Abstract
Aims: To identify immunostaining patterns that are predictive for p53 mutations and to investigate whether p53 mutations are associated with established risk factors for oral squamous cell carcinoma (OSCC).
Methods: Fifty five OSCCs were investigated for p53 protein expression by immunohistochemistry (IHC). Ten of these cases, including five p53 immunopositive and five p53 immunonegative cases, were subjected to microdissection of representative tumour areas followed by sequence analysis for the detection of TP53 mutations.
Results: Paired IHC and sequence analysis revealed that p53 immunoexpression in more than 25% of tumour cells was indicative of TP53 mutations, whereas p53 immunonegativity was not informative. Therefore, for p53 immunohistochemical interpretation, p53 immunonegative cases were excluded from the analysis and the cut off value for p53 immunoexpression was set at 25%. Of the OSCCs showing any p53 immunoexpression, 64% revealed staining in more than 25% of the tumour cells. p53 immunoexpression in more than 25% of the neoplastic cells was significantly associated with smoking but not with alcohol consumption. No significant association with smoking habits was found when OSCCs were dichotomised into p53 immunonegative and p53 immunopositive.
Conclusions: In OSCCs the following conclusions can be made: (1) p53 immunonegativity is not informative for TP53 mutations; (2) 25% p53 immunopositive cells appears to be a good cut off value to predict TP53 mutations; (3) p53 immunostaining patterns that appeared to be predictive for TP53 mutations were associated with the smoking habits of the patients.
Keywords: oral carcinoma, p53 staining pattern, TP53 mutation, tobacco smoking
Oral carcinogenesis is a multifactorial and multistep process. The specific molecular events involved, in addition to the sequence in which they usually occur, are only partially recognised.1,2 Epidemiological studies disclosed a significant association between oral squamous cell carcinoma (OSCC) and the smoking and alcohol drinking habits of the patients,3 but the specific molecular targets of these agents remain largely unknown.
TP53 is a tumour suppressor gene that encodes a phosphoprotein of 393 amino acids, with a molecular mass of 53 kDa. The p53 protein prevents cells from accumulating DNA damage by the induction of pathways leading to cell cycle arrest or apoptosis.4,5 The amount of p53 protein is increased in cells exposed to genotoxic stress, and because of its protective role it has been called “the guardian of the genome”.6 Mutated forms of p53 often lack these functions, and these represent the most frequently identified alterations in human cancers.7 Several studies suggest that p53 alterations are causally related to oral carcinogenesis8–10 and may precede well recognisable histological alterations.11,12 An association between tobacco smoking and TP53 mutation has been disclosed for some cancers of the upper aerodigestive tract.7,13 However, studies using immunohistochemistry (IHC) to investigate p53 alterations in OSCCs failed to find such an association.14–16 We hypothesised that part of the discrepancy among published data results from inappropriate categorisation of p53 IHC results, namely by dichotomising these into p53 negative and p53 positive, and assuming that these categories are synonymous with wild-type and mutant p53 protein, respectively. We expected that, upon identification of appropriate criteria for the categorisation of p53 IHC results, these discrepancies would be greatly diminished.
“The predictive value of a simple technique such as p53 immunohistochemistry to detect TP53 mutations is a clinically relevant issue”
Importantly, TP53 mutations have been associated with a poor response to certain treatment modalities,17,18 and recent trials indicate that gene therapy targeting TP53 could be a valuable complementary treatment in some of these cases.19,20 However, the methods presently available to detect TP53 mutations are still very laborious and involve high levels of expertise, preventing its current use in the routine laboratory.21 Therefore, the predictive value of a simple technique such as p53 IHC to detect TP53 mutations is a clinically relevant issue.
The aims of our present study were: (1) to identify p53 immunostaining patterns that are predictive of TP53 mutations; and (2) to correlate p53 staining patterns that appeared predictive of TP53 mutations with the smoking and alcohol drinking habits of the patients.
METHODS
Patients and tissues
Fifty five formalin fixed, paraffin wax embedded oral carcinomas, excised by surgery from patients who were seen at the department of oral and maxillofacial surgery and otorhinolaryngology from the Vrije Universiteit Medical Centre, Amsterdam, the Netherlands, were evaluated in this study for the expression of p53. Samples comprised 55 OSCCs, three of which were verrucous carcinomas.
Immunohistochemical analysis
Three sections of 4 μm were cut from formalin fixed, paraffin wax embedded tissues, and were mounted on to poly-L-lysine coated slides. Consecutive sections were used as negative controls of the IHC reaction and for haematoxylin and eosin (H&E) staining to confirm the diagnosis.
The streptavidin–biotin complex immunoperoxidase technique used has been described previously.11 Microwave antigen retrieval was performed in citrate at 100°C (3 × 5 minutes). A mouse monoclonal antibody recognising both mutant and wild-type p53 protein (clone D07; Dako, Glostrup, Denmark) was used as the primary antibody (1/500 dilution) and incubated overnight at 4°C. Negative controls consisted of phosphate buffered saline instead of primary antibody. A p53 immunopositive breast carcinoma was used as the positive control.
Quantification of IHC results
p53 immunonegativity was defined as the total absence of p53 immunostaining in the neoplastic cells. p53 immunopositive carcinomas were classified into four categories: 1, less than 25% positive cells within the immunopositive area/p53 staining restricted to the most peripheral (“basal”) cell layer of tumour nests; 2, between 25% and 50% positive cells within the immunopositive area; 3, between 50% and 75% positive cells within the immunopositive area; 4, between 75% and 100% positive cells within the immunopositive area. When different p53 staining patterns were present in one section, the ultimate score attributed to the tumour was always the highest score found in neoplastic areas.
Sequence analysis of exons 5–8 of the TP53 gene
Consecutive 9 μm sections (10 to 20, according to the size of tumour areas) of the same paraffin wax blocks used for IHC analysis were manually microdissected, guided by control H&E stained and by p53 IHC slides, to obtain areas with more than 75% of tumour cells for DNA extraction. Contamination between samples was avoided by following a strict protocol at all steps of analysis. DNA was extracted by proteinase K and phenol/chloroform extraction in the presence of a carrier (glycogen). DNA concentrations were calculated from the optical density at 260 nm.
Exons 5 to 8 of the TP53 gene were amplified individually from the extracted DNA by means of the polymerase chain reaction (PCR). The sizes of the amplified fragments were 287 bp (exon 5), 222 bp (exon 6), 237 bp (exon 7), and 237 bp (exon 8). PCR was performed in a 50 μl volume containing: 5 μl (containing at least 250 ng) of extracted DNA, 10mM Tris/HCl pH 8.3, 50mM KCl, 1.5mM MgCl2, 0.2mM dNTP, 2.5 U AmpliTaq DNA polymerase (Perkin Elmer-Cetus, Foster City, California, USA), 25 pmol of sense primer, and 25 pmol of antisense primer. The mixtures were overlaid with several drops of paraffin oil and incubated for four minutes at 95°C for DNA denaturation, followed by 40 cycles of amplification using a PCR processor (Bio-med, Therés, Germany). Each cycle included a denaturation step at 95°C for one minute, an annealing step at 60°C for one minute, and a chain elongation step at 72°C for two minutes. To ensure a complete extension of the amplified DNA, the final elongation step was prolonged with another five minutes at 72°C. A total of 5 μl of PCR product was finally analysed by electrophoresis on a 1.5% agarose gel, stained with ethidium bromide, and visualised under ultraviolet light.
PCR products were purified using the QIAquick PCR purification kit (Qiagen, Chatsworth, California, USA), following the manufacturer’s instructions. Extracted DNA was diluted in 30 μl of distilled water. Subsequently, the DNAs were directly sequenced by exon specific primers using the radioactive dideoxynucleotide method (AmpliCycle sequencing kit; Perkin Elmer, Norwalk, Connecticut, USA). Primers for exon 5 (antisense), exon 6 (sense), exon 7 (antisense), and exon 8 (sense) were end labelled with [33P]ATP (3000 Ci/mmol; Amersham Pharmacia Biotech, Roosendaal, The Netherlands). Cycle sequencing was performed in an MJ thermal cycler (PTC200; MJ Research, supplied by Biozym, Landgraaf, the Netherlands) as follows: two minutes at 95°C, followed by 28 cycles of one minute at 95°C, one minute at 58°C, and one minute at 72°C. Electrophoresis of the PCR products was carried out on a Genomyx Sequencer, using a 6% polyacrylamide denaturating gel (Genomyx Corp, Foster City, California, USA). The dideoxynucleotide lanes of several samples were grouped together (for example, all As, all Cs, etc) to facilitate mutation detection. Gels were exposed to film and the sequence on the autoradiographs was read by two independent investigators who did not know the p53 IHC patterns of the OSCCs.
Statistical analysis
For the purpose of statistical analysis, p53 staining patterns in carcinomas were divided into three groups, each with two categories, namely: I, p53 immunonegative versus p53 immunopositive cases; II, p53 immunoexpression in less than 25% of the neoplastic cells (excluding p53 immunonegative cases) versus p53 immunoexpression in more than 25% of the neoplastic cells; and III, p53 immunoexpression in less than 50% of the neoplastic cells (excluding p53 immunonegative cases) versus p53 immunoexpression in more than 50% of the neoplastic cells. The rationale for these categorisations is discussed later.
The smoking and alcohol drinking habits of the patients were each divided into two categories (smokers and non-smokers; drinkers and non-drinkers).
Statistical analysis was performed using SPSS 7.0 software and Fisher’s exact test. Stepwise discriminant analysis was used to evaluate whether smoking and alcohol use were independently associated with p53 status. Values were considered significantly different when the p value was less than 0.05.
RESULTS
p53 immunohistochemistry
Immunohistochemical staining for p53 was exclusively found in the nuclei of epithelial cells. None of the negative controls displayed brown staining in epithelial or other cells. Positive controls included in each experiment were consistently positive, indicating a successful IHC.
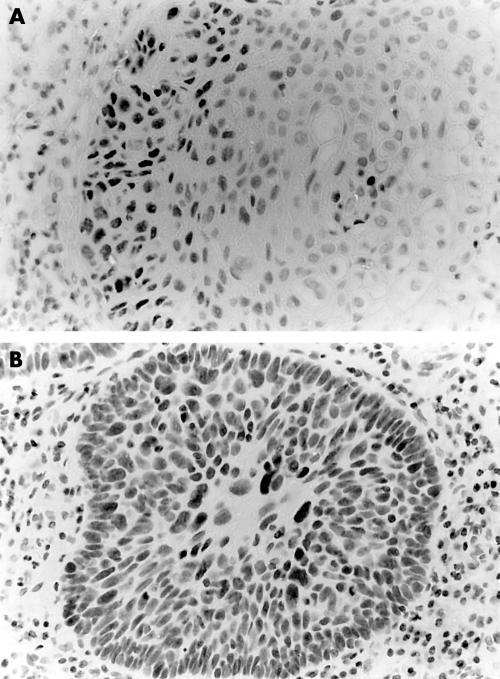
In oral carcinomas, the p53 staining pattern within the same tumour was often heterogeneous; 38% of the tumours showed different p53 scores in different areas, and in five of these cases p53 negative areas were found adjacent to p53 positive areas (table 1). Of the 55 oral carcinomas analysed, 22 showed no p53 staining, 12 showed less than 25% p53 positive cells (this group included all the verrucous carcinomas analysed), six cases showed between 25% and 50% positive cells, three cases showed between 50% and 75% positive cells, and 12 cases showed a clear staining in almost all of the tumour cells. In well differentiated tumours and verrucous carcinomas, p53 staining was often localised to the most peripheral layer(s) of the tumour lobules. Figure 1 shows representative examples of OSCCs showing p53 immunostaining in between 25% and 50% of the tumour cells (fig 1A) and in virtually all tumour cells (fig 1B).
Table 1.
Information concerning patients, tumours, and their p53 staining patterns
| Tn | Sex/age | Location | Smoking habits (cigarettes/day) | Drinking habits (U/day) | Grade | p53 immunostaining |
| T1 | M/81 | T | NS | <2 | M | Negative |
| T2 | F/45 | OL | >20 | >4 | M-P | Negative |
| T3 | M/60 | OL | NS | ND | W-M | Negative |
| T4 | F/55 | FM | >20 | >4 | W-M | Negative |
| T5 | M/58 | OL | unk | unk | M-P | Negative |
| T6 | M/70 | FM | 10–20 | >4 | M | Negative |
| T7 | F/63 | T | >20 | >4 | W-M | Negative |
| T8 | F/79 | T | NS | ND | W | Negative |
| T9 | M/71 | T/FM | 10–20 | 2–4 | W-M | Negative |
| T10 | M/52 | OL | >20 | unk | W | Negative |
| T11 | M/59 | T | NS | <2 | W-M | Negative |
| T12 | F/79 | T | NS | <2 | W | Negative |
| T13 | F/80 | T | NS | ND | M | Negative |
| T14 | M/72 | T | unk | >4 | M | Negative |
| T15 | M/63 | FM | 10–20 | 2–4 | W | Negative |
| T16 | M/59 | OL | >20 | >4 | W | Negative |
| T17 | M/68 | T | <10 | >4 | W-M | Negative |
| T18 | M/69 | T | 10–20 | 2–4 | W-M | Negative |
| T19 | M/67 | T | 10–20 | 2–4 | M | Negative |
| T20 | M/48 | T | unk | unk | W-M | Negative |
| T21 | F/28 | T | 10–20 | 2–4 | W-M | Negative |
| T22 | M/62 | T | >20 | 2–4 | M | Negative |
| T23 | M/59 | FM | >20 | >4 | M | <25% |
| T24 | F/105 | FM | NS | unk | VerrCa | <25% |
| T25 | F/66 | OL | NS | ND | W-M | <25% |
| T26 | F/57 | T | 10–20 | <2 | W | <25% |
| T27 | F/63 | T | >20 | >4 | W | <25% |
| T28 | F/73 | OL | NS | ND | W-M | <25%* |
| T29 | F/82 | T | unk | unk | M | <25%* |
| T30 | M/75 | T | 10–20 | >4 | W-M | <25% |
| T31 | F/81 | OL | NS | <2 | W | <25%* |
| T32 | F/76 | OL | NS | <2 | VerrCa | <25%* |
| T33 | F/86 | T | NS | ND | W-M | <25% |
| T34 | F/99 | OL | NS | unk | VerrCa | <25% |
| T35 | M/77 | OL | unk | unk | W | 25–50% |
| T36 | M/76 | OL | 10–20 | <2 | W-M | 25–50% |
| T37 | F/81 | T | NS | ND | W-M | 25–50% |
| T38 | F/56 | OL | unk | unk | M | 25–50% |
| T39 | F/87 | FM | unk | unk | M-P | 25–50% |
| T40 | F/69 | T | NS | ND | W-M | 25–50% |
| T41 | F/49 | FM | >20 | >4 | W-M | 50–75% |
| T42 | M/70 | T | 10–20 | >4 | W-M | 50–75% |
| T43 | F/50 | OL | unk | unk | M-P | 50–75% |
| T44 | M/58 | FM | >20 | >4 | M | 75–100% |
| T45 | F/55 | T | >20 | >4 | W-M | 75–100% |
| T46 | M/45 | OL | unk | unk | P | 75–100% |
| T47 | F/75 | T | NS | ND | M | 75–100% |
| T48 | F/47 | OC | >20 | 2–4 | M | 75–100% |
| T49 | M/56 | T | 10–20 | 2–4 | W-M | 75–100% |
| T50 | M/48 | OL | >20 | >4 | M | 75–100% |
| T51 | F/84 | FM | 10–20 | 2–4 | M | 75–100% |
| T52 | M/39 | T | >20 | 2–4 | W | 75–100% |
| T53 | F/51 | FM | 10–20 | >4 | W-M | 75–100%* |
| T54 | M/55 | T | 10–20 | 2–4 | W-M | 75–100% |
| T55 | M/81 | T | 10–20 | >4 | W-M | 75–100% |
*Coexistent areas with no p53 staining in the tumour cells.
Location of tumour: T, tongue; FM, floor of mouth; OL, other location (within the oral cavity); OC, oral cavity (location not specified). Grade (grade of differentiation): W, well differentiated SCC; W-M, well to moderately differentiated SCC; M, moderately differentiated SCC; M-P, moderately to poorly differentiated SCC; P, poor differentiated SCC; VerrCa, verrucous carcinoma.
ND, non-drinker; NS, non-smoker; U, units of alcoholic beverage; unk, unknown.
Figure 1.
(A) Oral squamous cell carcinomas (OSCC) showing p53 immunostaining in between 25% and 50% of the neoplastic cells (tables 1 and 2; T38). Note that mainly the peripheral layers of the tumour stains for p53 (p53 immunohistochemistry, counterstained with haematoxylin); (B) OSCC showing p53 immunostaining in almost all neoplastic cells (table 1; T53) (p53 immunohistochemistry, counterstained with haematoxylin).
TP53 mutational analysis in relation to p53 immunostaining patterns of oral carcinomas
To set criteria for p53 immunostaining patterns that are indicative of mutations we performed microdissection and sequence analysis of the TP53 hot spot region in 10 cases selected to represent various p53 immunostaining categories. These OSCCs were derived from patients with diverse smoking and drinking habits to discount possible confounding factors (table 2).
Table 2.
Demographic details of patients, risk factors for their OSCCs, and TP53 gene and p53 protein alterations
| Sequence of exons 5–8 of the TP53 gene | ||||||||
| Tn | Sex/age | Smoking habits (cig/day) | Alcohol habits (U/day) | p53 IHC | Mutation | Exon | Codon | AA change |
| T1 | M/81 | NS | <2 | Negative | CGA>TGA | 6 | 213 | Arg→Stop |
| T4 | F/55 | >20 | >4 | Negative | No | – | – | – |
| T12 | F/79 | NS | <2 | Negative | No | – | – | – |
| T17 | M/68 | <10 | >4 | Negative | Deletion 2bp | 5 | 145 | Frameshift |
| T19 | M/67 | 10–20 | 2–4 | Negative | CGA>TGA | 6 | 213 | Arg→Stop |
| T27 | F/63 | >20 | >4 | <25% | CGA>CGG* | 6* | 213* | Arg→Arg* |
| T31 | F/81 | NS | <2 | <25% | No | – | – | – |
| T38 | F/56 | unk | unk | 25–50% | CGC>CAC | 5 | 175 | Arg→His |
| T46 | M/45 | unk | unk | >75% | CAT>CGT | 6 | 193 | His→Arg |
| T52 | M/39 | >20 | 2–4 | >75% | TTT>CTT | 5 | 134 | Phe→Leu |
*Compatible with well described p53 polymorphism.
AA, amino acid; cig, cigarettes; IHC, immunohistochemistry; NS, non-smoker; OSCC, oral squamous cell carcinoma; unk, unknown; U, units of alcoholic beverage.
Mutations were found in exons 5 and 6, particularly at codon 213, and all but one (a deletion) consisted of transitions (table 2). No mutations were found in two of five p53 immunonegative cases (table 2; T4, T12). Among the remaining three p53 immunonegative cases we detected a frameshift mutation in one case (table 2; T17) and a nonsense mutation resulting in a stop codon and premature termination of the p53 protein in two cases (table 2; T1, T19; fig 2A). Among the two carcinomas showing less than 25% immunopositive cells, we could not detect mutations in one case (table 2; T31), whereas the other case revealed a silent mutation, which does not result in an amino acid change (table 2; T27). Missense mutations were demonstrated in all three carcinomas that showed p53 staining in more than 25% of tumour cells, including one case with p53 staining in between 25% and 50% of the tumour cells (table 2; T38) and two cases where p53 staining was present in more than 75% of the tumour cells (table 2; T46, T52; fig 2B). In summary, the cut off value of 25% p53 immunopositive cells was predictive for TP53 mutations, whereas p53 immunonegativity was non-informative.
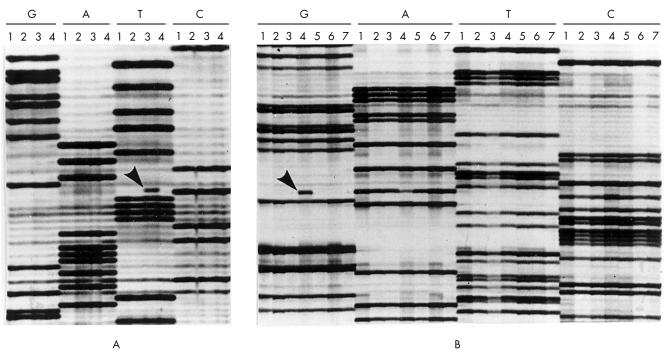
Figure 2.
Autoradiographs from part of the sequence of TP53 exon 6 in selected oral squamous cell carcinomas. Lanes are grouped together for the individual tumours to facilitate the identification of abnormal mutant bands (four tumours are shown in (A), and seven tumours are shown in (B)). (A) Shows a C to T transition at codon 213 in lane 3 (arrow). This mutation changed a codon that encodes arginine into a stop codon; no tumour cells in this carcinoma stained for p53 (table 2; T19). (B) Shows an A to G transition at codon 193 in lane 4 (arrow). This mutation results in a histidine to arginine change and the carcinoma showed p53 immunostaining in more than 75% of the tumour cells (table 2; T46).
p53 immunostaining patterns of oral carcinomas in relation to smoking and alcohol drinking habits of the patients
To compare our data with the published literature, p53 staining patterns were simply categorised as immunopositive or immunonegative. These were compared with the patient’s smoking and drinking habits and no significant associations were found (p = 0.8 and p = 0.7, respectively).
Additional analyses were performed where new criteria was set based on paired TP53 mutational analysis and p53 IHC. Immunonegative cases were excluded from the statistical analysis because this category is not informative—either wild-type TP53 or certain forms of mutated TP53 (for example, containing nonsense and frameshift mutations, in addition to homozygous deletions) can be present (our study).7,22 Two cut off values for p53 immunoexpression (25% and 50% p53 immunopositive cells, respectively) were established. We chose the cut off value of 25% based on our own (present and previous) data.11,23,24 The cut off value of 50% was used based on data of Kropveld et al,22 who showed a concordance between the presence (and type) of TP53 mutation and immunostaining in 96% of the carcinomas.
Using these criteria, the smoking habits of the patients were significantly associated with p53 immunostaining, using both 25% (p = 0.04) and 50% (p = 0.004) cut off values (table 3).
Table 3.
p53 expression pattern of oral carcinomas, according to smoking and alcohol drinking habits of the patients
| Tobacco | Alcohol | |||||
| p53 expression | Non-smoker (n=10*) | Smoker (n=17*) | p Value | Non-drinker (n=6*) | Drinker (n=19*) | p Value |
| <50% positive cells* (n=14) | 9 (64%) (90%) | 5 (36%) (29%) | 5 (42%) (83%) | 7 (58%) (37%) | ||
| 0.004 | 0.07 | |||||
| >50% positive cells (n=13) | 1 (8%) (10%) | 12 (92%) (71%) | 1 (8%) (17%) | 12 (92%) (63%) | ||
*Excluding p53 negative cases (see methods).
No association was found between the alcohol drinking habits of the patients and p53 immunostaining in the tumours using the cut off value of 25% immunopositive cells and excluding p53 negative cases (p = 0.6), whereas the 50% cut off value revealed a trend for association (p = 0.07; table 3).
In the patients studied, a strong association between tobacco smoking and alcohol drinking habits was present (p < 0.001). Stepwise discriminant analysis showed that only smoking was independently associated with p53 immunostaining (p < 0.01).
p53 immunostaining patterns of oral carcinomas obtained from patients who did not smoke and did not drink alcohol
Nine patients did not smoke and did not drink alcohol. Eight of these were women with relatively advanced age (range, 66–86 years; mean, 76). The p53 immunostaining patterns of these OSCCs were as follows: two OSCCs were p53 immunonegative, three OSCCs showed p53 immunostaining in less than 25% of the tumour cells, two showed p53 immunostaining in between 25% and 50% of the tumour cells, and one showed p53 immunostaining in virtually all tumour cells. The ninth case was a 60 year old man whose OSCC was p53 immunonegative (table 1).
DISCUSSION
In our study, we aimed to identify p53 immunoexpression patterns with predictive value for TP53 mutations and to test whether these mutations were related to the smoking and drinking habits of the patients.
In contrast to what has been previously assumed, p53 protein expression as detected by IHC does not always reflect the presence of mutant p53 protein, and neither does the absence of p53 staining preclude it.11,25 However, to our knowledge, few studies have tried to set a better criterion for the classification of p53 IHC results based on paired IHC and sequence analysis of the TP53 gene.22 In an attempt to improve the value of p53 IHC for the prediction of TP53 mutations, we sequenced the hot spot region of TP53 in a subset of OSCCs with defined p53 immunoexpression patterns. We then performed statistical analysis using different criteria for the categorisation of p53 IHC results to find out whether those differences could explain the discrepancy in the literature on the association between smoking habits and p53 alterations in the tumours.
“p53 protein expression as detected by immunohistochemistry does not always reflect the presence of mutant p53 protein, and neither does the absence of p53 staining preclude it”
Sequence analysis showed that, in OSCCs, p53 immunonegativity is not informative for the prediction of TP53 mutations because, in contrast to what might have been expected, TP53 mutations were detected in most of our sequenced p53 immunonegative cases. These findings are in agreement with other studies10,22,26–28 and justify our choice of exclusion of p53 immunonegative cases from the statistical analysis, because these cases would bias the results. The cut off value of 25% appeared to be a good predictor for TP53 mutations in this population. The silent mutation we detected in one OSCC expressing p53 in less than 25% of the tumour cells is a well described polymorphism, which is present in 3–10% of the population.29
Although the proportion of functionally relevant mutations outside the hot spot region of TP53 has been estimated to be less than 5% in lung and other cancers,21 in head and neck carcinoma 33% of the TP53 mutations have been found outside this region.22 Accordingly, it is likely that the proportion of TP53 mutations in our series of OSCCs is higher than that detected by mutational analysis. Nevertheless, we do not expect that this would dramatically influence our conclusions, because Kropveld and colleagues22 found a concordance between the presence (and type) of TP53 mutation at any exon and immunostaining in 96% of the head and neck carcinomas in their series when a cut off value of 50% was established.
In agreement with studies using similar categorisations of p53 IHC results,14–16 no significant association was found between p53 immunostaining in the neoplastic cells and the smoking habits of the patients, when all cases were analysed and categorised simply as p53 immunopositive or p53 immunonegative. However, using the new criteria of analysis, either with 25% or 50% cut off values, and with p53 immunonegative cases being excluded from the analysis, a significant association was found between p53 immunostaining and smoking habits. Although this association appeared to be more significant when 50% was used as the cut off value, the correlation between p53 immunostaining and TP53 mutation was higher when the 25% cut off was set (100% v 96%), and we have demonstrated a TP53 mutation in an OSCC showing immunostaining in between 25% and 50% of the tumour cells. It is worth noting that our previous data suggested that more than 25% immunopositive cells might be indicative of mutation, because all OSCCs that developed from premalignant lesions with suprabasal p53 staining showed p53 immunostaining in more than 25% of the tumour cells.11,23,24 Importantly, suprabasal p53 staining emerged as a marker for the malignant progression of oral leukoplakias in two independent populations.11,24 Studies where TP53 mutational analysis has been performed on carcinomas of the head and neck including OSCCs13,30,31 also demonstrated a significant association between the patients’ smoking habits and TP53 mutations. This suggests that, in these carcinomas, TP53 is an important molecular target of tobacco carcinogens, similar to that demonstrated for lung cancer.32
The role of alcohol (ab)use in oral carcinogenesis is widely accepted as a result of the convincing epidemiological data available, but the specific molecular targets of alcohol are still to be found. In contrast to tobacco, alcohol is not carcinogenic in vitro,33 but in patients with OSCC the two habits (smoking and drinking) are often associated, as shown in this and other studies.13,34 Moreover, stepwise discriminant analysis revealed an independent association with p53 immunostaining for tobacco, but not for alcohol, and this is in agreement with the results of others.31 Therefore, the trend found between alcohol drinking habits and p53 immunostaining may reflect the strong association between alcohol consumption and smoking habits in patients with OSCC, rather than indicating a causal relation between alcohol consumption and p53 alterations.
In our present study, nine patients are described who developed OSCC in the absence of smoking and alcohol drinking habits. Most of these tumours were obtained from women with advanced age and were not located in the floor of the mouth, which is a common location for OSCCs associated with the use of tobacco and alcohol. Previous studies performed on a subset of these patients35 showed that human papillomavirus DNA was not present in their tumours, arguing for another mechanism of carcinogenesis. Although the p53 status varied in these OSCCs, at least in one case, p53 mutation was very likely, because p53 immunoexpression was detected in almost all of the neoplastic cells. These findings suggest that p53 mutations may occur in the absence of (active) smoking or drinking habits. The p53 coding region contains more than 59 CpG dinucleotides, which are potential sites for the deamination of 5-methylcytosine, thereby causing endogenous mutations. Such mutations may confer a baseline risk for cancer, independent of known carcinogenic factors, and Greenblatt et al showed that C to T transitions at CpG sites account for 24% of all p53 mutations described.7 Interestingly, Brennan and colleagues30 showed that all p53 mutations found in head and neck carcinomas in five patients who did not smoke or drink alcohol occurred at sites containing CpG dinucleotides, potentially representing endogenous mutations. In addition, Nylander et al detected a new non-random 14 bp deletion in exon 8 of OSCCs collected in Sweden.36 Most of the patients carrying this deletion were women (80%), 60% of them were over 70 years at the time of diagnosis, and 66.7% of them did not smoke. However, only the association with female sex was significant (p = 0.02) in that study. One explanation advanced by the authors for the occurrence of that deletion was the close proximity of three GA dinucleotides in the area, which might have caused erroneous reannealing, with subsequent excision of the loop containing the sequence in between.
“The trend found between alcohol drinking habits and p53 immunostaining may reflect the strong association between alcohol consumption and smoking habits in patients with oral squamous cell carcinoma, rather than indicating a causal relation between alcohol consumption and p53 alterations”
In conclusion, this study calls attention to the criteria used in the interpretation of p53 IHC because this may explain the controversies found in the literature. While providing a link between carcinogen exposure (by tobacco smoking) and a molecular lesion (p53 alteration) in OSCCs, it also identifies a subgroup of patients (mainly older women) in whom no known carcinogenic factors could be identified, arguing for alternative mechanisms of oral carcinogenesis. These might include spontaneous mutations in the TP53 gene, either non-random deletions resulting from slipped mispairing during replication7,36 and/or deamination of cytosine, where age related methylation of CpG dinucleotides might play a role.7,30,37
Take home messages.
p53 immunonegativity is not informative for TP53 mutations in oral squamous cell carcinomas (OSCCs), which might explain some of the discrepancies in the literature
A cut off value of 25% p53 immunopositive cells appears to be predictive of TP53 mutations in OSCC
p53 immunostaining patterns that appeared to be predictive for TP53 mutations were associated with the smoking habits of patients with OSCC, but not with alcohol drinking
We identified a subgroup of patients (mainly older women) in whom no known carcinogenic factors could be identified, arguing for alternative mechanisms of oral carcinogenesis
These might include spontaneous mutations in the TP53 gene, either non-random deletions resulting from slipped mispairing during replication and/or deamination of cytosine, where age related methylation of CpG dinucleotides might play a role
Acknowledgments
The authors wish to thank T Tadema for laboratory facilities, M Tabor for advice on microdissection, and P van Diest for help in performing the statistical analyses.
Abbreviations
H&E, haematoxylin and eosin
IHC, immunohistochemistry
OSCC, oral squamous cell carcinoma
PCR, polymerase chain reaction
REFERENCES
- 1.Califano J, Van der Riet P, Westra W, et al. Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res 1996;56:2488–92. [PubMed] [Google Scholar]
- 2.Rosin MP, Cheng X, Poh C, et al. Use of allelic loss to predict malignant risk for low-grade oral epithelial dysplasia. Clin Cancer Res 2000;6:357–62. [PubMed] [Google Scholar]
- 3.La Vecchia C, Tavani A, Franceschi S, et al. Epidemiology and prevention of oral cancer. Oral Oncol 1997;33:302–12. [DOI] [PubMed] [Google Scholar]
- 4.Chang F, Syrjanen S, Syrjanen K. Implications of the p53 tumour-suppressor gene in clinical oncology. J Clin Oncol 1995;13:1009–22. [DOI] [PubMed] [Google Scholar]
- 5.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell 1997;88:323–31. [DOI] [PubMed] [Google Scholar]
- 6.Lane DP. p53, guardian of the genome. Nature 1992;358:15–16. [DOI] [PubMed] [Google Scholar]
- 7.Greenblatt MS, Bennet WP, Hollstein M, et al. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res 1994;54:4855–78. [PubMed] [Google Scholar]
- 8.Shin DM, Kim J, Ro JY, et al. Activation of p53 gene expression in premalignant lesions during head and neck tumorigenesis. Cancer Res 1994;54:321–6. [PubMed] [Google Scholar]
- 9.Lavieille JP, Brambilla E, Riva-Lavieille C, et al. Immunohistochemical detection of p53 in preneoplastic lesions and squamous cell carcinomas of the head and neck. Acta Otolaryngol (Stockh) 1995;115:334–9. [DOI] [PubMed] [Google Scholar]
- 10.Wood NB, Kotelnikov V, Caldarelli DD, et al. Mutation of p53 in squamous cell cancer of the head and neck: relationship to tumour cell proliferation. Laryngoscope 1997;107:827–33. [DOI] [PubMed] [Google Scholar]
- 11.Cruz IB, Snijders PJF, Meijer CJ, et al. p53 expression above the basal cell layer in oral mucosa is an early event of malignant transformation and has predictive value for developing oral squamous cell carcinoma. J Pathol 1998;184:360–8. [DOI] [PubMed] [Google Scholar]
- 12.Califano J, Westra WH, Koch W, et al. Unknown primary head and neck squamous cell carcinoma: molecular identification of the site of origin. J Natl Cancer Inst 1999;91:599–604. [DOI] [PubMed] [Google Scholar]
- 13.Field JK, Zoumpourlis V, Spandidos DA, et al. p53 expression and mutations in squamous cell carcinoma of the head and neck: expression correlates with patients use of tobacco and alcohol. Cancer Detect Prev 1994;18:197–208. [PubMed] [Google Scholar]
- 14.Matthews JB, Scully C, Jovanovic A, et al. Relationship of tobacco/alcohol use to p53 expression in patients with lingual squamous cell carcinomas. Eur J Cancer B Oral Oncol 1993;29B:285–99. [DOI] [PubMed] [Google Scholar]
- 15.Bongers V, Snow GB, Van der Waal I, et al. Value of p53 expression in oral cancer and adjacent normal mucosa in relation to the occurrence of multiple primary tumours. Eur J Cancer B Oral Oncol 1995;31B:392–405. [DOI] [PubMed] [Google Scholar]
- 16.Yan JJ, Tzeng CC, Jin YT. Overexpression of p53 protein in squamous cell carcinoma of buccal mucosa and tongue in Taiwan: an immunohistochemical and clinicopathological study. J Oral Pathol Med 1996;25:55–9. [DOI] [PubMed] [Google Scholar]
- 17.Ribeiro U Jr, Finkelstein SD, Safatle-Ribeiro AV, et al. p53 sequence analysis predicts treatment response and outcome of patients with esophageal carcinoma. Cancer 1998;83:7–18. [PubMed] [Google Scholar]
- 18.Matsuzoe D, Hideshima T, Kimura A, et al. p53 mutations predict non-small cell lung carcinoma response to radiotherapy. Cancer Lett 1999;135:189–94. [DOI] [PubMed] [Google Scholar]
- 19.Clayman GL, Frank DK, Bruso PA, et al. Adenovirus-mediated wild-type p53 gene transfer as a surgical adjuvant in advanced head and neck cancers. Clin Cancer Res 1999;5:1715–22. [PubMed] [Google Scholar]
- 20.Ganly I, Kirn D, Eckhardt SG, et al. A phase I study of Onyx-015, an E1B attenuated adenovirus, administered intratumorally to patients with recurrent head and neck cancer. Clin Cancer Res 2000;6:798–806. [PubMed] [Google Scholar]
- 21.Ahrendt SA, Halachmi S, Chow JT, et al. Rapid p53 sequence analysis in primary lung cancer using an oligonucleotide probe array. Proc Natl Acad Sci U S A 1999;96:7382–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kropveld A, Rozemuller EH, Leppers FGJ, et al. Sequencing analysis of RNA and DNA of exons 1 through 11 shows p53 gene alterations to be present in almost 100% of head and neck squamous cell cancers. Lab Invest 1999;79:347–53. [PubMed] [Google Scholar]
- 23.Cruz IB, Meijer CJLM, Snijders PJF, et al. p53 immunoexpression in non-malignant oral mucosa adjacent to oral squamous cell carcinomas: potential consequences for clinical management. J Pathol 2000;191:132–7. [DOI] [PubMed] [Google Scholar]
- 24.Cruz I, Napier SS, Van der Waal I, et al. Suprabasal p53 immunoexpression is strongly associated with high grade dysplasia and risk for malignant transformation in potentially malignant oral lesions from Northern Ireland. J Clin Pathol 2002;55:98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dowel SP, Ogden GR. The use of antigen retrieval for immunohistochemical detection of p53 over-expression in malignant and benign oral mucosa: a cautionary note. J Oral Pathol Med 1996;25:60–4. [DOI] [PubMed] [Google Scholar]
- 26.Xu L, Cheng Y-T, Huvos AG, et al. Overexpression of p53 in squamous cell carcinomas of head and neck without apparent gene mutations. Diagn Mol Pathol 1994;3:83–92. [DOI] [PubMed] [Google Scholar]
- 27.Coggi G, Bosari S, Roncalli M, et al. p53 protein accumulation and p53 gene mutation in esophageal carcinoma. A molecular and immunohistochemical study with clinicopathologic correlations. Cancer 1997;79:425–32. [DOI] [PubMed] [Google Scholar]
- 28.Rowley H, Sherrington P, Helliwell TR, et al. P53 expression and p53 gene mutation in oral cancer and dysplasia. Otolaryngol Head Neck Surg 1998;118:115–23. [DOI] [PubMed] [Google Scholar]
- 29.Carbone D, Chiba I, Mitsudomi T. Polymorphisms at codon 213 within the p53 gene. Oncogene 1991;6:1691–2. [PubMed] [Google Scholar]
- 30.Brennan JA, Jay OB, Wayne MK, et al. Association between cigarette smoking and mutation of the p53 gene in squamous-cell carcinoma of the head and neck. N Engl J Med 1995;332:712–17. [DOI] [PubMed] [Google Scholar]
- 31.Lazarus P, Stern J, Zwiebel N, et al. Relationship between p53 mutations incidence in oral cavity squamous cell carcinomas and patient tobacco use. Carcinogenesis 1996;17:733–9. [DOI] [PubMed] [Google Scholar]
- 32.Denissenko MF, Pao A, Tang M, et al. Preferential formation of benzo(a)pyrene adducts at lung cancer mutational hotspots in p53. Science 1996;274:430–2. [DOI] [PubMed] [Google Scholar]
- 33.Doll R, Peto R. The causes of cancer: quantitative estimation of avoidable risks of cancer in United States today. J Natl Cancer Inst 1981;66:1191–308. [PubMed] [Google Scholar]
- 34.Kabat GC, Chang CJ, Wynder EL. The role of tobacco, alcohol use, and body mass index in oral and pharyngeal cancer. Int J Epidemiol 1994;23:1137–44. [DOI] [PubMed] [Google Scholar]
- 35.Cruz IBF, Snijders PJF, Steenbergen RDM, et al. Age-dependence of human papillomavirus DNA presence in oral squamous cell carcinomas. Eur J Cancer B Oral Oncol 1996;32B:55–62. [DOI] [PubMed] [Google Scholar]
- 36.Nylander K, Schildt EB, Eriksson M, et al. A non-random deletion in the p53 gene in oral squamous cell carcinoma. Br J Cancer 1996;73:1381–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahuja N, Issa J-P J. Aging, methylation and cancer. Histol Histopatol 2000;15:835–42. [DOI] [PubMed] [Google Scholar]