Abstract
Aims: Tumour tissue microarray allows the analysis of hundreds of tumour samples simultaneously on a single microscope slide. However, the extremely small tissue samples taken from the original tissue may not always be representative of the entire tumour.
Methods: The reliability of this new technology was investigated by analysing HER-2 oncogene amplification by fluorescence in situ hybridisation (FISH) from representative slides of the whole tumour and small tissue core biopsies from 29 invasive breast tumours.
Results: The tissue microarray method had high accuracy; in only one of 29 cases (3.4%; 95% confidence interval, 0% to 10%) were the results discordant with whole tumour analysis.
Conclusion: Tumour microarray is a highly reliable method for analysing HER-2 oncogene amplification by FISH in human breast tumours.
Keywords: fluorescence in situ hybridisation, tissue microarray, breast carcinoma, HER-2
The HER-2 oncogene (also known as c-erbB2/HER-2/neu) is amplified in 20–30% of invasive breast carcinomas and accumulating data suggest that this biological marker has prognostic and possibly predictive value.1 Moreover, an effective new treatment for HER-2 positive metastatic breast cancer, consisting of an anti-HER-2 monoclonal antibody (trastuzumab, Herceptin™; Genentech, South San Francisco, California, USA),2 means that the determination of HER-2 status in breast tumours will become important in clinical practice. Retrospective examination of samples from tumour banks for providing clinical information is labourious and technically challenging, owing to the large number of samples to be evaluated.
Recently, Kononen et al developed a new technology for rapid molecular profiling of large numbers of cancer specimens in a single experiment.3–10 Tissue microarrays are constructed by bringing together very small cylindrical tissue sample disks from several tumours into a single paraffin wax block. Sections from these blocks can then be used in the analysis of genes or proteins by fluorescence in situ hybridisation (FISH) or by immunohistochemistry (IHC).
“An effective new treatment for HER-2 positive metastatic breast cancer, consisting of an anti-HER-2 monoclonal antibody, means that the determination of HER-2 status in breast tumours will become important in clinical practice”
The validity of tissue microarray technology in breast carcinoma has been investigated by IHC for biological markers such as progesterone and oestrogen receptors, and the HER-2 oncogene.6 The results showed that the analysis of two microarray disks is comparable to the analysis of a whole tissue section in more than 95% of cases.6 Because there is no standardisation for IHC methods, FISH is the most promising technique to evaluate HER-2 on microarrays. Accordingly, in our present study, we constructed several microarray disks (diameter, 1.5 mm) containing invasive breast carcinoma tissues; in addition, because small tissue samples might not always reflect the biological properties of the entire tumour, we investigated the false negative rate of this technique by examining 29 tumours, which were screened for their HER-2 status by FISH. These data were compared with those obtained by FISH using the entire tumour sample.
MATERIALS AND METHODS
Breast tumour tissue microarray disks
The breast tissue microarray was constructed as described previously.3,4 Briefly, small biopsies were retrieved from selected regions of donor tissue using stainless steel biopsy needles with an inner diameter of 1.5 mm (manual tissue puncher; Beecher Instruments, Silver Spring, Maryland, USA). Cores were inserted into a standard sized recipient array block. After construction of the array block, multiple 5 μm sections were cut and haematoxylin and eosin stained sections were reviewed by one pathologist (DL) to determine whether an invasive component of the tumour was present on the microarray for each sample.
FISH on formalin fixed tissue sections
Dual colour FISH was performed on sections of the formalin fixed samples using the PathVysion kit probe for HER-2 (spectrum orange labelled HER-2 and spectrum green labelled α satellite centromeric region for chromosome 17 (CEP17; Vysis Inc, Downers Grove, Illinois, USA)), according to the manufacturer's instructions. Briefly, slides were dewaxed, immersed in 0.2 mol/litre HCl for 20 minutes, incubated in 1 mol/litre sodium isothiocyanate solution at 80°C for 20 minutes, and immersed in a pepsin solution (0.125 mg/ml in 0.9% NaCl, pH 2.0) for 20 minutes at 37°C. The slides were then postfixed in 10% buffered formalin for 10 minutes, dehydrated through graded alcohol solutions (70%, 90%, and 100%), air dried, and denatured for five minutes at 73°C in 70% formamide/2× saline sodium citrate (SSC; 0.3 mol/litre sodium chloride and 0.03 mol/litre sodium citrate) solution. Slides were dehydrated again in 70%, 90%, and 100% ethanol and hybridised with 10 μl probe overnight at 37°C. The next day, slides were washed in a solution of 2× SSC/0.3% NP40 for two minutes at 73°C, and signals in tumour cells were scored with a Leica DMRB epifluorescence microscope (Leitz, Wetzlar, Germany) equipped with a triple bandpass filter (TRITC, fluorescein isothiocyanate, DAPI, Chroma, San Rafael, California, USA) using ×40 and ×100 objectives.
Analysis of interphase FISH
Criteria for gene amplification were tight clusters of HER-2 signals in multiple cells or a least two times more HER-2 signals than centromeric 17 (CEP17) signals/cell in at least 60 nuclei located in the invasive part of the tumour. Overlapping nuclei and nuclei lacking any hybridisation signals were excluded from analysis. HER-2 was not considered to be amplified if the ratio of HER-2 to CEP17 signals was ≤ 2.
RESULTS AND DISCUSSION
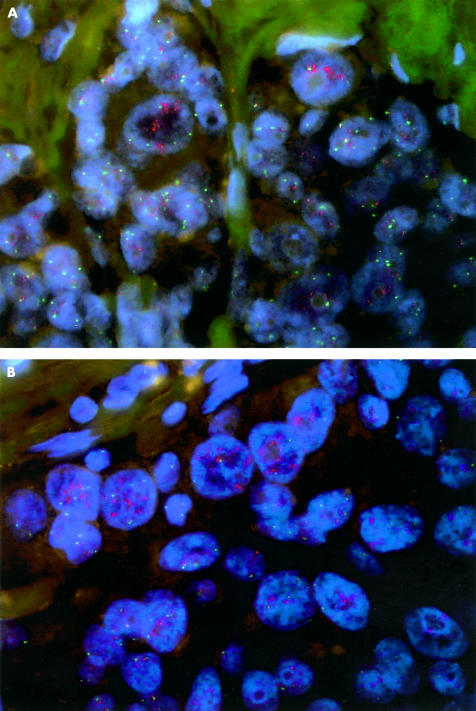
To estimate the reliability of tumour microarrays to define HER-2 oncogene amplification, we investigated the degree of agreement between FISH data obtained for HER-2 on whole tumour samples and on corresponding 1.5 mm microarray disks in 29 cases of invasive breast cancer. Disks were scored if at least 10% of the disk area contained tumour cells. We found that one of 29 cases (3.4%; 95% confidence interval, 0% to 10%) was discordant. In this case, the tumour was classified as HER-2 negative on the core disk and as HER-2 amplified on the whole tumour sample (table 1). These findings suggest a high correlation between the two investigation methods. The discrepancy might be attributed to tumour heterogeneity (fig 1A, B) and correlated to the data found in the literature; that is, that HER-2 status evaluated by IHC was discordant between the array disk and the whole section in one of 34 cases.6
Table 1.
HER-2 status assessed by fluorescent in situ hybridisation (FISH) in 29 breast cancer samples
| No. of cases | FISH on slides | FISH on disks |
| 9 | − | − |
| 1 | + | − |
| 19 | + | + |
Figure 1.
Dual colour fluorescence in situ hybridisation for the HER-2 (red) and α satellite centromeric region of chromosome 17 (green) in paraffin wax embedded breast specimens (DAPI counterstain). Heterogeneity and aneuploidy of invasive tumour cells: two different areas of the same tumour, one with aneuploidy and rare cells showing HER-2 amplification (A), and the other with several cells showing high HER-2 amplification (B).
“Cancer tissues frequently reveal heterogeneous expression patterns of molecular markers on standard full sections”
Another report found perfect concordance among 14 sets of FISH data obtained from tumour array sections and disaggregated nuclei from whole sections, using the same probe.3 Nevertheless, most studies have dealt with the ICH analysis of biological markers, and tissue core biopsies of 0.6 mm are taken from representative areas of paraffin wax embedded tissues arrayed on one recipient block. Such a diameter allows the analysis of 400–1000 tumour specimens simultaneously. By increasing the size of the core to 2 mm, the number of samples that can be analysed drops to 100–150. Cancer tissues frequently reveal heterogeneous expression patterns of molecular markers on standard full sections. Therefore, two or more samples from a single tumour might need to be analysed to assess accurately the global tumour status. Two studies reported that the analysis of two disks was comparable to screening whole tissue sections in more than 90% of cases,6,9 given that if one of the replicates was positive for the marker, the sample was scored as positive. The presence of a third core on the array reduced the risk of losing the case because of tissue damage, and allowed a majority decision (2 > 1) if one core differed from the other two.7 Finally, the analysis of proliferation index and tumour grade in more than 2000 bladder cancer specimens revealed that combined data from four cores/sample were representative of the whole tumour and were associated with the prognosis.10 Moreover, every individual core could provide meaningful data. In fact, increasing the diameter of the core samples to 2–4 mm to improve the representativity may not be optimal because finding heterogeneity within such a small region is often quite low. In contrast, punching multiple small cores (0.6 mm) from different regions captures the heterogeneity of the tumours more effectively.
In conclusion, although the limited number of cases evaluated in our study does not allow definitive conclusions to be made about the reliability of HER-2 evaluation by FISH using tumour tissue microarrays, this technology is a population level research tool useful for large retrospective studies, and is not intended for managing clinical diagnoses of individual cases. The recent development of a database model to manage clinical, pathology, and molecular data in high density microarray,11 together with automation of analysis and increased availability of this technique, will greatly accelerate the translation of basic research findings to clinical applications.
Take home messages.
The tissue microarray method had high accuracy; in only one of 29 cases were the results discordant with whole tumour analysis
Tumour microarray is a highly reliable method for analysing HER-2 oncogene amplification by fluorescent in situ hybridisation in human breast tumours
At the moment tumour microarray is a research tool useful for large retrospective studies, but improved technology may enable it to be used for clinical purposes in the future
Acknowledgments
D Gancberg and G Rouas are supported by grants from “Les Amis de l'Institut Bordet”, Brussels, Belgium. T Järvinen is supported by a grant from the Finnish Cancer Institute, Helsinki, Finland.
Abbreviations
FISH, fluorescence in situ hybridisation
IHC, immunohistochemistry
SSC, saline sodium citrate
REFERENCES
- 1.Toikkanen S, Helin H, Isola J, et al. Prognostic significance of HER-2 oncoprotein expression in breast cancer: a 30 year follow-up. J Clin Oncol 1992;10:1044–8. [DOI] [PubMed] [Google Scholar]
- 2.Cobleigh MA, Vogel CL, Tripathy D, et al. Multinational study of the efficacy and safety of humanized anti-HER-2 monoclonal antibody in women who have HER-2 overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol 1999;17:2639–48. [DOI] [PubMed] [Google Scholar]
- 3.Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumour specimens. Nat Med 1998;4:844–7. [DOI] [PubMed] [Google Scholar]
- 4.Bubendorf L, Kononen J, Koivisto P, et al. Survey of gene amplifications during prostate cancer progression by high-throughput fluorescence in situ hybridization on tissue microarrays. Cancer Res 1999;59:803–6. [PubMed] [Google Scholar]
- 5.Moch H, Schraml P, Bubendorf L, et al. High-throughput tissue microarray analysis to evaluate genes uncovered by cDNA microarray screening in renal cell carcinoma. Am J Pathol 1999;154:981–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camp LR, Charrette LA, Rimm DL. Validation of tissue microarray technology in breast carcinoma. Lab Invest 2000;80:1943–9. [DOI] [PubMed] [Google Scholar]
- 7.Hoos A, Urist MJ, Stojadinovic A, et al. Validation of tissue microarrays for immunohistochemical profiling of cancer specimens using the example of human fibroblastic tumors. Am J Pathol 2001;158:1245–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perrone EE, Theoharis C, Mucci NR, et al. Tissue microarray assessment of prostate cancer tumour proliferation in African–American and White men. J Natl Cancer Inst 2000;92:937–9. [DOI] [PubMed] [Google Scholar]
- 9.Gillett CE, Springall RJ, Barnes DM, et al. Multiple tissue core arrays in histopathology research: a validation study. J Pathol 2000;192:549–53. [DOI] [PubMed] [Google Scholar]
- 10.Nocito A, Bubendorf L, Tinner EM, et al. Microarrays of bladder cancer tissue are highly representative of proliferation index and histological grade. J Pathol 2001;194:349–57. [DOI] [PubMed] [Google Scholar]
- 11.Manley S, Mucci NR, De Marzo AM, et al. Relational database structure to manage high-density tissue microarray data and images for pathology studies focusing on clinical outcome. Am J Pathol 2001;159:837–43. [DOI] [PMC free article] [PubMed] [Google Scholar]