The advent of a novel category of anticancer drugs targeting individual genes is greatly affecting pathology and it appears that a new discipline—predictive molecular pathology—will become important in our field. For example, Herceptin (trastuzamab) treatment can only be used against tumours expressing the HER-2 oncogene1 and STI571 (Glivec) is highly efficient against tumours expressing c-kit (CD117).2 Because these target genes can be expressed in many different tumour types, oncologists are now increasingly demanding HER-2 and c-kit analyses for all of their patients so that they can potentially benefit from new drugs. The number of such analyses, often performed many years after the removal of the primary tumour, increases rapidly as more patients with cancer and clinicians become aware of the potential availability of new “miracle” drugs. Currently, the demand for retrospective c-kit and HER-2 analyses can be met using the tools of traditional molecular pathology.
However, what would happen if all living patients with cancer became aware of a new drug from which they could benefit if their tumour expressed one particular gene? The logistical problems connected to such a mass demand for analyses would be comparable to the situation on the sinking Titanic with a shortage of lifeboats more than 90 years ago. Alone in a small country like Switzerland (7000 000 inhabitants), there is an estimated number of > 150 000 living people with known potentially life threatening cancer. Tens of thousand of these patients would die of their disease before all of these tumours could be tested for a new therapeutic target gene if traditional testing methods were used. The situation will be further compounded by a multitude of additional drugs like Herceptin or Glivec appearing on the market very soon, each of which might require the testing of one or even several genes to identify responsive tumours.
Pathologists must now look for strategies that will allow them to perform unprecedented large scale molecular analyses. A few years ago, we developed a tool that may prove to be crucially important in this race against time. “Tissue microarray” (TMA) technology3,4 enables hundreds of tissue samples to be present on one microscope glass slide for simultaneous in situ analysis of DNA, RNA, and protein targets. Initial applications of the TMA method were in cancer research, where numerous studies have demonstrated the power of the approach.5–7 Considering the problem outlined above, it would be appealing to have tissue samples from all living patients with cancer in a tissue array format and to analyse all tissues simultaneously if a new gene specific treatment came on to the market.
“Tissue microarray” technology enables hundreds of tissue samples to be present on one microscope glass slide for simultaneous in situ analysis of DNA, RNA, and protein targets”
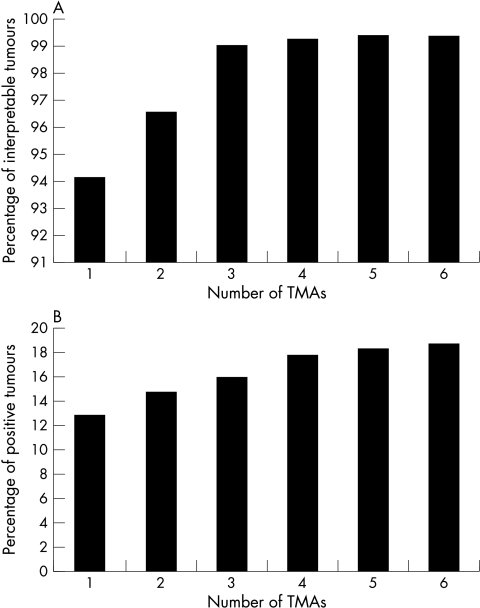
The crucial issue in a diagnostic TMA approach is how to achieve optimal or at least sufficient tumour representation on TMAs. In a previous issue of JCP this year, Gancberg et al provide data suggesting that HER-2 amplification could be reliably detected on TMAs.8 However, the number of tumours analysed in this important study is small and it appears that their set of tumours was selected (19 of 29 tumours amplified), so that perhaps borderline cases were lacking. To define criteria for diagnostic TMA applications we have recently analysed HER-2 overexpression/amplification in six replica TMAs, each containing a different tissue sample of a consecutive series of 786 breast cancers. The data suggest that a combination of two TMAs already results in up to 99% of interpretable tumours (fig 1A), whereas the frequency of positivity reaches the expected 17–20% only after combining the results of five to six TMAs (fig 1B). Because our fluorescent in situ hybridisation (FISH) data were obtained by estimation rather than by full scoring of FISH signals in 60 cells, it is possible that some amplified tumours were not detected because of conservative estimates. However, taking multiple samples for each tumour is the optimal way for dealing with potential tumour heterogeneity. The likelihood of detecting clinically relevant genetic heterogeneity by slightly increasing the sample diameter (1.5 mm) is small.
Figure 1.
Influence of number of arrayed samples on tissue microarray (TMA) data. (A) Influence of the number of samples analysed on interpretability of tumours (data from HER-2 immunohistochemistry; HercepTest, Dako). (B) Influence of the number of samples analysed on the percentage of positive tumours (data from HER-2 fluorescence in situ hybridisation; PathVysion, Vysis).
Apart from the concept of “microarray based predictive molecular pathology”, there are many other ways in which TMAs can be used in non-research oriented “routine” pathology laboratories. For example, the tedious work of testing new antibodies, comparing different antibodies for one target protein, and determining optimal staining conditions can best be done on small test TMAs containing several normal tissues and a selection of different tumour types. One of the most obvious and most powerful applications of the TMA technique in non-research laboratories is quality control. Here, the most frequently discussed weakness of the TMA method, the very small tissue samples (diameter, 0.6 mm), becomes an important strongpoint. The small size of arrayed tissue samples enforces maximal standardisation of both biology and processing (for example, fixation) of the tissue pieces used for control purposes. If consecutive sections of a TMA block are used it can be expected that staining variations between two slides are caused by differences in reagents or staining procedures.
The study of Packeisen et al,9 published in this issue, provides one example of using TMAs in quality control. In their innovative paper, they propose to manufacture special slides for immunohistochemistry (IHC) containing small sections of control tissues in a TMA format. Given the considerable workload and expense that results from the rigorous use of positive and negative control sections for all IHC incubations (including negative staining controls), this approach offers a great advantage over traditional methods. Although no exact numbers were provided in their paper, it is conceivable that slides with insufficient IHC staining were identified using this approach. The proposed method is likely to be adopted in many laboratories. It will be important, however, to ensure that slides containing control TMA sections are always freshly prepared. Using TMAs in many different studies, we have found that the age of slides is absolutely crucial for the outcome of IHC staining for many different antibodies (M Mirlacher et al, unpublished observations; 2002). Comparing IHC staining obtained in different laboratories is another important quality control application of TMAs. As yet, interlaboratory comparison studies have been limited to a small number of slides to reduce the workload for quality control purposes as much as possible. The TMA approach will allow interlaboratory studies to be expanded massively. For example, TMAs containing hundreds of tumours with outcome information could be distributed to participating laboratories and it would be possible not only to compare the analysis data between the laboratories but also to define, for each laboratory, to what extent their staining was related to prognosis or response to treatment.
“One of the most obvious and most powerful applications of the tissue microarray technique in non-research laboratories is quality control”
In summary, the TMA method is not only a valuable research tool. TMAs are highly practical for many purposes in diagnostic pathology. In particular, we anticipate that TMAs will become extremely important in the emerging field of predictive molecular pathology.
REFERENCES
- 1.Pegram MD, Lipton A, Hayes DF, et al. Phase II study of receptor-enhanced chemosensitivity using recombinant humanized anti-p185HER2/neu monoclonal antibody plus cisplatin in patients with HER2/neu-overexpressing metastatic breast cancer refractory to chemotherapy treatment. J Clin Oncol 1998;16:2659–71. [DOI] [PubMed] [Google Scholar]
- 2.Joensuu H, Roberts PJ, Sarlomo-Rikala M, et al. Effect of the tyrosine kinase inhibitor STI571 in a patient with a metastatic gastrointestinal stromal tumor. N Engl J Med 2001;344:1052–6. [DOI] [PubMed] [Google Scholar]
- 3.Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1998;4:844–7. [DOI] [PubMed] [Google Scholar]
- 4.Bubendorf L, Nocito A, Moch H, et al. Tissue microarray (TMA) technology: miniaturized pathology archives for high-throughput in situ studies. J Pathol 2001;195:72–9. [DOI] [PubMed] [Google Scholar]
- 5.Torhorst J, Bucher C, Kononen J, et al. Tissue microarrays for rapid linking of molecular changes to clinical endpoints. Am J Pathol 2001;159:2249–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simon R, Richter J, Wagner U, et al. High-throughput tissue microarray analysis of 3p25 (RAF1) and 8p12 (FGFR1) copy number alterations in urinary bladder cancer. Cancer Res 2001;61:4514–19. [PubMed] [Google Scholar]
- 7.Simon R, Nocito A, Hübscher T, et al. Patterns of her-2/neu amplification and overexpression in primary and metastatic breast cancer. J Natl Cancer Inst 2001;93:1141–6. [DOI] [PubMed] [Google Scholar]
- 8.Gancberg D, Di Leo A, Rouas G, et al. Reliability of the tissue microarray based FISH for evaluation of the HER-2 oncogene in breast cancer. J Clin Pathol 2002;55:315–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Packeisen J, Bürger H, Krech R, et al. Multi-tissue microarrays: a new approach for quality control in immunohistochemistry. J Clin Pathol 2002;55:613–15. [DOI] [PMC free article] [PubMed] [Google Scholar]