Abstract
Aims: To compare the performance of four media, singly and in combination, as direct plating media for the isolation of Salmonella enterica from human faeces.
Methods: Two thousand four hundred and nine routine, faecal samples received by four laboratories were inoculated on to xylose lysine desoxycholate (XLD), desoxycholate citrate (DCA), mannitol lysine crystal violet brilliant green (MLCB), and α-β chromogenic (ABC) agars using standardised protocols, reagents, and data collection. Isolates of presumptive salmonellae were identified using standard laboratory techniques and the results were analysed statistically.
Results: Direct plating recovered 46 of the 60 possible isolates of Salmonella spp recovered via enrichment broth. No isolates were recovered from direct plating that were not recovered via selenite enrichment. MLCB gave the highest isolation rate individually (84.8%) and amounts of competing flora (CF) did not affect the recognition of colonies. ABC proved highly specific, but insensitive, and isolation rates were adversely affected by any amount of CF. Isolation rates from XLD and DCA were only affected when the CF load was heavy. DCA was least specific, with only 9.01% of picks positive and greatest number of confirmatory tests. XLD and MLCB, in combination, gave the highest isolation rate.
Conclusions: Where the earlier results of direct plating may be advantageous, XLD and MLCB provide the optimal combination. For non-typhi salmonellae, MLCB is the best, single direct plating medium. For routine diagnostic work, XLD is most effective.
Keywords: salmonella, media, direct plating
A wide variety of selective, direct plating media has been used in the isolation of Salmonella enterica from human faeces. Xylose lysine desoxycholate (XLD) and desoxycholate citrate (DCA) agars are probably the two media most commonly used for this purpose in the UK.
Mannitol lysine crystal violet brilliant green agar (MLCB) is a highly selective medium for the isolation of salmonellae, which has been used principally in food and environmental microbiology. It is unsuitable for the isolation of S typhi, but its specificity may offer advantages in terms of easier recognition of suspect colonies, with fewer false positive picks.1
In addition, α-β chromogenic medium (ABC) is a DCA based chromogenic agar, selective for Salmonella spp, including S typhi, which may be more sensitive and specific than DCA.2 This medium uses a dual chromogen system, based on the ability of salmonellae to produce α-galactosidase in the absence of β-galactosidase. With the exception of this study, comparing ABC with DCA, we have been unable to find published studies of direct comparisons of these plating media in the isolation of Salmonella enterica.
“Mannitol lysine crystal violet brilliant green agar is a highly selective medium for the isolation of salmonellae, which has been used, principally, in food and environmental microbiology”
An evaluation of the performance of XLD, DCA, MLCB, and ABC, as direct plating media, was undertaken in four laboratories, adopting a standardised protocol. Data were collected to determine isolation rates of Salmonella spp on each medium, individually and in combination. Amounts of competing flora (CF) and confirmatory work generated were also assessed in each case.
MATERIALS AND METHODS
Media
XLD (LAB032; Lab M, Bury, UK), DCA (LAB065; Lab M), MLCB (LAB116; Lab M), and ABC (HAL001; Lab M) were prepared from single batches, in accordance with the manufacturer's recommendations, at a single site, to avoid interlaboratory variations. All media were subjected to full quality control procedures before distribution and were used within 14 days of preparation.
Samples
Two thousand four hundred and nine routine faecal samples, received from both hospital and community sources, were included in our study. Samples containing insufficient material for duplicate inoculation and those submitted for specific, limited examination were excluded.
Our study was carried out simultaneously at all four sites between July and September 1999.
Sample preparation
For formed samples, a pea sized portion of faecal material (approximately 1 g) was emulsified in 3 ml maximal recovery peptone saline diluent (MRD) (Oxoid CM 733; Oxoid, Basingstoke, UK)—that is, a 1/4 dilution. Liquid samples were diluted 1/4 vol/vol in MRD.3
Media inoculation and incubation
Using a sterile pipette, one drop (approximately 45 μl) of faecal suspension was inoculated on to each of the four plates and spread with sterile loops for single colonies. Whole plates were used throughout.
All plates were incubated (in separate racks) for 16–24 hours at 37°C in air.
A 1 ml aliquot of faecal suspension was inoculated into selenite broth to be incubated for 15–18 hours at 37°C before subculture to DCA.
Plate reading
Each set of plates was read independently by different fully qualified biomedical scientists previously familiarised with these media. One example of each morphological type of suspect colony for each plate was selected for further tests. Data on bacterial growth and potential salmonellae selected were recorded at the time of reading and verified at regular intervals by a separate senior member of staff.
Bacterial growth was estimated semiquantitatively on the basis of colony counting, as follows: one to 10 colonies, light growth (+); 11–50 colonies, moderate growth (++); and more than 50 colonies, heavy growth (+++). An assessment of CF was also made on this basis.
Identification
Isolates were identified as possible salmonellae by the absence of urease production. Urease negative isolates were then identified further by API 10S (Biomerieux, Stoke on Trent, UK) and serology. Those identified as Salmonella spp were referred to the Laboratory of Enteric Pathogens, Central Public Health Laboratory, London, for confirmation.
Statistical methods
Logistic regression was performed to determine whether the proportion of positive results (at any level) differed as a result of the presence of CF. This analysis was performed separately for each of the four agars and used the absence of CF as the baseline for comparison.
Chi squared tests were used to determine whether there was any interlaboratory variation and also to assess differences in the degree of +++ growth on the four agars. The isolation of salmonellae on each agar in comparison with the other agars was performed using McNemar's test, with a p value of 0.008 taken as the level of significance after correction for multiple comparisons.
RESULTS
Between July and September 1999, 2409 faecal samples were examined; the four laboratories examining 710, 811, 508, and 380 samples, respectively.
Results are summarised in tables 1–3. Salmonella spp were isolated from 46 samples by direct plating on any medium, an isolation rate of 1.9%. However, 60 samples were positive after selenite enrichment, an overall isolation rate of 2.5%. No samples were positive by direct plating alone, and no significant interlaboratory variation in isolation rates was found.
Table 1.
Salmonella spp isolations, presumptive isolates with per cent confirmed and API usage, by medium
| Medium | Salmonella isolations (% all direct isolates) n=46 | No. presumptive salmonellae selected (% positive) | No. API tests required |
| XLD | 33 (71.7) | 197 (16.8) | 105 |
| DCA | 31 (67.4) | 344 (9.0) | 212 |
| MLCB | 39 (84.8) | 393 (9.9) | 160 |
| ABC | 26 (56.5) | 90 (28.9) | 41 |
ABC, α-β chromogenic medium; DCA, desoxycholate citrate; MLCB, mannitol lysine crystal violet brilliant green; XLD, xylose lysine desoxycholate.
Table 2.
Growth quantitation of salmonella isolates by medium (%)
| Result | XLD | DCA | MLCB | ABC |
| + | 8 (24.2) | 1 (3.2) | 6 (15.4) | 4 (15.4) |
| ++ | 12 (36.4) | 9 (29.0) | 8 (20.5) | 11 (42.3) |
| +++ | 13 (39.4) | 21 (67.7) | 25 (64.1) | 11 (42.3) |
| Total | 33 | 31 | 39 | 26 |
ABC, α-β chromogenic medium; DCA, desoxycholate citrate; MLCB, mannitol lysine crystal violet brilliant green; XLD, xylose lysine desoxycholate. +, 1–10 colonies (light growth); ++, 11–50 colonies (moderate growth); +++, >50 colonies (heavy growth).
Table 3.
Competing flora by medium (%)
| Result | XLD | DCA | MLCB | ABC |
| − | 105 (4.4) | 169 (7.0) | 291 (12.0) | 83 (3.5) |
| + | 245 (10.2) | 425 (17.6) | 753 (31.3) | 255 (10.6) |
| ++ | 368 (15.3) | 400 (16.6) | 531 (22.0) | 214 (8.9) |
| +++ | 1691 (70.2) | 1415 (58.7) | 834 (34.6) | 1857 (77.1) |
| Total | 2304 | 2240 | 2118 | 2326 |
ABC, α-β chromogenic medium; DCA, desoxycholate citrate; MLCB, mannitol lysine crystal violet brilliant green; XLD, xylose lysine desoxycholate. +, 1–10 colonies (light growth); ++, 11–50 colonies (moderate growth); +++, >50 colonies (heavy growth).
The number of colonies of salmonellae was significantly greater on DCA and MLCB than on XLD or ABC (p < 0.01).
XLD and ABC allowed the heaviest growth of CF.
For ABC, a significant decrease in salmonella isolates was seen at all CF scores compared with a baseline of absent CF (+, p = 0.016; ++, p = 0.018; +++, p = 0.0001). For XLD and DCA, there was a significant decrease in isolates where CF scores were highest (+++) (p = 0.019 and p = 0.039, respectively). Fewer CF were isolated on MLCB, but whatever the CF score, there was no evidence that it had a detrimental effect on the numbers of salmonellae isolated.
Numbers of colonies selected as presumptive salmonellae and the percentage subsequently proving positive are presented in table 1, together with the numbers of API tests required, reflecting urease negative organisms growing through. This indicates the greater specificity of ABC and, to a lesser extent, XLD. However, ABC proved much less sensitive, isolating only 26 salmonellae.
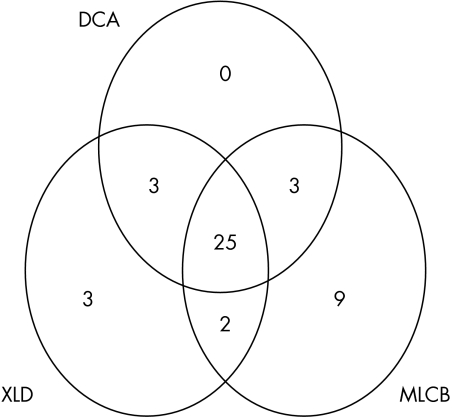
Given that ABC had significantly lower isolation rates than MLCB, the effects of plate combinations were only examined for the three remaining media (fig 1).
Figure 1.
Venn diagram showing numbers of salmonellae isolated on XLD (xylose lysine desoxycholate), DCA (desoxycholate citrate), and MLCB (mannitol lysine crystal violet brilliant green), individually and in combination.
The combination of the two best performing media, MLCB and XLD, compared with XLD alone, gave a 20% increase in positivity (95% confidence interval, 10.8 to 32.3).
DISCUSSION
Direct plating of faecal material on to selective agar has been an integral part of the microbiological investigation of enteric disease for many years, although several studies have shown that subculture from selective enrichment broth gives a higher yield of S enterica.4,5 The advantage of direct plating is that results could be available 24 hours before results from the enrichment method.
Persaud and Eykyn6 reported that, of 20 laboratories surveyed, 15 were using DCA and/or XLD as their primary plating medium, despite the lack of published evidence of their relative efficacy.
Our present study shows similar isolation rates for DCA and XLD, with large numbers of salmonellae growing on DCA. However, 147 extra colonies were selected from DCA, with double the number of API tests, illustrating the less specific colonial appearances of Salmonella spp on this medium.
“The advantage of direct plating is that results could be available 24 hours before results from the enrichment method”
It had been hoped that, by improving specificity, ABC would solve many of the problems associated with the more traditional media. Indeed, specificity was much improved, but isolation rates were so adversely affected by heavy growths of CF that this medium could not be recommended for direct isolation from faeces.
MLCB was first described in 19681 and has been used principally in food, environmental, and veterinary microbiology. It has not been used widely in human investigations because of its inhibitory effect on S typhi and S paratyphi A, although it does not inhibit most salmonellae, and lactose fermenting salmonellae will give typical colonial appearances.
CF were fewer on MLCB, but in those cases where CF contamination was heavy, the recognition of typical colonies did not appear to be impaired to the extent that isolation rates were reduced, and the yield from direct plating was 18.18% better than that seen for XLD; that is, 65% of possible salmonellae were detected.
The combination of XLD and MLCB resulted in a direct isolation rate of 75% of possible salmonellae, which compares favourably with previous studies, where direct isolation rates of 59% and 63.5% were reported.4,5
This is now the third study we are aware of that has demonstrated that selenite enrichment does not fail to isolate salmonellae detected by direct plating.4,5 Therefore, it seems that the only reason to continue direct plating for Salmonella spp would be a need for slightly earlier detection—for example, in outbreak situations. In these circumstances, MLCB would be the optimal single medium. A combination of MLCB and XLD could recover a further 10% of possible salmonellae at 24 hours, but the extra costs of a two plate combination might be difficult to justify.
Take home messages .
Selenite enrichment is the best method for isolating salmonellae; however, where the earlier results of direct plating may be advantageous, mannitol lysine crystal violet brilliant green (MLCB) and xylose lysine desoxycholate (XLD) provide the optimal combination
For non-typhi salmonellae, MLCB is the best single direct plating medium
For routine diagnostic work, XLD is most effective
Traditionally, it has been thought necessary to examine most diarrhoeal samples for both Salmonella spp (including S typih and S paratyphi) and Shigella spp by direct plating. DCA and XLD are examples of such multipurpose media.
However, enteric fever is not primarily a diarrhoeal illness and is now rare in the UK.7 Similarly, infection with Shigella spp has declined greatly over the past decade.8 Therefore, it may be appropriate to consider the use of more targeted techniques for the isolation of these organisms in specifically indicated circumstances.
Abbreviations
ABC, α-β chromogenic medium
CF, competing flora
DCA, desoxycholate citrate
MLCB, mannitol lysine crystal violet brilliant green
MRD, maximal recovery peptone saline diluent
XLD, xylose lysine desoxycholate
Footnotes
The authors present this study on behalf of the PHLS (Midlands) Bacterial Methods Evaluation Group.
REFERENCES
- 1.Inoue T, Takagi S, Ohnishi A, et al. Foodborne disease salmonella isolation medium (MLCB). Japanese Journal of Veterinary Science 1968;30(suppl):26. [Google Scholar]
- 2.Perry JD, Ford M, Taylor J, et al. ABC medium, a new chromogenic agar for selective isolation of Salmonella spp. J Clin Microbiol 1999;37:766–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nye KJ, Turner T, Coleman DJ, et al. A comparison of the isolation rates of Salmonella and thermophilic Campylobacter species after direct inoculation of media with a dilute faecal suspension and undiluted material. J Med Microbiol 2001;50:659–62. [DOI] [PubMed] [Google Scholar]
- 4.Forward KR, Rainnie BJ. Use of selenite enrichment broth for the detection of salmonellae from stool: a report of one year experience at a provincial public health laboratory. Diagn Microbiol Infect Dis 1997;29:215–17. [DOI] [PubMed] [Google Scholar]
- 5.Kelly S, Cormican M, Parke L, et al. Cost effective methods for isolation of Salmonella enterica in the clinical laboratory. J Clin Microbiol 1999;37:3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Persaud CA, Eykyn SJ. Stool culture—are you getting value for money? J Clin Pathol 1994;47:790–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anon. Salmonella typhi and Salmonella paratyphi laboratory reports. Cases only, England and Wales, 1980–2000. Laboratory reports to CDSC. London: PHLS Laboratory of Enteric Pathogens, March 2001.
- 8.Anon. Shigella laboratory reports, England and Wales, faecal isolates 1986–2000. Laboratory reports to CDSC. London: PHLS Laboratory of Enteric Pathogens, March 2001.