Abstract
Background: Retinoblastoma is the most common intraocular malignancy in childhood and is responsible for approximately 1% of all deaths caused by childhood cancer.
Aims/methods: Comparative genomic hybridisation was performed on 13 consecutive, histologically confirmed retinoblastomas to analyse patterns of chromosomal changes and correlate these to clinicopathological variables. Six cases were hereditary and seven cases were sporadic.
Results: In 11 of the 13 tumours chromosomal abnormalities were detected, most frequently gains. Frequent chromosomal gains concerned 6p (46%), 1q (38%), 2p, 9q (30%), 5p, 7q, 10q, 17q, and 20q (23%). Frequent losses occurred at Xq (46%), 13q14, 16q, and 4q (23%). High level copy number gains were found at 5p15 and 6p11–12. A loss at 13q14 occurred in three cases only. Relatively few events occurred in the hereditary cases (27) compared with the non-hereditary cases (70 events). The number of chromosomal aberrations in these 13 retinoblastomas showed a bimodal distribution. Seven tumours showed less than four chromosomal aberrations, falling into a low level chromosomal instability (CIN) group, and six tumours showed at least eight aberrations, falling into a high level CIN group. In the low level CIN group the mean age was half that seen in the high level CIN group, there were less male patients, and there were more hereditary and bilateral cases. Microsatellite instability was not detected in either of the two groups.
Conclusion: Despite the complex pattern of genetic changes in retinoblastomas, certain chromosomal regions appear to be affected preferentially. On the basis of the number of genetic events, retinoblastomas can be divided in low and a high level chromosomal instability groups, which have striking differences in clinical presentation.
Keywords: retinoblastoma, ocular tumour, chromosome, chromosomal instability, comparative genomic hybridisation
Retinoblastoma is the most common intraocular malignancy in childhood, being responsible for approximately 1% of all deaths caused by childhood cancer and for about 5% of blindness in children. According to the “two hit” hypothesis of Knudson,1 two complementary tumour inducing genetic events are necessary for retinoblast transformation. The first genetic change can occur in a germ cell (hereditary) or retinoblast (non-hereditary). It is only when the second genetic alteration occurs in a somatic target cell (a retinoblast), already carrying a “first hit”, that malignant transformation will occur.
Approximately 40% of patients with retinoblastoma have the hereditary form of the disease in which the first of two causative retinoblastoma gene (Rb) mutations is present in most or all cells in the body.2 Simple deletion of 13q is seen most often, although mosaicism of 13q deletion, ring chromosome 13, and interchanges between chromosome 13 and other chromosomes are also possible.3 These patients have an 85–90% risk of bilateral ocular disease, and a lifetime predisposition to non-ocular cancers throughout the body. The trait is transmitted in an autosomal-dominant fashion with 80–90% penetrance, giving children of parents with a hereditary retinoblastoma a 45% risk of developing retinoblastoma.2 The other 60% of patients with retinoblastoma have the non-hereditary (sporadic) form of retinoblastoma, which requires two subsequent hits to inactivate the Rb gene in a retinoblast. This sporadic form is unilateral, not transmittable, and does not predispose the patient second primary tumours.4 In addition to Rb inactivation, other genetic changes have been detected in retinoblastomas, such as isochromosomes of both 17q and 6p, extra copies of 1q, monosomy 16, and abnormalities of the short arm of chromosome 1 (1p).5,6 Recurrent gains have recently been described at 1q, 2p, and 6p, and a loss at 16q.7–9
“According to the “two hit” hypothesis of Knudson, two complementary tumour inducing genetic events are necessary for retinoblast transformation”
Comparative genomic hybridisation (CGH) is a cytogenetic technique that allows the detection of chromosomal losses (such as monosomies and deletions) and gains (such as trisomies and gene amplifications) in all tumour chromosomes in a single experiment, using only a small amount of tumour material, and without the need for cell culturing.10 The onset of genomic instability is a key step in carcinogenesis, and in colorectal cancer—for example, two major types of genomic instability have been proposed.11 Genomic instability at the chromosomal level gives rise to frank aneuploidy. Alternatively, genomic instability can occur at the DNA level—for example, when mismatch repair fails—causing frequent mutations (mutator phenotype), which can be demonstrated as microsatellite instability. In our present study, chromosomal instability in terms of chromosomal gains and losses was investigated by CGH in a series of 13 retinoblastomas. In addition, genomic instability at the DNA level was studied by microsatellite instability analysis.
MATERIAL AND METHODS
Patient data
We studied 13 histologically confirmed consecutive retinoblastomas from 13 patients. Table 1 shows the clinical data of these patients. There were five male and eight female patients, with a mean age of 14.2 months (range, 0–45) at date of enucleation. Six patients had a hereditary form of retinoblastoma and seven patients had a non-hereditary form. In eight patients, retinoblastoma occurred unilaterally and in five patients bilaterally. In patients with a bilateral tumour, only one tumour was studied. After enucleation, biopsies of the tumour were stored at −70°C. All 13 ocular specimens were studied histologically. According to the original histological reports, nine tumours were undifferentiated, whereas four tumours showed some degree of differentiation, as assessed by the presence of Flexner-Wintersteiner rosettes; in two tumours, superficial tumour infiltration in the optic nerve was present, until the level of the lamina cribrosa. None of the cases showed invasion of the choroidea.
Table 1.
Clinical data of the 13 patients with retinoblastoma
| No. | Age at diagnosis | Sex | H/N | Site | Grade of differentiation | Ingrowth in optic nerve | Follow up |
| 1 | 16 months | M | N | Uni | UD | – | NED 8 years |
| 2 | Birth | M | N | Uni | UD | – | Died after 2 months |
| 3 | 6 months | F | H | Bi | WD | – | NED, 8 years |
| 4 | 1 month | M | H | Bi | WD | – | Secondary pinealoblastoma, died 41 months |
| 5 | 45 months | F | N | Uni | WD | – | NED, 7 years |
| 6 | 4 months | M | H | Bi | UD | – | NED, 3 years |
| 7 | 6 months | F | N | Uni | UD | + | NED, 3 years |
| 8 | 12 months | M | N | Uni | UD | + | NED, 3 years |
| 9 | 5 days | F | H | Bi | WD | – | NED, 2 years |
| 10 | 38 months | F | N | Uni | UD | – | NED, 2 years |
| 11 | 23 months | F | N | Uni | UD | – | NED, 1 year |
| 12 | 25 months | F | H | Uni | UD | – | NED, 1 year |
| 13 | 9 months | F | H | Bi | UD | – | NED, 1 year |
Bi, bilateral; F, female; H, hereditary; M, male; N, non-hereditary; NED, no evidence of disease; UD, undifferentiated; Uni, unilateral; WD, well differentiated.
Tissue
Normal DNA for CGH was obtained from peripheral blood lymphocytes of a healthy female donor, and tumour DNA was extracted from 20 sections (10 μm) of frozen biopsy specimens, using the Qiagen DNA isolation kit, according to the manufacturer’s recommendations.
Comparative genomic hybridisation
CGH was performed as described previously.12 In brief, normal metaphase spreads were obtained by culturing lymphocytes from a healthy female donor, according to standard procedures.
Tumour DNA was labelled with biotin-16-dUTP (Roche/Boehringer, Mannheim, Germany) and the reference DNA with digoxigenin-11-dUTP (Roche/Boehringer) by nick translation. The concentration of DNAse I was adjusted so that the length of the labelled DNA fragments was 0.5–1.5 kb.
The hybridisation of the DNA on metaphase spreads was performed as follows: 300 ng of biotin labelled tumour and 300 ng of digoxigenin labelled normal DNA, together with 40 μg unlabelled Cot-1 DNA (Gibco BRL, Breda, the Netherlands), were ethanol precipitated and the pellet was dissolved in 6 μl 50% formamide, 10% dextrane sulfate in 2× saline sodium citrate (SSC; 0.3M sodium chloride, 0.03M sodium citrate, pH 7.0). This probe mixture was denatured at 80°C for 10 minutes. Normal metaphase slides were denatured at 72°C for six minutes in a coplin jar containing 70% formamide in 2× SSC, pH 7.0. The probe mixture was mounted on to the slides, sealed with a cover slip, and hybridised for three days in a humid incubator at 40°C.
The slides were washed for five minutes in 2× SSC at room temperature, three times for five minutes in 0.1× SSC at 45°C, and once for five minutes in 0.05% Tween 20 in TN (100mM Tris/HCl, pH 7.6, 150mM sodium chloride). The slides were then pre-incubated for 10 minutes at 40°C in blocking solution (0.5% blocking reagent (Roche/Boehringer) in TN), followed by a 60 minute incubation at 40°C with 12.5 μg/ml fluorescein isothiocyanate (FITC) conjugated avidin (Sigma Aldrich, Zwijndrecht, Netherlands) and 4 μg/ml tetramethyl rhodamine isothiocyanate (TRITC) conjugated sheep antidigoxigenin (Roche/Boehringer) in blocking solution. The slides were washed three times for five minutes at room temperature in 0.05% Tween 20 in TN and dried in an ethanol series. Finally, the slides are mounted with 20 μl antifade solution (Vectashield; Vector Laboratories, Burlingame, California, USA) containing 0.35 μg/μl DAPI.
The slides were analysed on a Leica DM-RA microscope (Leica, Cambridge, UK) equipped with filter sets for DAPI, FITC, and TRITC, using a ×100 objective. The images were captured with a monochrome CCD camera (Cohu 4913; Cohu Inc, San Diego, California, USA) and processed by Cytovision CGH software (Cyto Vision 3.5; Applied Imaging, Newcastle upon Tyne, UK). The average green to red fluorescence ratios of chromosomes of 10 to 15 well selected metaphases were plotted along ideograms of the corresponding chromosomes, together with the 95% confidence interval. Deviations from normal were interpreted as gains and losses when the 95% confidence interval did not contain 1.0. Small, distinct loci showing a green to red fluorescence ratio greater than 1.5 were regarded as high level amplifications. Chromosomes 19, 1p32-ter, and 16p were excluded from the analysis because the presence of repetitive sequences may interfere with a reliable interpretation of the green to red fluorescence ratio in these areas.
Microsatellite instability
Microsatellite instability was studied using the Roche HNPCC microsatellite instability test. The reference panel markers used were: BAT25, BAT26, D2S123, D5S346, and D17S250.
Normal (non-tumour) tissue from the enucleated eye of each patient was used as a control.
Statistics
The Mann-Whitney U test was used for comparing means of continuous variables between the two groups. For significance of differences in the distribution of categorical variables, cross tables were analysed with the two sided Fisher exact test; p values < 0.05 were considered significant.
RESULTS
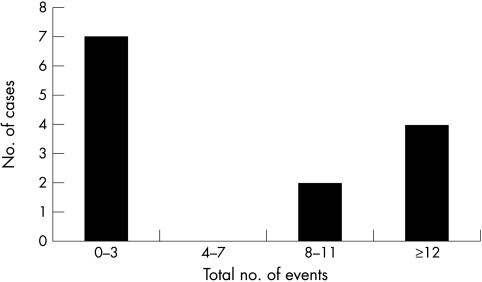
CGH analysis revealed chromosomal abnormalities in 11 of 13 cases. Table 2 lists the results for individual retinoblastomas and fig 1 shows an overview of the affected regions. As shown in fig 2, the number of chromosomal aberrations showed a bimodal frequency distribution. In one group of patients with a low level of chromosomal instability (CIN), tumours had only none to three events, whereas in the group with a high level of chromosomal instability there were eight or more events. Table 3 summarises the details on these two groups. The mean age in the low level CIN group is about half that of the high level group (p = 0.07). In the low level CIN group, there were fewer male patients, and more hereditary and bilateral cases. Because of the small number of case, these differences were not significant.
Table 2.
Chromosomal aberrations detected by comparative genomic hybridisation in 13 retinoblastomas
| No. | Hereditary (H) or non-hereditary (N) | CGH gains | CGH losses |
| 1 | N | 9q, Xp, Xq | – |
| 2 | N | 4q, 5p, 5q, 6q, 8q, 9p, 10q, 11p, 12q, 13q, 14q, 18q | – |
| 3 | H | 6p | Xq |
| 4 | H | – | – |
| 5 | N | 1q, 4p, 5p, 6p, 6q, 17q, 18p, 18q, 21q (5p15 amplif) | 8q, 10q, 13q, 15q, 16p, 16q, 18q, Xp, Xq |
| 6 | H | 8q | Xq |
| 7 | N | 6p (6p11–12 amplif) | 13q |
| 8 | N | 2p, 8p, 10q | – |
| 9 | H | – | – |
| 10 | N | 1q, 6p, 7p, 7q, 9q, 10p, 10q, 11p, 11q, 13q, 17q, 20q, 22q | 3p, 3q, 4p, 4q, 8p, 8q, 14q, Xp, Xq |
| 11 | N | 1q, 2p, 6p, 7q, 9q | 4q, 10q, 16q, Xq |
| 12 | H | 1q, 2p, 5p, 6p, 11q, 20q | 4q, 13q, 16p, 16q, Xq |
| 13 | H | 1p, 1q, 2p, 7q, 9q, 12q, 17q, 20q | 17p |
The figures in bold are high level copy number gains.
Figure 1.
Overview of affected chromosomal regions in the 13 retinoblastomas as detected by comparative genomic hybridisation. Lines left of each chromosome represent losses, and lines on the right represent gains. Thick solid lines represent high level copy number gains (amplifications).
Figure 2.
The number of chromosomal aberrations detected by comparative genomic hybridisation in 13 retinoblastomas shows a bimodal distribution, with seven cases showing low level chromosomal instability (CIN) and six cases showing high level CIN.
Table 3.
Comparison of comparative genomic hybridisation results and clinicopathological variables between retinoblastomas with low level and those with high level chromosomal instability (CIN)
| Low level CIN (<6 events) n=7 | High level CIN (≥6 events) n=6 | p Value | |
| Events (mean) | 1.7 | 14.2 | 0.02 |
| Gains (mean) | 1.3 | 9.5 | 0.03 |
| Losses (mean) | 0.4 | 4.7 | 0.02 |
| Age (mean, in months) | 10.1 | 23.3 | NS |
| Male to female ratio | 1.3:1 | 5:1 | NS |
| Hereditary (%) | 57 | 33 | NS |
| Bilateral (%) | 57 | 17 | NS |
| Undifferentiated (%) | 57 | 83 | NS |
| Death of disease (%) | 14 | 17 | NS |
NS, not significant.
Overall, gains were seen more frequently than losses, on average 5.0 and 2.4 for each case, respectively. A mean of 7.5 events (range, 0–14) was found. The most frequent losses were at Xq (46%), 13q14, 16q, and 4q (23%). Frequent gains were found at 6p (46%), 1q (38%), 2p and 9q (30%), 5p, 7q, 10q, 17q, and 20q (23%). High level copy number gains were detected in two cases, at chromosome regions 5p15 and 6p11–12, respectively.
A 13q loss, very localised at band 13q14 only, occurred in only three cases (one of which was hereditary whereas the other two were non-hereditary) and a 13q gain was seen in two cases (both of them being non-hereditary).
In the six hereditary cases, 27 events occurred (mean, 4.5)—19 gains and eight losses—whereas in the seven non-hereditary cases there were 70 events (mean, 10)—47 gains and 23 losses. However, these differences also were not significant. Four patients had a well differentiated tumour. In two of these tumours no events occurred. In one patient, only a gain at 6p and a loss at Xq were found. In the fourth patient, nine gains and nine losses were detected. This means that most patients with a well differentiated tumour belonged to the low level CIN group.
Five (hereditary) patients had a bilateral tumour. In two of these tumours no events were found, in two other patients, two events each (gains at 6p or 8q, losses at Xq) were found, and in the fifth patient 10 gains and one loss were found. Most patients with a hereditary, bilateral retinoblastoma belonged to the low level CIN group.
Overall, two patients (cases 2 and 4) died of disease during an average follow up period of 39.3 months (range, 2–96). The first patient still had residual disease after surgery with outgrowth of the tumour at the next visit. Palliative treatment was instituted on request of the parents. The tumour of this patient had 12 gains. The other patient developed a secondary tumour (a pinealoblastoma) of which he died 41 months after detection of the retinoblastoma. The retinoblastoma of this patient showed no detectable chromosomal aberrations.
In none of the 13 retinoblastoma patients was microsatellite instability detected.
DISCUSSION
CGH is the method of choice for analysing chromosomal instability. It may serve as a first line analysis in tumour genetics, pointing to regions of interest for further, more detailed, molecular genetic studies, such as fluorescence in situ hybridisation, loss of heterozygosity analysis, and DNA sequencing. It is a relatively fast screening technique that can point at specific chromosomal regions that might play a role in the pathogenesis or progression of tumours.12 Until now, few CGH studies have been reported in retinoblastoma.7–9
With respect to specific chromosomal changes, Mairal and colleagues7 reported over representation of 1q, 2p, and 6p, and loss of 9p, 13q14, and 16q. In addition, they detected high level copy number gains at 2p23–25 and 1q21. Similar findings have been reported by others.4,7–9 Our results are partly in agreement with this (for example, 1q, 2p, 6p, 13q14), although there are also differences (for example, gain at 9q, loss at Xq).
In our study, we found a loss at 13q14 (the site of the Rb gene) in only three patients; two of these patients had a non-hereditary retinoblastoma and one a hereditary retinoblastoma. Two other patients had a gain at 13q14. The finding of a normal chromosome 13 in patients with retinoblastoma has been reported before. In these patients, changes in other chromosomes such as chromosome 1 and 17 have been reported.6,13
Although in absolute numbers our series was only small (13 patients), given the low incidence of retinoblastoma, it was still a valuable study. According to the number of chromosomal aberrations found, two groups could be distinguished, one group with low level CIN and one group with high level CIN. Even though the groups are small, some striking differences were seen. Patients in the low level CIN group were younger, there were more male patients, and more hereditary, bilateral tumours were seen than in the high level CIN group. Similar findings were reported recently by Herzog et al,8 who found, in a study of unilateral retinoblastomas, that older children (36.2–84.1 months) had a higher level of CIN than younger children (2.0–9.8 months).
“In our study, we found a loss at 13q14 (the site of the Rb gene) in only three patients”
Cytogenetic analysis of retinoblastomas with regard to chromosome 6 has been reported by Cano et al.14. In 14 of the 34 tumours studied, additional copies of 6p were found. The nature of these 6p imbalances were isochromosome 6p, translocations of 6p to other chromosomes, tetrasomy 6, and additional derived 6q. It was suggested that the presence of additional copies of 6p is associated with an undifferentiated histology and invasion of the optic nerve, which are both indicators of a bad prognosis. In our study, six of the 13 patients had a gain at 6p. Four of these patients had an undifferentiated tumour and in only one case there was (superficial) tumour infiltration in the optic nerve (patient 7). Nevertheless, all six patients are alive without evidence of disease.
Take home messages.
The pattern of genetic changes in retinoblastomas is extremely complex
Nonetheless, certain chromosomal regions appear to be affected preferentially
Relatively few events occurred in the hereditary cases (27) compared with the non-hereditary cases (70 events)
On the basis of the number of genetic events, retinoblastomas can be divided in low and a high level chromosomal instability groups, which have striking differences in clinical presentation
In the low level CIN group the mean age was half that seen in the high level CIN group, there were less male patients, and there were more hereditary and bilateral cases
Microsatellite instability was not detected in either of the two groups
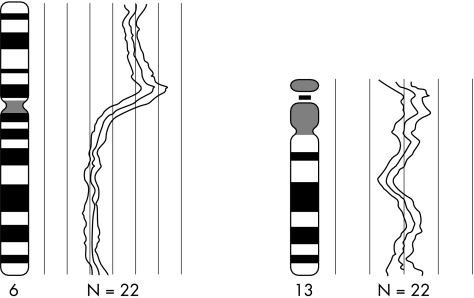
Six of the 13 tumours in our present study had a gain at 6p21.3–22, which is in line with the observations of Chen et al.9 In addition, we found high level amplification at 6p12.3 in one tumour (fig 3), suggesting an important gene is present at this site. The gain at 2p could correspond to the NMYC gene, which has been reported to be of prognostic relevance in neuroblastoma, another embryonic neuroectodermal tumour.15,16 However, in our study (and in other studies) no relation could be found between the over representation of 2p and prognosis in retinoblastoma.
Figure 3.
Green to red fluorescence ratios of chromosomes 6 and 13 (both measured as an average over 22 hybridised chromosomes) of retinoblastoma case number seven. These two chromosomes carried the only abnormalities present in this tumour, showing a gain of the complete short arm of chromosome 6, including a small region with amplification on bands 6p11–12, and a small loss on 13q14–21, which is the site of the retinoblastoma locus.
The bimodal distribution of chromosomal changes in retinoblastomas is an intriguing observation, which could point to fundamentally different mechanisms of genomic instability in the high and low level CIN tumours. In colorectal cancer a similar situation occurs, where microsatellite unstable tumours with a failing DNA mismatch repair system show only minor chromosomal changes in contrast to microsatellite stable tumours, which are often frankly aneuploid. However, in our present study, microsatellite instability was detected in none of the retinoblastomas, either low or high level CIN, indicating that other mechanisms may confer genomic instability in low level CIN retinoblastomas. In addition, CIN status (either low or high level) was associated with several clinicopathological variables. Low level CIN retinoblastomas were more frequently hereditary and bilateral, whereas high level CIN tumours occurred more frequently in older, male patients, and were more often undifferentiated.
These observations support the hypothesis that two biologically different categories of retinoblastomas exist, which seem to have a different clinical behaviour.
Acknowledgments
We thank R Broekhuizen and C Postma for their technical assistance.
Abbreviations
CGH, comparative genomic hybridisation
CIN, chromosomal instability
FITC, fluorescein isothiocyanate
Rb, retinoblastoma gene
SCC, saline sodium citrate
TRITC, tetramethyl rhodamine isothiocyanate
REFERENCES
- 1.Knudson AG. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A 1971;68:820–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harbour JW. Overview of RB gene mutations in patients with retinoblastoma. Implications for clinical genetic screening. Ophthalmology 1998;105:1442–7. [DOI] [PubMed] [Google Scholar]
- 3.Mao W, Lin X, Ma Q, et al. Lymphocyte chromosome survey in 80 patients with retinoblastoma. Eye Science 1989;5:7–13. [PubMed] [Google Scholar]
- 4.Potluri VR, Helson L, Ellsworth RM, et al. Chromosomal abnormalities in human retinoblastoma. A review. Cancer 1986;58:663–71. [DOI] [PubMed] [Google Scholar]
- 5.Chaum E, Ellsworth RM, Abramson DH, et al. Cytogenetic analysis of retinoblastoma: evidence for multifocal origin and in vivo gene amplification. Cytogenet Cell Genet 1984;38:82–91. [DOI] [PubMed] [Google Scholar]
- 6.Gardner HA, Gallie BL, Knight LA, et al. Multiple karyotypic changes in retinoblastoma tumor cells: normal chromosome no. 13 in most tumors. Cancer Genet Cytogenet 1982;6:201–11. [DOI] [PubMed] [Google Scholar]
- 7.Mairal A, Pinglier E, Gilbert E, et al. Detection of chromosome imbalances in retinoblastoma by karyotypic analysis coupled to comparative genomic hybridization. Genes Chromosomes Cancer 2000;28:370–9. [PubMed] [Google Scholar]
- 8.Herzog S, Lohmann DR, Buiting K, et al. Marked differences in unilateral isolated retinoblastomas from young and older children studied by comparative genomic hybridization. Hum Genet 2001;108:98–104 [DOI] [PubMed] [Google Scholar]
- 9.Chen D, Gallie BL, Squire JA. Minimal regions of chromosomal imbalance in retinoblastoma detected by comparative genomic hybridization. Cancer Genet Cytogenet 2001;129:57–63. [DOI] [PubMed] [Google Scholar]
- 10.Hermsen MAJA, Meijer GA, Baak JPA, et al. Comparative genomic hybridisation: a new tool in cancer pathology. Hum Pathol 1996;27:342–9. [DOI] [PubMed] [Google Scholar]
- 11.Cahill DP, Kinzler KW, Vogelstein B, et al. Genetic instability and darwinian selection in tumours. Trends Cell Biol 1999;9:57–60. [PubMed] [Google Scholar]
- 12.Weiss MM, Hermsen MAJA, Meijer GA, et al. Comparative genomic hybridisation. Mol Pathol 1999;52:243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Squire J, Gallie BL, Phillips RA. A detailed analysis of chromosomal changes in heritable and non-heritable retinoblastoma. Hum Genet 1985;70:291–301. [DOI] [PubMed] [Google Scholar]
- 14.Cano J, Oliveros O, Yunis E. Phenotype variants, malignancy, and additional copies of 6p in retinoblastoma. Cancer Genet Cytogenet 1994;76:112–15. [DOI] [PubMed] [Google Scholar]
- 15.Doz F, Peter M, Schleiermacher G, et al. N-MYC amplification, loss of heterozygosity on the short arm of chromosome 1 and DNA ploidy in retinoblastoma. Eur J Cancer 1996;32:645–9. [DOI] [PubMed] [Google Scholar]
- 16.Tsuda H, Shimosato Y, Upton MP, et al. Retrospective study on amplification of N-myc and c-myc genes in pediatric solid tumors and its association with prognosis and tumor differentiation. Lab Invest 1988;59:321–7. [PubMed] [Google Scholar]