Abstract
Aim: In Okinawa, a subtropical island located between the East China Sea and Pacific Ocean, 2000 km south of mainland Japan, the incidence of oral squamous cell carcinoma is 1.5 times higher than that seen in mainland Japan, and a large number of these patients have been reported to be infected with the Epstein-Barr virus (EBV). This study aimed to gain a better understanding of the pathogenesis of this malignancy in this area by carrying out genomic analysis of EBV.
Methods: Fifty four patients with oral squamous cell carcinoma reported from 1997 to 1999 in Okinawa were compared with 21 and 20 patients from Kitakyushu and Kumamoto in Kyushu, mainland Japan, respectively. Diagnosis was confirmed by conventional histological examination of paraffin wax sections. EBV was detected by non-isotopic in situ hybridisation (NISH) and the polymerase chain reaction (PCR) (Bam HI-F, EBV nuclear antigen 2 (EBNA2), and latent membrane protein 1 (LMP-1) regions). Sequence analysis of the PCR products was also carried out.
Results: In Okinawa, 25 patients were found to be infected with EBV type A by analysing the 3′ sequence divergence of the EBNA2 genes. Six patients were positive for EBV type B, and eight for both type A and B. Therefore, type A virus infection was demonstrated in 33 of 54 patients, and type B in 14 of 54. In total, 39 of 54 patients were infected with EBV. However, the “f” variant was shown in only one patient, who was also infected with type A virus. In contrast, 97.0% of EBV type A infected patients showed a 30 bp deletion of the LMP-1 gene, but those infected with EBV type B did not. Sequence analysis of the type A virus EBNA2 gene revealed slight variations of the sequence (mutations)—48991G→T and 48998C→A—in 18 of 33 cases compared with the B95-8 strain, and in 14 cases, in addition to these, a further mutation of 48917T→C was demonstrated; in the single remaining case, only one mutation at 49137A→G was detected. The mutations at 48991 (G→T), and 49137 (A→G) are associated with amino acid changes Arg→Met and Thr→Ala, respectively. In contrast, no mutation was seen in the EBNA2 DNA from the 14 cases of type B virus when compared with that of the Jijoye strain. In Kitakyushu and Kumamoto, only 10 of 41 patients (six in Kitakyushu and four in Kumamoto) were infected with EBV. Among them, nine patients were infected with type A virus, and only one patient from Kitakyushu was infected with type B virus. The 48991G→T and 48998C→A mutations of the EBNA2 region were demonstrated in type A virus, but the 48917T→C and 49137A→G mutations were not when compared with the B95-8 strain. In the case of type B virus no mutation was noted. A 30 bp deletion was found in these nine cases of type A, but not in type B. The sequence analysis of EBV type A in Okinawa, Kitakyushu, and Kumamoto showed slight variations when compared with B95–8, but EBV type B LMP-1 did not when compared with the Jijoye strains.
Conclusion: In Okinawa, EBV infection was frequently demonstrated in oral squamous cell carcinoma (p < 0.001). However, in mainland Japan there was no significant correlation between EBV and oral squamous cell carcinoma. In Okinawa, EBV type B infection is approximately 10 times more common than in the mainland. However, in these areas—Okinawa, Kitakyushu, and Kumamoto—the frequency of the “f ” variant was very low, whereas a high incidence of a 30 bp deletion of LMP-1 was noted. The number of EBV (including type A and/or B) infected oral squamous cell carcinomas in Okinawa was about three times higher than that seen in the mainland, although the frequency of oral squamous carcinoma was only 1.5 times higher than that seen in the mainland. A high prevalence of type B virus infection and slight differences in the EBNA2 gene sequence in the type A virus might influence the frequency of this carcinoma in Okinawa.
Keywords: Epstein-Barr virus subtypes, Epstein-Barr virus nuclear antigen 2, latent membrane protein 1, oral squamous cell carcinomas
Epstein-Barr virus (EBV) causes widespread infection; it is known to be the causative agent of infectious mononucleosis, and is associated with Burkitt's lymphoma, nasopharyngeal carcinoma, peripheral T cell lymphoma, and other cancers.1–4 Recently, 7–10% of gastric carcinomas were also reported to be infected with EBV.5–7 Furthermore, there are two subtypes of EBV, A (type 1) and B (type 2), which differ in their ability to transform B cells (a process known as immortalisation).8, 9 EBV nuclear antigen 2 (EBNA2) function is associated with this phenomenon.10–14 Rickinson et al reported that the type B virus is much less efficient at immortalisation of B cells than the type A virus.15 This may be one reason why the type B virus had long remained undiscovered. EBV type A virus is predominant in developed countries.16 The type B virus was recently discovered in patients with Burkitt's lymphoma in central Africa and New Guinea, and in one study was found in 40% of patients with Burkitt's lymphoma.17 Type B virus differs in the BamHI YH region encoding EBNA2,17, 18 and because of its restricted geographical distribution a close association with Burkitt's lymphoma was suggested. However, recent investigations have revealed that the type B virus is also frequently found in normal populations in the USA, and not only in the endemic area of Burkitt's lymphoma.19 Sixbey et al reported type B virus infection in 40% of healthy individuals in the USA,20 and Scully et al reported that the occurrence of B type virus infection in human immunodeficiency virus positive subjects was higher than that seen in the general community.21
In mainland Japan, type A virus has been reported to be predominant. Kunimoto et al reported that 37 of 79 cases of tonsillitis contained type A virus, and only one type B.22 Tomita and colleagues23 reported that type B virus was detected in one case in 37 cases of malignant lymphoma of the sinonasal region, compared with 22 of 37 cases with type A virus infection. Furthermore, Sidagis et al reported 73 cases of EBV related gastric carcinoma in mainland Japan, of which 51 cases were infected with type A virus, and only four with type B.24
“In mainland Japan, type A virus has been reported to be predominant”
In Okinawa, a subtropical island located between the East China Sea and Pacific Ocean, 2000 km south of mainland Japan, the incidence of oral squamous cell carcinoma is 1.5 times higher than that seen in mainland Japan.25 We recently reported that 46 of 60 cases of oral squamous cell carcinoma were positive for EBV by the polymerase chain reaction (PCR).26 In that paper we reported our molecular epidemiological analysis of EBV subtypes in oral squamous cell carcinoma by determining the 3′ sequence divergence of the EBV EBNA2 gene by PCR. The 30 bp deletion of the cytoplasmic C-terminal domain of the latent membrane protein 1 (LMP-1) gene and the BamHI “f” variant were also analysed.
MATERIALS AND METHODS
Tissue samples and cell lines
Fresh samples of oral squamous cell carcinoma (n = 54) from Okinawa (table 1) were obtained from the department of oral surgery, Ryukyu University Hospital. These were all patients with squamous cell carcinoma coming to surgery from April 1997 to March 1999. The tumours were grouped into well, moderately, and poorly differentiated types according to the World Health Organisation classification.27 Most of the patients in our study were farmers, fishermen, or government employees. There were no miners or heavy industrial workers. Forty two patients were heavy smokers and eight were heavy drinkers. We compared these patients with those from Kitakyushu city and Kumamoto city in Kyushu in mainland Japan. The Okinawa prefecture (population, 1.27 million) is in southern Japan and is subtropical. Kitakyushu city (population, 1.01 million) and Kumamoto city (population, 0.64 million) are located in Kyushu in mainland Japan. Samples of oral squamous cell carcinoma from the department of oral surgery, University of Occupational and Environmental Health in Kitakyushu (n = 21) and Kumamoto University in Kumamoto (n = 20) in the same period from 1997 to 1999 were examined (tables 2,3). The cell lines B95-8 and Raji served as positive controls for type A, and the cell line Jijoye was used for type B.
Table 1.
List of patients with oral squamous cell carcinoma in Okinawa, histological differentiation, and detection of Epstein-Barr virus (EBV)
| Patient | Age (years) and sex | Location of the tumour | Stage and differentiation | EBV type | NISH | LMP | EBNA | 30 bp | ||
| 1 | 42 | M | O | IV | W | − | − | − | − | |
| 2 | 42 | M | M | I | W | − | − | − | − | |
| 3 | 48 | M | M | III | W | A+B | + | + | + | + |
| 4 | 70 | M | M | IV | W | A+B | + | + | + | + |
| 5 | 58 | M | F | III | W | A+B | + | + | + | − |
| 6 | 69 | M | T | IV | W | A+B | + | + | + | + |
| 7 | 57 | M | T | I | W | A | + | + | + | + |
| 8 | 67 | M | O | IV | W | A | + | + | + | + |
| 9 | 60 | M | T | IV | W | − | − | − | − | |
| 10 | 59 | M | M | I | W | − | − | − | − | |
| 11 | 77 | M | O | III | W | A | + | + | + | + |
| 12 | 43 | M | T | III | W | A | + | + | + | − |
| 13 | 75 | M | T | IV | W | A | + | + | + | + |
| 14 | 66 | M | F | II | W | A | + | + | + | + |
| 15 | 53 | M | B | I | W | A | + | + | + | + |
| 16 | 80 | F | T | I | W | A | + | + | + | + |
| 17 | 39 | M | L | II | W | − | − | − | − | |
| 18 | 70 | M | T | II | W | A | + | + | + | + |
| 19 | 47 | F | T | II | W | A | + | + | + | + |
| 20 | 60 | M | T | III | W | A | + | + | + | + |
| 21 | 78 | F | M | III | W | A+B | + | + | + | + |
| 22 | 70 | M | T | I | W | B | + | + | + | − |
| 23 | 57 | M | T | II | W | − | − | − | − | |
| 24 | 79 | M | T | II | W | A | + | + | + | + |
| 25 | 78 | M | T | III | W | − | − | − | − | |
| 26 | 65 | M | O | III | W | A | + | + | + | + |
| 27 | 80 | M | F | II | W | B | + | + | + | − |
| 28 | 64 | M | T | III | W | A | + | + | + | + |
| 29 | 86 | M | M | IV | M | A | + | + | + | + |
| 30 | 70 | M | T | O | M | A | + | + | + | + |
| 31 | 69 | M | T | I | M | − | − | − | − | |
| 32 | 55 | M | B | I | M | A | + | + | + | + |
| 33 | 60 | M | L | IV | M | A | + | + | + | + |
| 34 | 71 | M | B | II | M | A | + | + | + | + |
| 35 | 59 | M | T | II | M | A | + | + | + | + |
| 36 | 50 | M | T | II | M | A | + | + | + | + |
| 37 | 39 | M | B | I | M | − | − | − | − | |
| 38 | 70 | M | T | I | M | − | − | − | − | |
| 39 | 78 | M | T | IV | M | − | − | − | − | |
| 40 | 73 | M | F | III | M | A | + | + | + | + |
| 41 | 60 | M | O | II | M | B | + | + | + | − |
| 42 | 80 | M | O | I | M | B | + | + | + | − |
| 43 | 68 | M | L | II | M | B | + | + | + | − |
| 44 | 65 | M | F | III | M | A+B | + | + | + | + |
| 45 | 60 | M | O | III | M | A+B | + | + | + | + |
| 46 | 77 | M | F | IV | M | A+B | + | + | + | + |
| 47 | 47 | F | B | II | M | − | − | − | − | |
| 48 | 67 | M | F | III | M | B | + | + | + | − |
| 49 | 67 | M | O | IV | M | − | − | − | − | |
| 50 | 48 | M | L | II | M | − | − | − | − | |
| 51 | 80 | M | F | III | P | A | + | + | + | + |
| 52 | 57 | M | O | III | P | A | + | + | + | + |
| 53 | 63 | M | O | II | P | A | + | + | + | + |
| 54 | 61 | M | L | IV | P | − | − | − | − | |
Location: B, buccal mucosa; F, mouth floor; L, lower gum; M, maxilla; O, oropharynx; T, tongue.
Differentiation, degree of the histological differentiation of the tumour: M, moderately differentiated; P, poorly differentiated; W, well differentiated. NISH, non-isotopic in situ hybridisation of EBV encoded RNA 1; LMP, immunohistochemical demonstration of latent membrane protein 1 antigen; EBNA, immunohistochemical demonstration of the EBV nuclear antigen 2; 30 bp, 30 bp deletion of LMP-1.
Table 2.
List of patients with oral squamous cell carcinoma in Kitakyushu, histological differentiation and detection of Epstein-Barr virus (EBV)
| Patient | Age (years) and sex | Location of the tumour | Diff | EBV type | NISH | LMP | EBNA | 30 bp | |
| 1 | 52 | M | T | W | A | + | + | + | + |
| 2 | 73 | M | M | W | A | + | + | + | + |
| 3 | 57 | M | T | W | − | − | − | − | − |
| 4 | 48 | M | T | W | − | − | − | − | − |
| 5 | 52 | F | B | W | − | − | − | − | − |
| 6 | 58 | F | T | W | − | − | − | − | − |
| 7 | 62 | M | F | M | − | − | − | − | − |
| 8 | 54 | F | M | M | A | + | + | + | + |
| 9 | 52 | M | T | M | A | + | + | + | + |
| 10 | 55 | M | T | M | − | − | − | − | − |
| 11 | 73 | F | T | M | A | + | + | + | + |
| 12 | 83 | M | M | M | − | − | − | − | − |
| 13 | 62 | M | T | M | B | + | + | + | − |
| 14 | 52 | M | F | M | − | − | − | − | − |
| 15 | 74 | M | T | M | − | − | − | − | − |
| 16 | 90 | F | U | M | − | − | − | − | − |
| 17 | 83 | M | M | M | − | − | − | − | − |
| 18 | 43 | M | T | M | − | − | − | − | − |
| 19 | 53 | M | F | M | − | − | − | − | − |
| 20 | 70 | M | B | M | − | − | − | − | − |
| 21 | 45 | M | F | P | − | − | − | − | − |
Location: B, buccal mucosa; F, mouth floor; L, lower gum; M, maxilla; O, oropharynx; T, tongue. Diff, degree of the histological differentiation of the tumour: M, moderately differentiated; P, poorly differentiated; W, well differentiated. NISH, non-isotopic in situ hybridisation of EBV encoded RNA 1; LMP, immunohistochemical demonstration of latent membrane protein 1 antigen; EBNA, immunohistochemical demonstration of the EBV nuclear antigen 2; 30 bp, 30 bp deletion of LMP-1.
Table 3.
List of patients with oral squamous cell carcinoma in Kumamoto, histological differentiation and detection of Epstein-Barr virus (EBV)
| Patient | Age (years) and sex | Diff | EBV type | NISH | LMP | EBNA | 30 bp | |
| 1 | 72 | M | W | − | − | − | − | − |
| 2 | 66 | F | W | − | − | − | − | − |
| 3 | 70 | M | W | − | − | − | − | − |
| 4 | 74 | F | W | − | − | − | − | − |
| 5 | 60 | M | W | A | + | + | + | + |
| 6 | 56 | M | W | A | + | + | + | + |
| 7 | 87 | F | W | A | + | + | + | + |
| 8 | 68 | M | W | − | − | − | − | − |
| 9 | 78 | F | W | − | − | − | − | − |
| 10 | 71 | M | M | − | − | − | − | − |
| 11 | 74 | F | M | − | − | − | − | − |
| 12 | 79 | F | M | − | − | − | − | − |
| 13 | 77 | M | M | − | − | − | − | − |
| 14 | 72 | M | M | − | − | − | − | − |
| 15 | 56 | M | M | − | − | − | − | − |
| 16 | 50 | M | M | − | − | − | − | − |
| 17 | 84 | F | M | − | − | − | − | − |
| 18 | 59 | M | M | − | − | − | − | − |
| 19 | 63 | M | M | − | − | − | − | − |
| 20 | 55 | F | P | A | + | + | + | + |
Diff, degree of the histological differentiation of the tumour: M, moderately differentiated; P, poorly differentiated; W, well differentiated. NISH, non-isotopic in situ hybridisation of EBV encoded RNA 1; LMP, immunohistochemical demonstration of latent membrane protein 1 antigen; EBNA, immunohistochemical demonstration of the EBV nuclear antigen 2; 30 bp, 30 bp deletion of LMP-1.
Morphological investigation
Samples were fixed in 10% phosphate buffered formalin, routinely processed in paraffin wax, and sectioned at 4 μm. Haematoxylin and eosin staining were performed on 4 μm dewaxed sections. For the immunohistochemical staining, dewaxed sections were pretreated with 3% H2O2 for 20 minutes, washed, and blocked with a non-immune goat serum for 30 minutes. Antibodies to EBV EBNA2 (clone PE2) and EBV LMP-1 (clone CS1-4) were obtained from Dako (Dako A/S, Glostrup, Denmark). Slides were then incubated with diluted (1/100) primary antibodies for 25 minutes, washed three times, and incubated with biotinylated second antibody with avidin and biotinylated horseradish peroxidase complex (Dako, Kyoto, Japan). DAB (3′,3-diaminobenzidine; Dako) was used as a chromogen.
Non-isotopic in situ hybridisation (NISH) for EBV EBER1
NISH for EBV was performed using the Dako DNA ISH detection kit for EBV encoded RNA 1 (EBER1) on dewaxed 4 μm sections. NISH was carried out according to the manufacturer's instructions.
Sample DNA preparation
DNA was extracted from fresh specimens of oral squamous cell carcinoma after treatment with proteinase K (1 μg/ml) freshly prepared in 10μM Tris/HCl (pH 8.0), 10μM EDTA, and 1% sodium dodecyl sulfate at 37°C overnight. The DNA was then purified by repeated extraction using phenol/chloroform/isoamyl alcohol (49/49/2), followed by ethanol precipitation.
The 110 bp β globin gene was detected in all samples according to the method of Saiki et al using their primers (PCO3 and PCO4).28
Analysis of the EBV subtype
Analysis of the EBV subtype was carried out by determining the 3′sequence divergence of the EBNA2 gene by means of PCR, as described by Borisch et al.1 The EBNA2 region was amplified according to the methods of Borisch and colleagues1 and Wu et al,29 using primers gen 1 and gen 2 (table 4). A 1 μg sample of genomic DNA was used for the PCR. The reaction mixture contained 10mM Tris/HCl, pH 8.3, 50mM KCl, 1.5mM MgCl2, 0.4mM of each dNTP, 200μM of each primer, and 2.4 units of Taq DNA polymerase (Cetus-Takara, Otsu, Japan). After the first denaturation at 99°C for five minutes, PCR was carried out for a total of 35 cycles: denaturation at 96°C for 30 seconds, annealing at 55°C for one minute, and extension at 72°C for one minute. After the PCR reaction, the PCR product was mixed with EBNA2A and EBNA2B specific primers for nested PCR to separate the subtypes A and B.
Table 4.
List of primers and probes29
| Primers and probes | Nucleotide sequence positions |
| EBNA2 | |
| gen 1: 5′ AGGGATGCCTGGACACAA3′ | 44410–48827 |
| gen 2: 5′ GTGCTGGTGCTGCTGGTGG3′ | 49410–49391 |
| EBNA2 subtype 1 | |
| A1: 5′ TCTTGATAGGGATCCGCTAGGATA 3′ | 48839–48862 |
| A2: 5′ ACCGTGGTTCTGGACTATCTGGATC 3′ | 49335–49311 |
| Probe: 5′ CTCTGTCACAACCGAGGCTTACC 3′ | 49048–49070 |
| EBNA2 subtype 21 | |
| B1: 5′ CATGGTAGCCTTAGGACATA 3′ | |
| B2: 5′ AGACTTAGTTGATGCCCTAG 3′ | |
| Probe: 5′ AGGCCTACTCTTCCTCAACCCAG 3′ | |
| BamHI-F | |
| F1: 5′ GGAACTGAGCCAGTAGGATA 3′ | 149–173 |
| F2: 5′ AATGTTCTGCAGGGTAACGG 3′ | 980–960 |
| LMP-130 | |
| F1: 5′ GTGGGGGTCGTCATCATCTC 3′ | 168190–168209 |
| R2: 5′ CGGAAGAGGTTGAAAACAAA 3′ | 168350–168331 |
| Probe: 5′ GGCGGGCCCTGGTCACCTCC 3′ | 168311–168330 |
EBNA2, Epstein-Barr virus nuclear antigen 2; LMP-1, latent membrane protein 1.
Sequencing of the EBNA2 type A and B genes
The PCR products of EBNA2, type A and B, were extracted from the agarose gel. The extracted DNA was cloned into a T-vector that was prepared from Bluescript (Stratagene, La Jolla, California, USA) according to the method of Marchuk et al.31 The Bluescript plasmid is digested with EcoRV, and incubated with Taq (1 unit/μg plasmid/20 μl volume) using standard buffer conditions (10mM Tris/HCl, pH 8.3, 50mM KCl, 1.5mM MgCl2, and 200 μg/ml bovine serum albumin) in the presence of 2 dTTP for two hours at 70°C. The sequence analysis was carried out using a Hitachi SQ 5500 DNA sequencer (Hitachi, Tokyo, Japan).
BamHI “f” variant analysis
The “f” variant analysis was based on the presence of an extra enzyme site at the BamHI-F fragment region of EBV DNA. According to the method of Wu et al,29 a PCR technique was used to detect the EBV BamHI “f” variant. PCR was carried out using a primer pair designed from the BamHI-F region (BamHI-F1 and BamHI-F2; table 4). The PCR conditions were the same as those described in the above section on the analysis of EBV subtype. After the PCR, 10 μl aliquots of the PCR products were digested by BamHI (10 U/μl) restriction enzyme (BRL, Grand Island, New York, USA) at 37°C for two hours. The products were analysed from the extra BamHI-F site by electrophoresis in a 1.5% agarose gel stained with ethidium bromide.
Analysis of the C-terminal region of LMP-1 by PCR
The C-terminal cytoplasmic regions of the EBV type A and B LMP-1 genes were amplified using the primer pair F1/R2 (table 4).30, 32–34 The PCR amplification was carried out using a 1 μg sample of genomic DNA. The reaction mixture contained 200μM of primer pair, 10mM Tris/HCl, pH 8.3, 50mM KCl, 1.5mM MgCl2, 0.01% gelatin, 0.4mM of each dNTP, and 2.4 units of Taq DNA polymerase. The reaction mixture was heated to 94°C for five minutes to denature the DNA–DNA hybrid. Each amplification cycle comprised one minute of denaturation at 94°C, one minute of annealing at 61°C, and one minute of extension at 72°C. The amplification was run for 50 cycles. The PCR products were subjected to 2% agarose gel electrophoresis. The DNA was extracted from the agarose gel and cloned into the T-vector. Sequence analysis was carried out using the Hitachi SQ5500 DNA sequencer (Hitachi, Tokyo, Japan).
Negative controls for PCR analysis
To prevent false positive results, the sample preparation and the PCR were carried out by three authors independently (MH, KT, JM). Furthermore, distilled water was used as a negative control and no positive reaction was obtained.
Statistical analysis
Statistical analysis was performed using the Mantel-Haentzel χ2 method (Statistical Analysis System; SAS Institute Inc, Cary, North Carolina, USA). Significance was set at p < 0.05.
RESULTS
Morphological investigations, immunohistochemistry, and NISH
Twenty eight of the 54 Okinawan cases were well differentiated carcinomas. Twenty two and four of the 54 cases were moderately and poorly differentiated types, respectively (table 1). The mean (SD) age of the patients was 59 (2.8) years; 50 were men and four were women. One case was stage 0, 11 were stage I, and 42 were stage II–IV. Immunohistochemically, 36 of the 54 cases were positive for the LMP-1 (fig 1) and EBNA2 antigens. By means of NISH, EBV encoded RNA 1 (EBER1) was demonstrated in cancer cells (fig 2) from 36 of the 54 patients, all of whom were also positive for the LMP-1 and EBNA2 antigens immunohistochemically (table 1). There was no significant correlation between EBV infection and the histological differentiation of the carcinoma.
Figure 1.
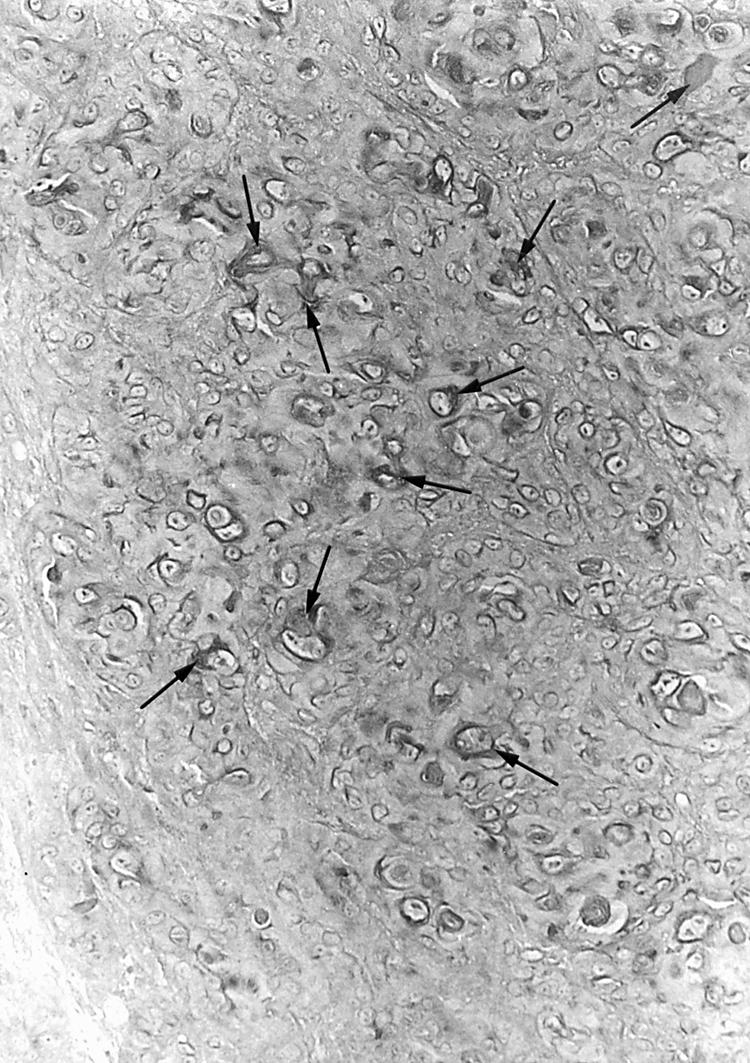
Immunohistochemical demonstration of the latent membrane protein 1 antigen. A large number of cancer cells were positive (arrows). Original magnification, ×200.
Figure 2.
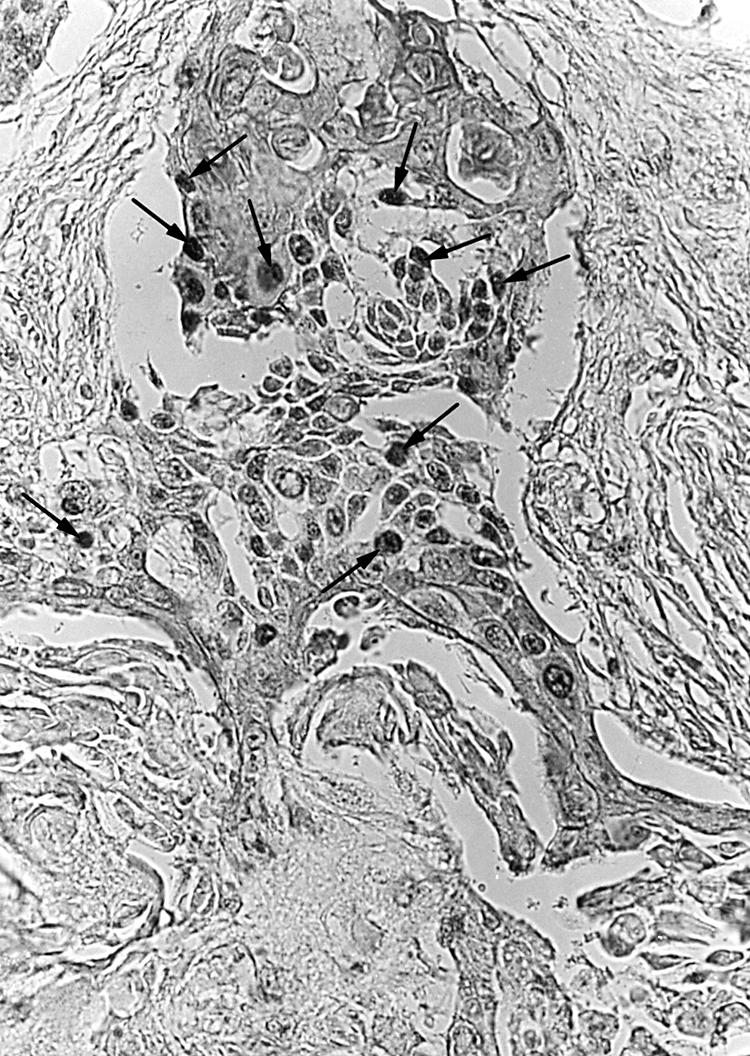
Demonstration of Epstein-Barr virus encoded RNA 1 in squamous cell carcinoma cells by non-isotopic in situ hybridisation. Numerous positive signals were seen (arrows). Original magnification, ×200.
In Kitakyushu and Kumamoto, six of 21 and nine of 20 cases were well differentiated carcinomas, respectively (tables 2,3). The mean (SD) age of the patients in Kitakyushu and Kumamoto was 61.5 (13.4) and 68.6 (10.2), respectively. All cases were stage I or II. Five and eight patients in Kitakyushu and Kumamoto, respectively, were women. Immunohistochemically, six patients in Kitakyushu and four in Kumamoto were positive for the LMP-1 and EBNA2 antigens, and these 10 patients were also positive for EBER1 by means of the NISH assay.
Detection of EBV EBNA2 DNA and EBV subtype analysis
Thirty nine of the 54 Okinawan patients were positive for EBV EBNA2 DNA using the PCR method (fig 3A; table 1). The analysis of EBV subtype was performed by determining the 3′ sequence divergence of EBNA2 using the designated PCR primers. The presence of a 496 bp product represents the type A strain, and a 150 bp the type B strain. EBV DNA samples from the B95-8 and Jijoye cell lines were used as controls for type A and B, respectively. Twenty five patients were positive for the type A strain alone, and six for type B. Eight patients were positive for both types A and B (table 1). Thus, in total 39 of the 54 patients were infected with EBV, of which 33 and 14 patients were positive for the type A and B viruses, respectively. EBV infection in the oral squamous cell carcinoma was frequently demonstrated (p < 0.001). However, no significant correlation was found between EBV type B infection and oral squamous cell carcinoma.
Figure 3.
Demonstration of PCR products of Epstein-Barr virus (EBV) type A and B EBV nuclear antigen 2. Bands of 496 bp from EBV type A (*) and 150 bp from EBV type B (arrowhead) were demonstrated in patients from (A) Okinawa, (B) Kitakyushu, and (C) Kumamoto.
In the patients from Kitakyushu (n = 21) and Kumamoto (n = 20), 10 (six from Kitakyushu and four from Kumamoto) were positive for EBV EBNA2 DNA by PCR (fig 3B,C; tables 2,3). Nine patients were infected with type A, and only one patient (from Kitakyushu) was infected with type B, as assessed by analysis of the EBNA2 region.
In the mainland, there was no significant correlation between the detection of EBV DNA and oral squamous cell carcinoma.
Sequence analysis of EBNA2 DNA
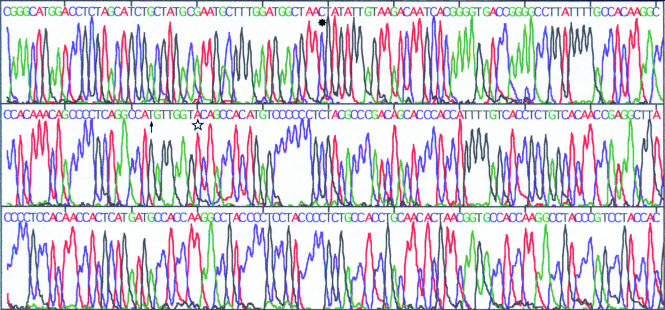
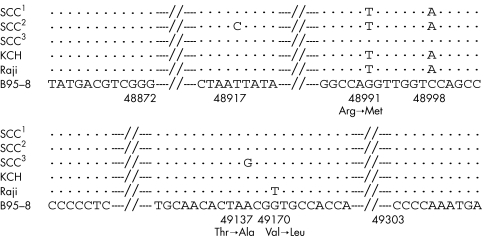
The 496 bp products of the 33 cases of type A virus EBNA2 DNA and the 150 bp products of the 14 cases of type B virus EBNA2 DNA in the 41 EBV infected patients from Okinawa were analysed by a DNA sequencer. EBV DNA from the B95-8,35, 36 Raji, and Jijoye18 cell lines was used as control DNA. In 18 of the 33 type A virus positive cases, two mutations (sequence variations) were found (48991G→T and 48998C→A) when compared with B95-8 DNA (fig 4). In 14 patients, an additional, third mutation (48917T→C) was demonstrated (fig 5). In the remaining one type A positive patient, only one mutation (49137A→G) was found. The mutations at 48991 (G→T) and at 49137 (A→G) were associated with amino acid changes—Arg→Met and Thr→Ala—respectively, but the mutations at 48917 and 48998 were not associated with amino acid changes (base exchanges only). The 48991G→T and 48998C→A mutations were also found in the Raji EBNA2 region. Furthermore, in Raji EBNA2, another mutation at 49140 was also seen when compared with the B95-8 strain, and this mutation was associated with the amino acid change Val→Leu (fig 6).
Figure 4.
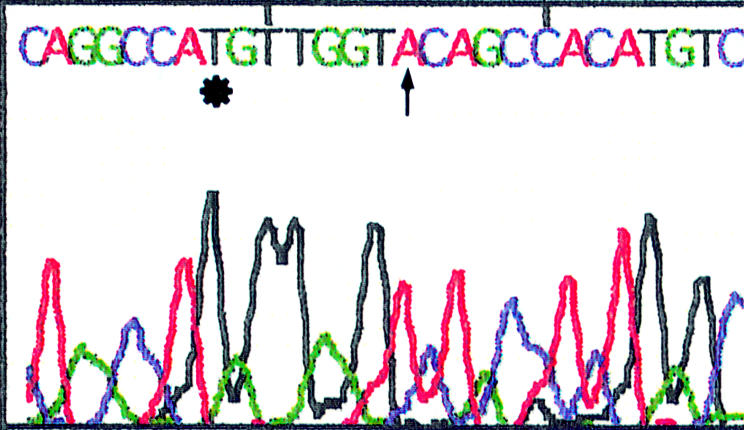
Sequence analysis of the PCR product (Epstein-Barr virus (EBV) type A nuclear antigen 2). Changes of T→G at position 48991 (*) and A→C at position 48998 (↑) were demonstrated in 18 of the patients when compared with the B95-8 strain.
Figure 5.
Sequence analysis of the PCR product (Epstein-Barr virus (EBV) type A nuclear antigen 2). There were three mutations: at 48917 (T→C; *), at 48991 (G→T; ↑), and at 48998 (C→A; ) when compared with the B95-8 strain.
Figure 6.
Schematic diagram of the Epstein-Barr virus (EBV) nuclear region 2 (EBNA2) of EBV type A virus. SCC1: the sequence of the type A virus EBNA2 region seen in 18 Okinawan patients. SCC2: the sequence of the type A virus EBNA2 region seen in 14 Okinawan patients. SCC3: the sequence of the type A virus EBNA2 region seen in the remaining Okinawan patient. KCH: the sequence of the type A virus EBNA2 region of the Kumamoto and Kitakyushu patients. Raji: the sequence of the EBNA2 region of the Raji strain. B95-8: the sequence of EBNA2 region of the B95-8 strain.
In contrast, in the 14 cases of type B virus, no mutation was demonstrated in the 150 bp PCR products when compared with Jijoye EBNA2 DNA (data not shown).
In nine cases of type A virus from Kitakyushu and Kumamoto, the 48991G→T and 48917C→A mutations were also noted, but the 48137A→G mutation was not. Furthermore, in the one type B virus case from Kitakyushu, no mutation was seen.
BamHI “f” variant analysis
The “f” variant analysis was based on the presence of an extra BamHI restriction enzyme site in the F fragment region of EBV DNA. The PCR amplified product of the prototype BamHI-F fragment is 831 bp in size. However, after cleavage with the BamHI restriction enzyme, two fragments of 702 bp and 129 bp in size were yielded for the “f” variant. Only one EBV type A infected Okinawan case (case 6 in table 5) was “f” variant. All other cases (Okinawa, Kitakyushu, and Kumamoto cases) belonged to the prototype BamHI-F type.
Table 5.
Numbers of Epstein-Barr virus (EBV) subtypes and “f” variants in Okinawa
| EBV positive cases | EBV subtypes | “f” variant | ||
| 39 | A | 25 | F | 38 |
| B | 6 | f | 1 | |
| A+B | 8 | |||
Analysis of the LMP-1 C-terminal region
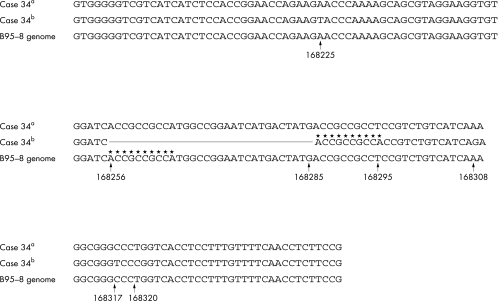
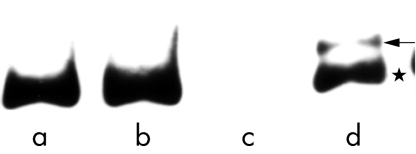
The 30 bp deletion in the LMP-1 gene resulted in a 131 bp PCR (as opposed to 161 bp in the wild-type gene) in the EBV type A and B LMP-1 genes. In the 33 EBV type A positive Okinawan patients, 32 had the 30 bp deletion of the LMP-1 gene (position, 168256–85; table 1; fig 7), whereas the 30 bp deletion was not demonstrated in patients infected with type B virus. In the patients with the 30 bp deletion, the T at position 168295 was changed to A. Then the repeated sequence ACCGCCGCCA (at 168286–95), which was the same as the sequence at 168256–65, was noted and a 30 bp deletion at 168256–85 was demonstrated. Furthermore, additional mutations at 168225, 168308, 168317, and 168320 were seen when compared with the EBV DNA sequence from the B95-8 cell line. The mutations at 168225, 168317, and 168320 resulted in amino acid changes, Ser→Thr, Gly→Asp, and Glu→Arg, respectively. In contrast, the sequence of the 161 bp product was the same as that from the B95-8 cell line. Interestingly, both the 161 bp and the 131 bp products of the type A virus were demonstated in patients 34 from Okinawa (figs 7, 8). Although it is possible that this tumour favoured the different EBV type A infection, the virus could represent changes over a long period of time. In contrast, nine EBV type A positive patients from Kitakyushu and Kumamoto harboured the 30 bp deletion of the LMP-1 gene, although the one type B infected case did not (tables 2,3). However, the PCR products of both the type A and type B virus LMP-1 genes from Kitakyushu and Kumamoto showed no additional sequence variations when compared with the B95-8 (type A) and Jijoye (type B) strains.
Figure 7.
Schematic diagram of the latent membrane protein 1 (LMP-1) region of Epstein-Barr virus (EBV) type A. Case 34a: the sequence of the 161 bp product of case 34. Case 34b: the sequence of the 131 bp product of case 34. B95-8: the sequence of the B95-8 LMP-1 region. The straight line indicates the 30 bp deletion from position 168256 to 188285; the line of asterisks indicates the repeated sequence.
Figure 8.
Southern blot hybridisation of PCR products after amplification of the latent membrane protein 1 (LMP-1) gene (case 34). Lane a (case 32), a 131 bp product was amplified; lane b (case 33), a 131 bp product was amplified; lane c, negative control (distilled water); lane d (case 34), both 161 (←) and 131 (*) bp products were amplified.
DISCUSSION
It has been reported that the incidence of EBV infection varies geographically. Taiwan, the nearest neighbour of Okinawa, and southern China (which historically had close relations with Okinawa, including some migration) have been reported as high EBV infection areas. In Taiwan, Wu et al reported that the type A virus was detected in 25 of 30 cases of nasopharyngeal carcinoma, and the remaining five were infected with both the type A and type B viruses.29 In southern China (Shanghai, People's Republic of China), Chen et al reported that only one of 16 nasopharyngeal carcinoma biopsies was infected with the type B virus, and the other 15 cases were infected with the type A virus.37
A high frequency of the BamHI “f” variant of EBV was reported from southern China,38, 39 and this “f” variant might be associated with the development or maintenance of nasopharyngeal carcinoma. However, in Okinawa, the incidence of type B virus infection was much higher than that reported from southern China; in our series, 14 of the total 39 EBV infected carcinomas from Okinawa were infected with the type B virus. In addition, the “f” variant form was found in only one case, so that the frequency of “f” variant positive cases also differs greatly from that seen in Taiwan29 and southern China. Again, in contrast to Okinawa, in mainland Japan the incidence of type B virus infection (1.3–5%) was slightly lower than that of Taiwan and southern China.22–24, 40 In our present study, only one of 41 cases from Kitakyushu in the mainland had type B infection. The incidence of type B virus infection in Okinawa (34.1%) was nearer to that seen in Taiwan (16.7%) than to that seen in mainland Japan and southern China, but was still about twice as high as that seen in Taiwan. However, the incidence of the “f” variant in mainland Japan has been reported to be very low (1.4%),24 and is similar to that seen in Okinawa. In our present study, the EBNA2 gene of the type A virus in Okinawa had non-silent mutations (sequence variations) at positions 48991 and 49173. In contrast, the type A virus EBNA2 gene in Kitakyushu and Kumamoto had only the 48991 mutation when compared with the B95-8 strain. The interaction of EBNA2 with C-promoter binding factor 1 or recombination signal binding protein Jκ (RBPJκ),10 is thought to be associated with the tumorigenecity of the EBNA2 gene. These mutations, associated with the amino acid changes seen in the Okinawan strain type A virus EBNA2, might have an influence on the interaction between the EBNA2 protein and RBPJκ. However, Ling and colleagues11 and Henkel and colleagues10 reported that mutant EBNA2, having mutations at amino acids 323 and 324 of the EBNA2 transcript, failed to have such an influence. The present amino acid changes were located at positions 165 and 180, different from those reported by Ling et al,11 and an influence on such an interaction was obscure. LMP-1 has also been reported to be a viral oncogene.12 The transmembrane domains, together with the C-terminal cytoplasmic domain, are required for the maximal activation of NF-κB.41, 42 Furthermore, Hu et al reported a 30 bp deletion and point mutations in the C-terminal cytoplasmic region of the LMP-1 gene in Chinese nasopharyngeal carcinomas,32 and this 30 bp deletion mutant transformed rodent cells more efficiently.43 However, to date there are no in vivo data indicating that 30 bp deletion mutants show altered function when compared with EBV strains without the 30 bp deletion.44–47 In mainland Japan, Mori et al also reported the 30 bp deletion of the LMP-1 gene in 91.7% of EBV related gastric carcinoma and 83.3% of throat washings from healthy adults.48 In our present study, the LMP-1 genes of the type A virus from the mainland all showed the 30 bp deletion and in Okinawa the 30 bp deletion was seen in 97.0% of the EBV type A infected patients. The 30 bp deletion mutants are common in Okinawa, similar to mainland Japan. Furthermore, there were no sequence variations of the LMP-1 genes in the mainland EBV when compared with the B95-8 strain.
“In Okinawa, the incidence of type B virus infection was much higher than that reported from southern China”
As reported previously, human papillomavirus (HPV) infection in oral squamous cell carcinoma, especially well differentiated cases, in Okinawa is higher than that seen in mainland Japan.26 HPV might affect the histological differentiation of squamous cell carcinoma.26 The number of well differentiated cases in Okinawa was higher than that seen in the mainland. However, recently the number of such cases has decreased greatly. In general, the prevalence of smoking in Okinawa has not been particularly high, although most of the patients in our series from Okinawa and the mainland were heavy smokers, and there is no clear correlation between smoking and EBV infection. Furthermore, only eight patients in our present series were heavy drinkers. However, no correlation between alcohol drinking and EBV infection has been demonstrated. Thus, in Okinawa, both EBV and HPV might be associated with the tumorigenecity of the oral mucosa. However, recently HPV infection has been decreasing,49 and the role of EBV in tumorigenicity should be studied further. In Okinawa, the subtropical climate and hygiene conditions (water supply and sewage disposal, etc) might influence the incidence of EBV. The high incidence of type B infection may reflect in part the previous history of the region. More than 100 years ago Okinawa was an independent kingdom, and had close relations with China, Taiwan, and South East tropical Asian countries.
In conclusion, the rate of EBV infection in oral squamous cell carcinomas in Okinawa was about three times higher than that seen in mainland Japan, although the incidence of these carcinomas was only 1.5 times higher in Okinawa than in the mainland. High EBV type B infection and the slight differences in the EBNA2 gene might influence the incidence of oral squamous cell carcinoma in Okinawa.
Take home messages.
In Okinawa, Epstein-Barr virus (EBV) infection was associated with oral squamous cell carcinoma (p < 0.001), although this was not the case in mainland Japan
In Okinawa, EBV type B infection is approximately 10 times more common than in the mainland
In Okinawa and the mainland the frequency of the “f ” variant was very low, whereas a high incidence of a 30 bp deletion of LMP-1 was noted
The number of EBV infected oral squamous cell carcinomas in Okinawa was about three times higher than that seen in the mainland, although the frequency of oral squamous carcinoma was only 1.5 times higher
A high prevalence of type B virus infection and slight differences in the EBV nuclear antigen 2 gene sequence in the type A virus might influence the frequency of this carcinoma in Okinawa
Abbreviations
EBV nuclear antigen 2, EBER1, EBV encoded RNA 1
EBNA2, EBV nuclear antigen 2
EBV, Epstein-Barr virus
HPV, human papillomavirus
LMP-1, latent membrane protein 1
NISH, non-isotopic in situ hybridisation
PCR, polymerase chain reaction
RBPJκ, recombination signal binding protein Jκ
REFERENCES
- 1.Borisch B, Hennig I, Laeng RH, et al. Association of the subtype 2 of the Epstein-Barr virus with T-cell non-Hodgkin's lymphoma of the midline granuloma type. Blood 1993;82:858–64. [PubMed] [Google Scholar]
- 2.Pallesen G, Hamilton-Dutoit SJ, Zhou XG. The association of Epstein-Barr virus (EBV) with T cell lymphoproliferation and Hodgkin's disease. Two new developments in the EBV field. Adv Cancer Res 1993;62:179–239. [DOI] [PubMed] [Google Scholar]
- 3.Weiss LM, Strickler JG, Warnke RA, et al. Epstein-Barr virus DNA in tissue of Hodgkin's disease. Am J Pathol 1987;129:86–91. [PMC free article] [PubMed] [Google Scholar]
- 4.Zur Hausen H, Schulte-Holthausen H, Kein G, et al. EBV DNA in biopsies of Burkitt's lymphoma and anaplastic carcinomas of the nasopharynx. Nature 1970;228:1056–8. [DOI] [PubMed] [Google Scholar]
- 5.Fukayama M, Hayashi Y, Iwasaki Y, et al. Epstein-Barr virus-associated gastric carcinoma and Epstein-Barr virus infection of the stomach. Lab Invest 1994;71:73–81. [PubMed] [Google Scholar]
- 6.Shibata D, Tokunaga M, Uemura Y, et al. Association of Epstein-Barr virus with undifferentiated gastric carcinomas with intense lymphoid infiltration. lymphoepithelioma-like carcinoma. Am J Pathol 1991;139:469–74. [PMC free article] [PubMed] [Google Scholar]
- 7.Tokunaga M, Land CE, Uemura Y, et al. Epstein-Barr virus in gastric carcinoma. Am J Pathol 1993;143:1250–4. [PMC free article] [PubMed] [Google Scholar]
- 8.Liebowitz D, Kieff E. Epstein-Barr virus. In: Roizman B, Whitley RJ, Lopez C, eds. The human herpesviruses. New York: Raven Press, 1993:107–72.
- 9.Skare J, Farley J, Strominger JL, et al. Transformation by Epstein-Barr virus requires DNA sequences in the region of BamHI fragments Y and H. J Virol 1985;55:286–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henkel T, Ling PD, Hayward SD, et al. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein Jκ. Science 1994;265:92–5. [DOI] [PubMed] [Google Scholar]
- 11.Ling PD, Rawlins DR, Hayward SD. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc Natl Acad Sci U S A 1993;90:9237–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang D, Lebowitz D, Kieff E. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 1985;43:831–40. [DOI] [PubMed] [Google Scholar]
- 13.Wang F, Gregory CD, Rowe M, et al. Epstein-Barr virus nuclear antigen 2 specifically induces expression of the B-cell activation antigen CD 23. Proc Natl Acad Sci U S A 1987;84:3452–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimber-Strobl U, Kremmer E, Grässer F, et al. The Epstein-Barr virus nuclear antigen 2 interacts with an EBNA2 responsive cis-element of the terminal protein 1 gene promoter. EMBO J 1993;12:167–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rickinson AB, Young LS, Rowe M. Influence of the Epstein-Barr virus nuclear antigen EBNA 2 on the growth phenotype of virus-transformed B cells. J Virol 1987;61:1310–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Epstein MA, Achong BG, Barr YM. Virus particles in cultured lymphoblasts from Burkitt's lymphoma. Lancet 1964;i:702–3. [DOI] [PubMed] [Google Scholar]
- 17.Zimber U, Adldinger HK, Lenoir GM, et al. Geographical prevalence of two types of Epstein-Barr virus. Virology 1986;154:56–66. [DOI] [PubMed] [Google Scholar]
- 18.Adldinger HK, Delius H, Freese UK, et al. A putative transforming gene of Jijoye virus differs from that of Epstein-Barr virus prototypes. Virology 1985;141:221–34. [DOI] [PubMed] [Google Scholar]
- 19.Young LS, Yao QY, Rooney CM, et al. New type B isolates of Epstein-Barr virus from Burkitt's lymphoma and from normal individuals in endemic areas. J Gen Virol 1987;68:2853–62. [DOI] [PubMed] [Google Scholar]
- 20.Sixbey JW, Shirley P, Chesney PJ, et al. Detection of a second widespread strain of Epstein-Barr virus. Lancet 1989;ii:761–5. [DOI] [PubMed] [Google Scholar]
- 21.Scully TB, Apolloni A, Hurren L, et al. Coinfection with A- and B-type Epstein-Barr virus in human immunodeficiency virus-positive subjects. J Infect Dis 1990;162:643–8. [DOI] [PubMed] [Google Scholar]
- 22.Kunimoto M, Tamura S, Tabata T, et al. One-step typing of Epstein-Barr virus by polymerase chain reaction: predominance of type 1 virus in Japan. J Gen Virol 1992;73:455–61. [DOI] [PubMed] [Google Scholar]
- 23.Tomita Y, Ohsawa M, Mishiro Y, et al. The presence and subtype of Epstein-Barr virus in B and T cell lymphomas of the sino-nasal region from the Osaka and Okinawa districts of Japan. Lab Invest 1995;73:190–6. [PubMed] [Google Scholar]
- 24.Sidagis J, Ueno K, Tokunaga M, et al. Molecular epidemiology of Epstein-Barr virus (EBV) in EBV-related malignancies. Int J Cancer 1997;72:72–6. [DOI] [PubMed] [Google Scholar]
- 25.Statistics and Information Department of Secretariat. Vital Statistics of Japan. 1995. In: Age adjusted death rates from malignant neoplasms by site, each prefecture. Tokyo: Ministry of Health and Welfare of Japan, 1995:440.
- 26.Tsuhako K, Nakazato I, Miyagi J, et al. Comparative study of oral squamous cell carcinoma in Okinawa, Southern Japan and Sapporo in Hokkaido, Northern Japan; with special reference to human papillomavirus and Epstein-Barr virus infection. J Oral Pathol Med 2000;29:70–9. [DOI] [PubMed] [Google Scholar]
- 27.Pindborg JJ, Reichart PA, Smith CJ, et al. World Health Organization International Histological Classification of Tumours. Histological typing of cancer and precancer of the oral mucosa, 2nd ed. Berlin: Springer, 1997.
- 28.Saiki RK, Scharf S, Faloona F, et al. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 1985;230:1350–4. [DOI] [PubMed] [Google Scholar]
- 29.Wu S-J, Lay J-D, Chen C-L, et al. Genomic analysis of Epstein-Barr virus in nasal and peripheral T-cell lymphoma: a comparison with nasopharyngeal carcinoma in an endemic area. J Med Virol 1996;50:314–21. [DOI] [PubMed] [Google Scholar]
- 30.Vasef MA, Kamel OW, Chen Y-Y, et al. Detection of Epstein-Barr virus in multiple sites involved by Hodgkin's disease. Am J Pathol 1995;147:1408–15. [PMC free article] [PubMed] [Google Scholar]
- 31.Marchuk D, Drumm M Saulino A, et al. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res 1990;19:1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu L-F, Zabarovsky ER, Chen F, et al. Isolation and sequencing of the Epstein-Barr virus BNLF-1 gene (LMP1) from a Chinese nasopharyngeal carcinoma. J Gen Virol 1991;72:2399–409. [DOI] [PubMed] [Google Scholar]
- 33.Knecht H, Bachmann E, Brousset P, et al. Deletions within the LMP1 oncogene of Epstein-Barr virus are clustered in Hodgkin's disease and identical to those observed in nasopharyngeal carcinoma. Blood 1993;82:2937–42. [PubMed] [Google Scholar]
- 34.Takano Y, Kato Y, Saegusa M, et al. The role of the Epstein-Barr virus in the oncogenesis of EBV(+) gastric carcinomas. Virchows Arch 1999;434:17–22. [DOI] [PubMed] [Google Scholar]
- 35.Baer R, Bankier AT, Biggin MD, et al. DNA sequence and expression of the B95–8 Epstein-Barr virus genome. Nature 1984;310:207–11. [DOI] [PubMed] [Google Scholar]
- 36.Dambaugh T, Hennessy K, Chamnankit L, et al. U2 region of Epstein-Barr virus DNA may encode Epstein-Barr nuclear antigen 2. Proc Natl Acad Sci U S A 1984;81:7632–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen X, Pepper SV, Arrand JR. Prevalence of the A and B types of Epstein-Barr virus DNA in nasopharyngeal carcinoma biopsies from Southern China. J Gen Virol 1992;73:463–6. [DOI] [PubMed] [Google Scholar]
- 38.Lung ML, Lam WP, Sham J, et al. Detection and prevalence of the “f” variant of Epstein-Barr virus in Southern China. Virology 1991;185:67–71. [DOI] [PubMed] [Google Scholar]
- 39.Lung ML, Chang GC. Detection of distinct Epstein-Barr virus genotypes in NPC biopsies from Southern Chinese and Caucasians. Int J Cancer 1992;52:34–7. [DOI] [PubMed] [Google Scholar]
- 40.Miyashita T, Kawaguchi H, Asada M, et al. Epstein-Barr virus type B in patient with T-cell lymphoma. Lancet 1991;337:1045–7. [DOI] [PubMed] [Google Scholar]
- 41.Huen DS, Henderson SA, Croom-Carter D, et al. The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-κB and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene 1995;10:549–60. [PubMed] [Google Scholar]
- 42.Mitchell T, Sugden B. Stimulation of NF-κB-mediated transcription by mutant derivatives of the latent membrane protein of Epstein-Barr virus. J Virol 1995;69:2968–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu L-F, Chen F, Zheng X, et al. Clonability and tumorgenicity of human epithelial cells expressing the EBV encoded membrane protein LMP1. Oncogene 1993;8:1575–83. [PubMed] [Google Scholar]
- 44.Berger C, McQuain C, Sullivan JL, et al. The 30-bp deletion variant of Epstein-Barr virus-encoded latent membrane protein-1 prevails in acute infectious mononucleosis. J Infect Dis 1997;176:1370–3. [DOI] [PubMed] [Google Scholar]
- 45.Munch M. Epstein-Barr virus strain characterization. Acta Pathol Microbiol Immunol Scand 1998;106:425–33. [DOI] [PubMed] [Google Scholar]
- 46.Sandvej K, Gratama JW, Munch M, et al. Sequence analysis of the Epstein-Barr virus (EBV) latent membrane protein-1 gene and promoter region: identification of four variants among wild type EBV isolates. Blood 1997;90:323–30.EB [PubMed] [Google Scholar]
- 47.Yeh TS, Li SN, Wu CJ, et al. Sequence variations between two Epstein-Barr virus LMP-1 variants have no effect on the activation of NF-kappa B activity. DNA Cell Biol 1997;16:1311–19. [DOI] [PubMed] [Google Scholar]
- 48.Mori S, Itoh T, Tokunaga M, et al. Deletions and single-base mutations within the carboxy-terminal regions of the latent membrane protein 1 oncogene in Epstein-Barr virus-related gastric cancers of southern Japan. J Med Virol 1999;57:152–8. [DOI] [PubMed] [Google Scholar]
- 49.Miyagi J, Tsuhako K, Kinjo T, et al. Recent striking changes in histological differentiation and rate of human papillomavirus infection in squamous cell carcinoma of the lung in Okinawa, a subtropical island in southern Japan. J Clin Pathol 2000;53:676–84. [DOI] [PMC free article] [PubMed] [Google Scholar]