrane receptors whose intracellular domain is released by proteolytic cleavage at three sites (S1, S2 and S3).3,4,10,11 S1 cleavage occurs within the secretory pathway so that a processed heterodimeric form is transported to the cell surface. After ligand binding to the receptor Notch, two proteases acting sequentially mediate the activation of Notch. First, cleavage occurs at an extracellular site (S2, 12 amino acids outside the transmembrane domain) by metalloproteinase TACE/ADAM17.10 The resultant carboxyterminal product is called NEXT (Notch EXtracellular Truncation) and is required for the S3-cleavage performed by presenelin within the transmembrane region. The S3 cleavage releases the cytoplasmic domain of Notch (NICD), which translocates into the nucleus and binds to the transcription factor CBF1/RBP-Jκ. In the absence of NICD, CBF1/RBP-Jκ acts as a transcriptional repressor.12 The binding of NICD to CBF1/RBP-Jκ converts CBF1/RBP-Jk from a transcriptional repressor to a transcriptional activator and is sufficient to induce expression of target genes. Downstream targets of Notch signaling include basic helix-loop-helix (bHLH) proteins like HES-1 and HES-5.13,14 They are able to antagonize other bHLH factors like MyoD that affect differentiation.15 Using the methods and experiments described in this study, we show that Notch and Jagged-1 are upregulated and that activation of Notch occurs early during liver regeneration of rat liver. The findings from cell culture experiments with primary rat hepatocytes and the effects of interfering with expression of Notch and Jagged-1 during liver regeneration (described in this study) reveal potential regulatory effects of Notch and Jagged during the regenerative process.
Material and Methods
RNA Isolation and Real-Time PCR Analysis
Tissue (50 mg) frozen in liquid nitrogen added to 1 ml TRIzol (Invitrogen, CA) was used to isolate total RNA. DNase I digestion and reverse transcription reactions (Superscript II RNase H− Reverse Transcriptase, Invitrogen, CA) were performed according to the manufacturer’s protocol. The following primers (designed with Primer Express, Applied Biosystems) and reaction conditions were used for semiquantitative real-time polymerase chain reaction (PCR) using SYBR®-Green technique: Notch mRNA was detected using primers 5′CACCCATGAC-CACTACCCAGTT3′ and 5′CCTCGGACCAATCA-GAGATGTT3′, which amplified a 186-bp fragment; Jagged-1 mRNA was amplified with 5′AACTGGTAC-CGGTGCGAA3′ and 5′TGATGCAAGATCTCCCT-GAAAC3′ primers that generated a 190-bp fragment. For detection of HES-1, 5′CGACACCGGACAAACCA-AA3′ and 5′GAATGTCTGCCTTCTCCAGCTT3′ primers were used to amplify a 174-bp fragment. HES-5 was detected by 5′ACCGCATCAACAGCAGCATT3′ and 5′AGGCTTTGCTGTGCTTCAGGT3′ primers amplifying a 135-bp product. As internal control, a 105-bp β-actin fragment was amplified with 5′AG-GCATCCTCACCCTGAAGTA3′ and 5′CACACG-CAGCTCATTGTAGA3′ oligonucleotides. The standard conditions used for real-time PCR were as follows: 50°C for 10 minutes and 95°C for 2 minutes followed by 50 cycles of 15 s denaturation at 95°C, 45 s annealing/ elongation at 58°C or 60°C. SYBR® Green signal was measured in each step. Baseline was set between cycles 3 and 15. Each 96-well plate carried the same standard curve with β-actin. Mean fold gene expression was calculated with Applied Biosystems software regarding to the standard-curve and in addition with the 2 (Delta Delta CCT) Method. Data are expressed as mean ± SEM. The Student’s t test was performed for evaluation of significant differences between partial hepatectomy and Sham animal groups. Significance was determined at P < .05.
BrdU Incorporation and Detection in Cultured Primary Hepatocytes
Primary hepatocytes were exposed to 5-bromo-2′-deoxyuridine (Kit from Amersham Biosciences, Piscataway, NJ) using the recommended dilution of 1:1,000 in hepatocyte growth medium (HGM) for 24 hours. After incubation, cells were washed with PBS and fixed with methanol/glacial acetic acid solution (3:1, v/v) and air dried. The detection of BrdU was performed as instructed by the manufacturer’s protocol using a horse anti-mouse purified secondary antibody. For final staining, aminoethylcarbazole (AEC)-peroxidase substrate kit (Vector Laboratories, Burlingame, CA) was used and cells were counterstained with Shandon hematoxylin. Positive and negative stained nuclei were counted under the microscope.
BrdU Incorporation Into Hepatic Cells After Partial Hepatectomy
BrdU was injected intraperitoneally within 1 hour after partial hepatectomy and every 24 hours thereafter. We pursued this approach in order to be able to assess the cumulative label of all hepatocytes that had progressed through the cell cycle in a given animal up to the point of sacrifice. The amount of BrdU was injected at 50 mg/kg per rat.
Silencing Jagged-1 and Notch-1 Using a siRNA-Vector and In Vivo Transfection
For silencing experiments in vivo, psiRNA-hH1neo kit (InvivoGen, San Diego, CA) was used containing the human H1 promoter. Specific sequences for Jagged-1 (AAG GAG TAT CAG TCC CGC GTC) and Notch-1 (AAG TGG GAC CTG CCT GAA TGG) silencing were selected after general recommendations (Ambion Inc., Austin, TX). A scrambled sequence was used as a negative control (AAT CGC ATA GCG TAT GCC GTT). The synthesized oligonucleotides were designed to contain a loop sequence -TCAAGAG- to transcribe a dsRNA with hairpin structure. The annealed oligonucleotides were ligated into the BbsI-digested silencing RNA (siRNA) plasmid and competent E. coli GT116 were transformed. Plasmid expressing E. coli were selected and grown up in LB-broth containing 50 μg/ml kanamycin. Plasmids were isolated using Maxi-Prep (BioRad, Hercules, CA) and controlled by sequencing. Two days before PHx, 250 μg of plasmid was delivered into the liver using InvivoGene SHUTTLE (Q-Biogene, Montreal, QC) by injection into the superior mesenteric vein. Silencing of Notch-1 and Jagged-1 was controlled by immunohistochemical detection of Notch-1- and Jagged-1-protein in liver sections. The control animals were transfected with the scrambled-psiRNA vector as a control vector. Uptake and distribution in the liver was controlled using the native psiRNA vector and after immunohistochemical detection of the plasmid using anti-β-Galactosidase antibody (Cortex Biochem, San Leandro, CA). The plasmid was mainly taken up by periportal and midzonal hepatocytes, and much less in other cells like endothelial or bile duct cells. The cells were stained positive during the whole time period after PHx.
Results
Expression Pattern of Notch-1 and Jagged-1 in Regenerating Liver
In order to investigate the Notch signaling pathway in liver regeneration, we analyzed the expression of Notch-1 and Jagged-1 using the two-thirds partial hepatectomy model in adult rats. We performed semi-quantitative real-time PCR to examine the expression of Notch and Jagged-1 mRNA in normal and regenerating liver. We found that Notch mRNA was down-regulated between 6 hours and 2 days after partial hepatectomy (P ≤ 0.01 partial hepatectomy vs. sham) and returned to normal levels at day 7 (Fig. 1A). Conversely, Jagged-1 gene expression (Fig. 1B) was moderately increased 2.13 and 3 fold at 3 and 6 hours after partial hepatectomy, respectively (P < .02 partial hepatectomy vs. sham). After 12 hours, Jagged-1 mRNA was down-regulated through day 2 and returned to normal levels by day 4. Notch protein was very weakly expressed and barely detectable in whole cell lysates. Because of this low expression of Notch-1 and the localization of Notch-1 and Jagged-1 in cell membrane, we isolated and analyzed levels of Notch and Jagged proteins in plasma membrane preparations via Western blot (Fig. 2). Notch-1 protein levels increased up to day 2–4 after partial hepatectomy, and was followed by a decrease (Fig. 2). Western blot analyses of sham operated rats did not show significant changes in Notch-1 protein levels. Jagged-1 protein was well expressed in plasma membrane proteins of normal liver and showed a strong increase through day 4 after partial hepatectomy, remaining slightly elevated at day 7 (Fig. 2). We also investigated changes in other isoforms of Notch and Jagged and we analyzed the protein extracts for Notch-2 (M20, 25-255, Santa Cruz, CA) and Jagged-2 (R19, Santa Cruz, CA). We were not able to detect Notch-2 protein, and Jagged-2 protein was extremely weak without visible changes in protein levels after partial hepatectomy (data not shown). Due to the higher protein levels of Notch and Jagged-1, we assumed that these isoforms may be more important for cell–cell interactions in the liver and focused the study on Notch and Jagged-1. (The names Notch and Jagged to be used further in this study, refer to Notch-1 and Jagged-1, unless otherwise indicated.)
Fig. 1.
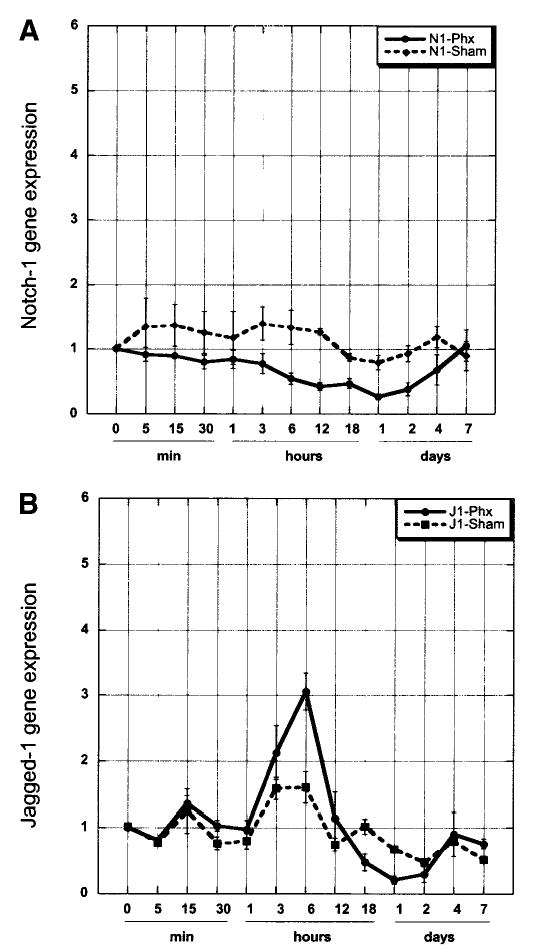
Real-time PCR analysis of Notch (A) and Jagged (B) gene expression in normal, sham operated, and partially hepatectomized (partial hepatectomy) rat liver. Data are normalized to expression in normal liver (actual expression of normal liver samples was CtNotch = 26; CtJagged = 27.9). Data represent the mean value ± SEM (n ≥ 3).
Fig. 2.
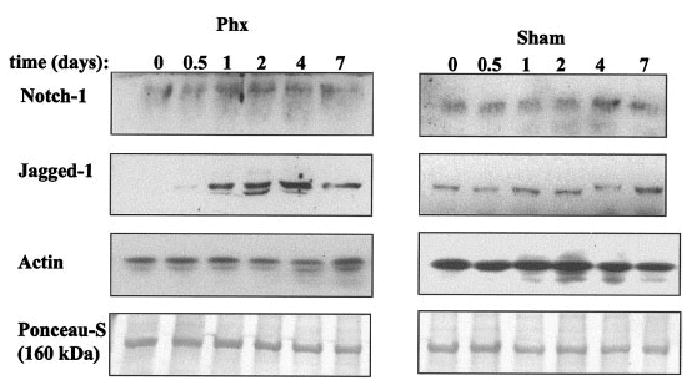
Detection of Notch and Jagged expression by Western blot analysis. Plasma membrane proteins were isolated from normal and partial hepatectomy or sham liver. Monoclonal antibody against Notch (hamster, Upstate) or polyclonal antibody against Jagged16 were used to analyze Notch (A) and Jagged (B) expression. (C) Jagged detection in whole cell lysate of normal (t = 0), partial hepatectomy, and sham rat livers. PLEASE NOTE: Due to the very low level of expression of both Notch and Jagged in normal and sham operated liver, a longer time exposure was used for the sham samples. The t = 0 sample of both sham and partial hepatectomy is the same tissue sample. It appears over-expressed in the sham samples only due to the longer exposure applied to the sham samples to allow detection of the protein.
Localization of Notch and Jagged Proteins in Rat Liver
We investigated the cellular distribution of both Notch receptor and Jagged ligand in rat liver by immunohistochemical staining of sections of formalin-fixed and paraffin-embedded liver tissue. In normal liver, we observed a high level of Notch in bile duct and endothelial cells, as well as staining of the plasma membrane in hepatocytes (Fig. 3A). Staining for Notch was enhanced at day 4 after partial hepatectomy in periportal hepatocytes (shown in Fig. 3B and 3C at low and high magnification). Staining for Jagged in normal liver (Fig. 3D) was seen predominantly in biliary epithelium with weaker staining seen in hepatocytes. Enhanced staining for Jagged was observed at day 4 in both biliary epithelium and periportal hepatocytes (shown in Fig. 3B, 3C, 3E, and 3F at low and high magnification). There were no changes in slivers of sham-operated animals (data not shown).
Fig. 3.
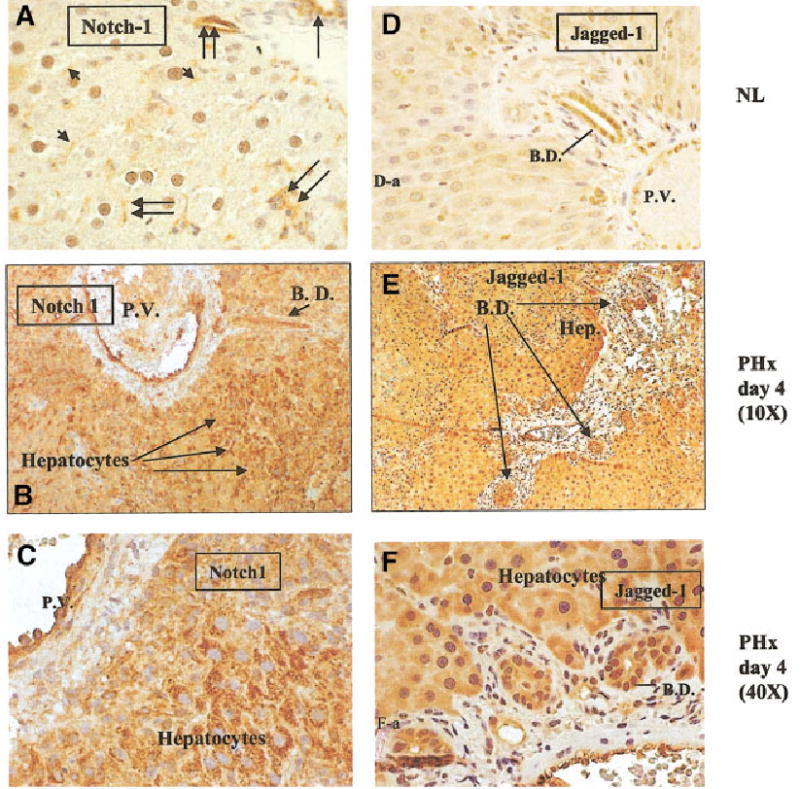
Time course of Notch and Jagged expression in regenerating liver determined by immunohistochemical staining. (A) Normal liver, magnification at 40×. Notch staining is shown on bile ductules (single long arrow), sinusoidal endothelial and small vessel endothelial cells (double arrows), and hepatocyte plasma membranes (short arrows). (B and C) Regenerating liver, 4 days after partial hepatectomy. Figure 3B was taken at a magnification of 10× and Fig. 3C at a magnification of 40×. Both pictures demonstrate staining of Notch in endothelial cells and periportal hepatocytes. (D) Normal liver (magnification: 20×). Jagged staining predominantly in bile ductules with weaker staining seen in hepatocytes. (E and F) Regenerating liver. Magnifications at 10× and 40×, respectively. Strong immunoreactivity for Jagged is seen in bile ductules and periportal hepatocytes. B.D. = bile ductile; P.V. = portal vein.
During the upregulated expression of Jagged- and Notch-proteins in the periportal regions at day 4 after partial hepatectomy, Notch and Jagged were clearly colocalized on the membrane of hepatocytes (Supplemental Figs. 1A and 1B). There was no significant colocalization of Notch and Jagged seen in slivers of sham operated animals (Fig. 4B).
Fig. 4.
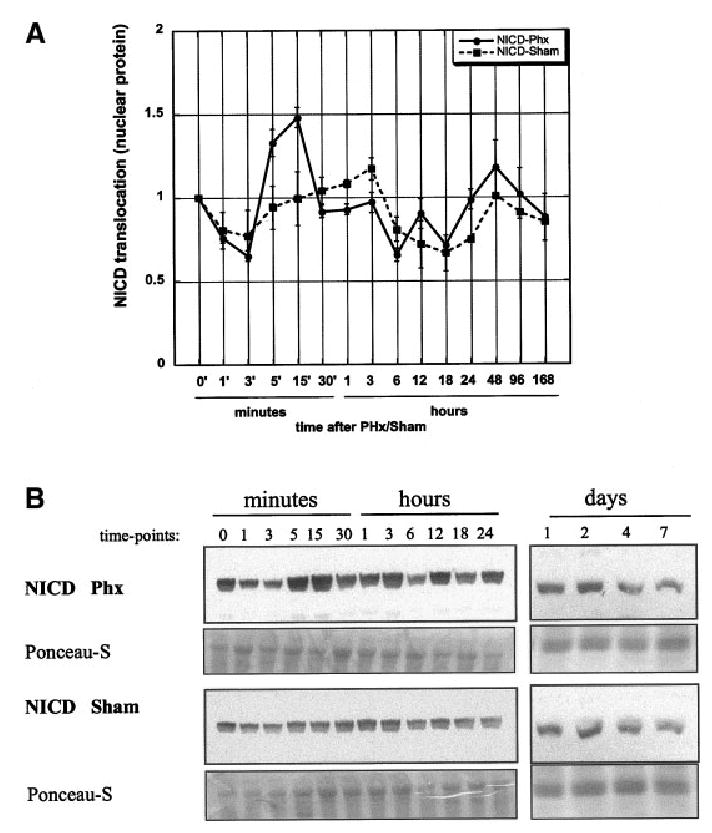
Detection of cytoplasmic domain of Notch (NICD) in nuclear protein (NP) extracts by Western blot analysis (rabbit polyclonal antibody, Upstate). (A) Densitometric analysis of Western blots for NICD in nuclear protein (NP). Data are shown as mean ± SEM (n = 3). (B) Representative Western blot of NICD detection in NP of rat liver. Ponceau-S stain of a band at 176 kDa are used as loading control. Numbers indicate time elapsed after operation in minutes, hours, and days.
Activation of Notch After Partial Hepatectomy
To determine the activation of Notch and the translocation of NICD to the nucleus after partial hepatectomy, we isolated nuclear proteins from normal and regenerating livers. Detection of NICD was pursued using antibody specific to the carboxy-terminal intracytoplasmic portion of Notch, which is cleaved off after ligand binding and migrates to the nucleus (see Introduction). Our results show an increased translocation of NICD starting 5–15 minutes after partial hepatectomy, which indicates an early activation of Notch (Fig. 4). Quantification of the results of the Western blots showed modest fluctuations around the control levels from 30 minutes until day 7. No significant changes were seen in the sham-operated animals. These changes were also observed by detection of NICD via immunofluorescence. Figure 5 shows immunofluorescence for NICD in normal liver (Fig. 5A-a and 5A-b) and liver at 15 minutes after partial hepatectomy (Fig. 5B-a and 5B-b), at the time with the highest concentration of Notch in the nucleus (from the data of Fig. 5). Green fluorescence for NICD was clearly detectable in the nucleus 15 minutes after partial hepatectomy. Cytoplasmic or membrane associated NICD was the predominant localization seen in hepatocytes of sham-operated livers (Fig. 5C-a and 5C-b) and normal liver (Fig. 5A-a and 5A-b).
Fig. 5.
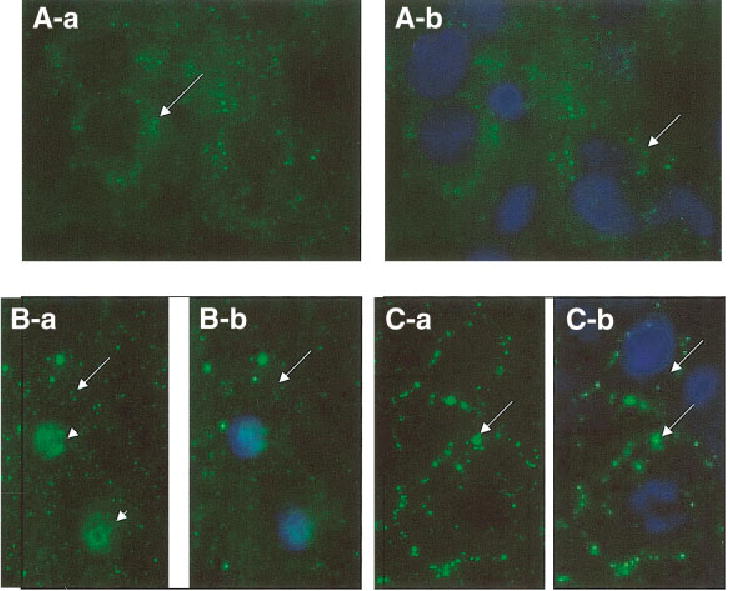
Detection of localization of the intra-cytoplasmic domain of Notch (NICD) in normal liver (A-a and A-b), liver at 15 minutes after partial hepatectomy (B-a and B-b), or sham operation (C-a and C-b). In normal liver and in sham-operated animals, NICD is localized only on the cytoplasm or the plasma membrane. There is no green fluorescence in the nuclei. Green fluorescence is seen in the nuclei at 15 minutes after partial hepatectomy. The nuclei were counter-stained with Hoechst dye shown in Fig. A-b, B-b, and C-b, to serve as comparison with the corresponding (a) figures in order to facilitate visual localization of the nuclei. Cytoplasmic and membrane localization of NICD is shown by long arrows. Nuclear localization (seen only in B-a) is shown by short arrows.
Early Upregulation of Notch Target Gene HES-1 During Liver Regeneration
In order to further explore the implication of the above findings suggesting early activation of Notch after partial hepatectomy, we investigated the expression of target genes of Notch. We used real-time PCR analysis to determine the expression of the genes HES-1 and HES–5. HES-1, which can be found in many tissues (13), was well expressed in normal liver compared to HES-5 (cycle threshold: CtHES1 = 24.5; CtHES5 = 35). After Notch activation, expression of HES-1 was upregulated early after partial hepatectomy and reached a maximum at 1 hour (P < .04 vs. control) (Supplemental Figure 2A). This time frame of change in HES-1 expression correlates well with the early nuclear translocation of NICD after partial hepatectomy shown above. The later decrease of NICD in the nuclei (after 3 h) also correlates with the subsequent down-regulation of HES-1 gene expression. The expression returned to normal levels by 12 hours after partial hepatectomy. Although the expression changes of HES-5 after partial hepatectomy were less striking than those found in HES-1, HES-5 expression showed a minor increase at 1–6 hours, followed by a decrease of 85% between 12 hours and 48 hours (Supplemental Figure 2B). Data from RNA from sham-operated livers do not show significant changes in either HES-1 or HES-5.
BrdU Uptake in Rat Hepatocytes Treated With Soluble-rrJagged Protein
Isolated rat hepatocytes from normal livers were cultured in low density on collagen coated culture dishes in HGM in the absence or presence of growth factors HGF and epidermal growth factor (EGF). After plating, cells were exposed to 0.2 μg/ml or 2 μg/ml recombinant rat Jagged soluble protein (R&D Systems, Minneapolis, MN). Control cultures were treated with the same volume of carrier solution (0.1% BSA in 1× PBS). BrdU was added to the cultures 24 before harvesting. At the end of the BrdU exposure, cultures were fixed and stained for BrdU. The results are shown in Supplemental Figure 3A. We found that rr-Jagged treatment increased the percentage of BrdU positive nuclei in a dose-dependent fashion with the highest effect at the 48-h time point in the absence of growth factors. Measurable increase was also seen at day 4 of culture, when (in the presence of HGF plus EGF) hepatocytes are at the peak of proliferation. We also determined the presence of Notch protein in the hepatocyte cultures at 24 and 48 hours. The results are shown in Supplemental Figure 3B. Notch protein was present at both 24 and 48 hours in the cultures of hepatocytes. These findings provide direct evidence that endogenous Notch signaling may act as mitogenic or growth enhancing signal for hepatocytes.
Effects of Silencing RNA for Notch and Jagged on Hepatic Cell Proliferation During Liver Regeneration
In order to directly assess the effects of Notch and Jagged on liver regeneration, rats were injected with silencing RNA vectors for Notch or Jagged 2 days before performing partial hepatectomy. Injections with siRNA-vector containing a scrambled sequence were used as control (see Materials and Methods). DNA synthesis was monitored by injecting BrdU 1 hour after partial hepatectomy and every 24 hours thereafter. We pursued this approach in order monitor the cumulative labeling of hepatocytes and other cell types from the performing of partial hepatectomy to the time of sacrifice in each animal. BrdU incorporation was monitored using anti-BrdU immunohistochemistry and assessing the percent of BrdU labeled nuclei as per established procedures (see Materials and Methods). The results are shown in Table 1. Silencing RNA for Notch and Jagged (injected separately) each suppressed hepatocyte proliferation at day 2, 3, and 4 after partial hepatectomy. The results were statistically significant (P < .05) for all values in comparison to the animals injected with “scramble” RNA vector. Because the animals were injected daily, the number of BrdU labeled nuclei continued to rise in both the control (injected with vehicle solution) animals and the animals injected with “scramble” RNA vector from day 2 to day 4 after partial hepatectomy, whereas the percent of BrdU labeled nuclei in the livers of animals injected with the silencing RNA vectors remained generally unchanged.
Table 1.
Effect of Notch-1 and Jagged-1 Silencing on Proliferation of Hepatocytes In Vivo
| Treatment | Days Post-Phx | Mean of BrdU Positive Nuclei | Standard Error |
|---|---|---|---|
| Normal liver | 2 | 35.40 | 5.4 |
| Scra-siRNA | 2 | 40.70 | 3.6 |
| J1-siRNA | 2 | 7.30* | 3.8 |
| N1-siRNA | 2 | 24.70† | 3.6 |
| Normal liver | 3 | 59.60 | 7.6 |
| Scra-siRNA | 3 | 43.00 | 4.3 |
| J1-siRNA | 3 | 25.90† | 3.9 |
| N1-siRNA | 3 | 29.95‡ | 2.2 |
| Normal liver | 4 | 68.50 | 3.2 |
| Scra-siRNA | 4 | 63.15 | 4.2 |
| J1-siRNA | 4 | 29.9* | 2.4 |
| N1-siRNA | 4 | 35.4* | 4.2 |
Effect of notch and jagged silencing RNA on hepatocyte proliferation after partial hepatectomy.
P-value of significance against Scra-siRNA treatment.
P < .0005;
P < .005;
P < .05.
Despite the change in BrdU labeling induced by either Notch or Jagged silencing RNA, there was no statistically significant change in liver weight between the different groups (data not shown). This probably reflects other compensatory mechanisms that may correct liver weight during regeneration when hepatocyte proliferation is inhibited. We also examined the amount of Notch and Jagged proteins at day 4 after partial hepatectomy by immunohistochemistry, comparing animals injected with Notch- or Jagged-silencing RNA versus those injected with “scramble” RNA. The results are shown in Fig. 5. Immunohistochemical staining in the periportal regions for both Notch and Jagged in the animals injected with silencing RNA was decreased compared to the animals injected with the “scramble” RNA.
Discussion
Liver regeneration is a very complex process influenced by a great variety of growth factors, cytokines, and cell–cell interactions.2 We investigated the role of the Notch/ Jagged signaling pathway in the regenerative process using the standard model of two-thirds partial hepatectomy. The baseline expression of Notch and Jagged in the different cell types in the liver revealed widespread expression, with higher expression on the hepatocytes and the biliary epithelium, but also expression of Notch on endothelial cells of the sinusoids and small vessels. Our findings with the rat are comparable to what has been described in human liver.7,8
At the beginning of regeneration, Notch is minimally expressed in hepatocytes as detected by immunohistochemistry (Fig. 3A). The activated form of Notch (NICD) reaches peak levels in hepatocyte nuclei very early, at 15 minutes after partial hepatectomy. The expression of the Notch-dependent gene Hes1 peaks at 1 hour, suggesting induction from the transport of the NICD to the nucleus. It is not clear whether hepatocyte Notch is activated by Jagged expressed in hepatocytes or by Jagged expressed in other cell types. The increase in nuclear content of NICD at 15 minutes after partial hepatectomy may be occurring in any of the cells expressing Notch in normal liver, i.e., hepatocytes, sinusoidal cells, and biliary epithelium. The morphology of the nuclei and the cells shown in Fig. 6 demonstrate that at least some of the cells positive for NICD immunofluorescence are hepatocytes. The nuclear presence of NICD may be easier to demonstrate in hepatocytes given the overall much larger size of the hepatocyte nuclei compared to other hepatic cell types.
Fig. 6.
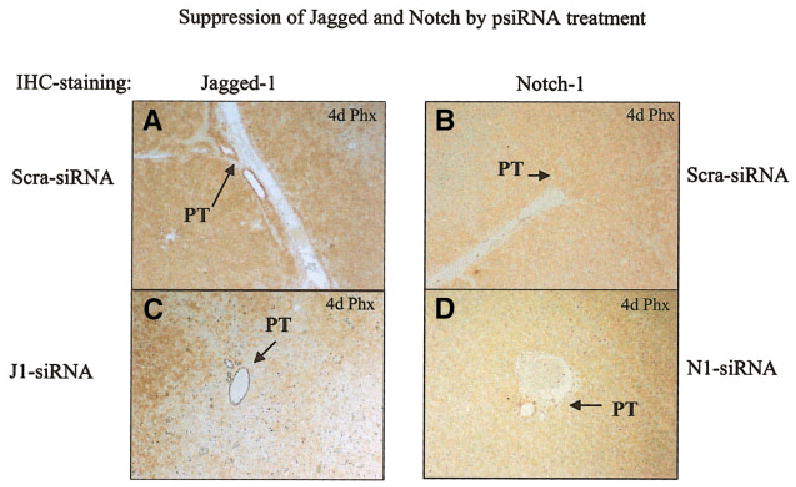
Immunohistochemical stains for Notch and Jagged proteins in animals injected with “scramble” RNA (scra-siRNA), Jagged-1 silencing RNA (J1-siRNA), and Notch1 silencing RNA (N1-siRNA). The animals were injected 2 days before partial hepatectomy. The sections were performed on livers at 4 days after partial hepatectomy (6 days after the RNA injections). There is evident decrease in the expression of both Jagged and Notch in the periportal regions of the hepatic lobules in the animals injected with the silencing RNA for either of the two genes. The portal triads in each section are shown by arrows. PT: portal triad.
An important finding of our study in relation to liver regeneration is that a significant increase occurs after partial hepatectomy in protein levels of both Notch and Jagged, mainly in periportal hepatocytes. Multiple previous studies have indicated that hepatic regeneration proceeds from the periportal to the pericentral regions, with a wave of mitoses,17 expression of metalloproteinases,18 and TGF-β1.19 Our findings demonstrate that there is a dramatic increase in expression of Notch and Jagged in the periportal regions, albeit at later time points. The increase in both Notch and Jagged, as shown by the Western blot in Fig. 2, occurs primarily from 0.5 to 7 days. This change may affect proliferation of any of the cells expressing the two proteins. In the case of hepatocytes, however, it is also likely that Jagged and Notch reside on the same cells (hepatocytes) and that their colocalization on the plasma membrane of the hepatocytes, as shown in Supplemental Fig. 1, may be stimulating a pathway operating through a juxtacrine loop. It is also possible that expression of Notch and Jagged in hepatocytes may mediate events in adjacent cells. Although hepatocyte proliferation in the rat peaks at 24 and 48 hours, proliferation of endothelial cells starts at day 3 and proceeds until day 6.20 Endothelial cells express Notch and it is possible that expression of Jagged may affect proliferation or other events dependent on Notch signaling in endothelial cells. Recently, some interesting studies revealed that Notch decreases proliferation of endothelial cells by downregulation of VEGFR-2 via HESR-121 and activation of Notch signaling enhances cessation of proliferation and formation of vessel-like structures in a three-dimensional angiogenesis model.22 Notch, which is expressed in endothelial cells in the liver, would also have a role in revascularization and thereby take part in remodeling of the hepatic micro-architecture during liver regeneration.23 The Notch receptor expressed at the endothelial cells may be stimulated by its ligand Jagged that is highly expressed in proliferating hepatocytes. At 72–144 hours after partial hepatectomy, sinusoidal endothelial cells start to infiltrate the avascular clusters of proliferating hepatocytes.20,24,25 Given the findings from other studies, the presence of Jagged on hepatocytes may cause a decrease in endothelial cell proliferation and promote formation of mature sinusoids, a hallmark of return to a quiescent liver status. Existing literature also suggests that after Notch cleavage, the extracellular domain can be transferred into hepatocytes by trans-endocytosis and thereby increase Notch content of hepatocytes.26 An early activation of Notch in sinusoidal cells by Jagged of hepatocytes would thereby activate gene expression in sinusoidal cells but also affect Notch signaling in hepatocytes due to additional intracellular cell-autonomous Notch-Jagged association.27 More studies focusing on specific cell populations are required to assess these possibilities.
A decrease in expression of Notch and Jagged induced by silencing RNA before partial hepatectomy had significant effects on the rate of proliferation of hepatocytes, as shown in Table 1. This finding is also complementary to our other observation in Supplemental Fig. 8, in which it is shown that treatment of hepatocytes with 2 μg/ml soluble rr-Jagged protein increases the BrdU uptake in hepatocytes in culture. The known specific interaction of rr-Jagged with Notch should result in an induction of HES-1. We detected, using real-time PCR, that HES-1 gene expression was induced by a factor of 11 at 1 hours after treatment of 48-h cultured hepatocytes with rr-Jagged (data not shown). The results in Fig. 6 and Supplemental Fig. 1 and Table 1 demonstrate that, whatever the precise mechanism and signaling pathways, activation of Notch in hepatocytes enhances hepatocyte proliferation and that this pathway is important during liver regeneration. Presence of Jagged is equally important in that regard. The findings with silencing RNA are specific and not seen when “scramble” siRNA vector was used as control. Despite the observed effects on hepatocyte proliferation, there was a slight (10%–25%) but not significant decrease in liver weight between the control, “scramble,” and silencing RNA treated groups. Liver weight is not a sensitive end-point for changes in kinetics of cell proliferation during liver regeneration. Previous studies have shown that treatment of the live with a variety of mito-inhibitory drugs or irradiation does not substantially affect the final liver weight, due to compensatory contribution of hepatocyte cellular hypertrophy in the absence of hepatocyte proliferation.2
While the changes in Notch protein as shown by both Western blot and immunohistochemistry during different time points in regeneration are easily demonstrable, the changes in Notch mRNA do not parallel in magnitude the changes seen in Notch protein. This suggests that the increase in Notch protein is not so much due to transcriptional changes in Notch gene but due to increased efficiency of Notch protein synthesis and decrease in its degradation. Similar changes in protein content have been demonstrated for other signaling molecules, such as beta-catenin.28
The role of Notch/Jagged signaling in bile duct cell proliferation and duct assembly is not clear from these studies. Bile duct epithelium was positive for both Notch and Jagged protein. Lemaigre29 pointed out that Notch actually controls interactions between blood vessels and the mesenchyme and that the impact of Jagged mutations in bile duct morphogenesis is only indirect due to dys-morphogenesis of periductal structures. Studies on Alagille syndrome affected patients revealed that bile ducts are not congenitally lacking but that the ductal paucity develops progressively after birth,30 suggesting the idea that Notch pathway is required to maintain a differentiated phenotype of bile duct cells. Again, much remains to be understood about the mechanisms by which Notch and Jagged regulate biliary epithelium development and growth.
In summary, our studies present evidence that Notch and Jagged signaling pathways are activated and play an important role in cell proliferation during liver regeneration after partial hepatectomy. The precise sequence of events and the cellular pathways and types affected need to be better understood. Evidence from many other systems of tissue development, however, suggest that these changes are likely to be important. Further studies are needed to pursue the impact of the Notch and Jagged signaling in specific hepatic cell types.
Supplementary Material
Acknowledgments
The authors thank Donna Beer Stolz, Mark A. Ross, Wendy M. Mars, Thomas Lehmann, Peter Pediaditakis, and Karen Mule for their superior intellectual and technical support at different stages throughout the study.
Footnotes
Dr. Michalopoulos is a founding member, consultant and stockholder of Kytaron, Inc.
Supported by NIH grants CA30241 and CA35373.
References
- 1.Grisham J. A morphologic study of deoxyribonucleic acid synthesis and cell proliferation in regenerating liver; autoradiography with thymidine-H3. Cancer Res. 1962;22:842–849. [PubMed] [Google Scholar]
- 2.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60 –66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 3.Baron M, Aslam H, Flasza M, Fostier M, Higgs JE, Mazaleyrat SL, Wilkin MB. Multiple levels of Notch signal regulation (review) Mol Membr Biol. 2002;19:27–38. doi: 10.1080/09687680110112929. [DOI] [PubMed] [Google Scholar]
- 4.Baron M. An overview of the Notch signalling pathway. Semin Cell Dev Biol. 2003;14:113–119. doi: 10.1016/s1084-9521(02)00179-9. [DOI] [PubMed] [Google Scholar]
- 5.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770 –776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 6.Louis AA, Van Eyken P, Haber BA, Hicks C, Weinmaster G, Taub R, Rand EB. Hepatic jagged1 expression studies. Hepatology. 1999;30:1269 –1275. doi: 10.1002/hep.510300512. [DOI] [PubMed] [Google Scholar]
- 7.Nijjar SS, Wallace L, Crosby HA, Hubscher SG, Strain AJ. Altered Notch ligand expression in human liver disease: further evidence for a role of the Notch signaling pathway in hepatic neovascularization and biliary ductular defects. Am J Pathol. 2002;160:1695–1703. doi: 10.1016/S0002-9440(10)61116-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nijjar SS, Crosby HA, Wallace L, Hubscher SG, Strain AJ. Notch receptor expression in adult human liver: a possible role in bile duct formation and hepatic neovascularization. Hepatology. 2001;34:1184 –1192. doi: 10.1053/jhep.2001.29399. [DOI] [PubMed] [Google Scholar]
- 9.Krantz ID, Piccoli DA, Spinner NB. Clinical and molecular genetics of Alagille syndrome. Curr Opin Pediatr. 1999;11:558 –564. doi: 10.1097/00008480-199912000-00015. [DOI] [PubMed] [Google Scholar]
- 10.Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, et al. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 11.Mumm JS, Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- 12.Dou S, Zeng X, Cortes P, Erdjument-Bromage H, Tempst P, Honjo T, Vales LD. The recombination signal sequence-binding protein RBP-2N functions as a transcriptional repressor. Mol Cell Biol. 1994;14:3310–3319. doi: 10.1128/mcb.14.5.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 14.Ohtsuka T, Sakamoto M, Guillemot F, Kageyama R. Roles of the basic helix-loop-helix genes Hes1 and Hes5 in expansion of neural stem cells of the developing brain. J Biol Chem. 2001;276:30467–30474. doi: 10.1074/jbc.M102420200. [DOI] [PubMed] [Google Scholar]
- 15.Kuroda K, Tani S, Tamura K, Minoguchi S, Kurooka H, Honjo T. Delta-induced Notch signaling mediated by RBP-J inhibits MyoD expression and myogenesis. J Biol Chem. 1999;274:7238 –7244. doi: 10.1074/jbc.274.11.7238. [DOI] [PubMed] [Google Scholar]
- 16.Columbano A, Ledda-Columbano GM, Coni PP, Faa G, Liguori C, Santa Cruz G, Pani P. Occurrence of cell death (apoptosis) during the involution of liver hyperplasia. Lab Invest. 1985;52:670 –675. [PubMed] [Google Scholar]
- 17.Rabes HM. Kinetics of hepatocellular proliferation as a function of the microvascular structure and functional state of the liver. Ciba Found Symp. 1977:31–53. doi: 10.1002/9780470720363.ch3. [DOI] [PubMed] [Google Scholar]
- 18.Kim TH, Mars WM, Stolz DB, Michalopoulos GK. Expression and activation of pro-MMP-2 and pro-MMP-9 during rat liver regeneration. Hepatology. 2000;31:75–82. doi: 10.1002/hep.510310114. [DOI] [PubMed] [Google Scholar]
- 19.Jirtle RL, Carr BI, Scott CD. Modulation of insulin-like growth factor-II/ mannose 6-phosphate receptors and transforming growth factor-beta 1 during liver regeneration [published erratum appears in J Biol Chem 1991 Dec 25;266:24860] J Biol Chem. 1991;266:22444 –22450. [PubMed] [Google Scholar]
- 20.Modis L, Martinez-Hernandez A. Hepatocytes modulate the hepatic microvascular phenotype. Lab Invest. 1991;65:661–670. [PubMed] [Google Scholar]
- 21.Taylor KL, Henderson AM, Hughes CC. Notch activation during endothelial cell network formation in vitro targets the basic HLH transcription factor HESR-1 and downregulates VEGFR-2/KDR expression. Microvasc Res. 2002;64:372–383. doi: 10.1006/mvre.2002.2443. [DOI] [PubMed] [Google Scholar]
- 22.Liu ZJ, Shirakawa T, Li Y, Soma A, Oka M, Dotto GP, Fairman RM, et al. Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis. Mol Cell Biol. 2003;23:14 –25. doi: 10.1128/MCB.23.1.14-25.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D’Amore PA, Ng YS. Won’t you be my neighbor? Local induction of arteriogenesis. Cell. 2002;110:289 –292. doi: 10.1016/s0092-8674(02)00869-3. [DOI] [PubMed] [Google Scholar]
- 24.Ross MA, Sander CM, Kleeb TB, Watkins SC, Stolz DB. Spatiotem-poral expression of angiogenesis growth factor receptors during the revascularization of regenerating rat liver. Hepatology. 2001;34:1135–1148. doi: 10.1053/jhep.2001.29624. [DOI] [PubMed] [Google Scholar]
- 25.Wack KE, Ross MA, Zegarra V, Sysko LR, Watkins SC, Stolz DB. Sinusoidal ultrastructure evaluated during the revascularization of regenerating rat liver. Hepatology. 2001;33:363–378. doi: 10.1053/jhep.2001.21998. [DOI] [PubMed] [Google Scholar]
- 26.Parks AL, Klueg KM, Stout JR, Muskavitch MA. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development. 2000;127:1373–1385. doi: 10.1242/dev.127.7.1373. [DOI] [PubMed] [Google Scholar]
- 27.Sakamoto K, Ohara O, Takagi M, Takeda S, Katsube K. Intracellular cell-autonomous association of Notch and its ligands: a novel mechanism of Notch signal modification. Dev Biol. 2002;241:313–326. doi: 10.1006/dbio.2001.0517. [DOI] [PubMed] [Google Scholar]
- 28.Monga SP, Pediaditakis P, Mule K, Stolz DB, Michalopoulos GK. Changes in WNT/beta-catenin pathway during regulated growth in rat liver regeneration. Hepatology. 2001;33:1098 –1109. doi: 10.1053/jhep.2001.23786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemaigre FP. Development of the biliary tract. Mech Dev. 2003;120:81–87. doi: 10.1016/s0925-4773(02)00334-9. [DOI] [PubMed] [Google Scholar]
- 30.Van Eyken P, Sciot R, Callea F, Van der Steen K, Moerman P, Desmet VJ. The development of the intrahepatic bile ducts in man: a keratin-immunohistochemical study. Hepatology. 1988;8:1586 –1595. doi: 10.1002/hep.1840080619. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


