Abstract
Rapid advances in molecular biological techniques have made it possible to study disease pathogenesis at a genomic level. T cell receptor (TCR) gene rearrangement is an important event in T cell ontogeny that enables T cells to recognise antigens specifically, and any dysregulation in this complex yet highly regulated process may result in disease. Using techniques such as Southern blot hybridisation, polymerase chain reaction, and flow cytometry it has been possible to characterise T cell proliferations in malignancy and in diseases where T cells have been implicated in the pathogenesis. The main aim of this article is to discuss briefly the process of TCR gene rearrangement and highlight the disorders in which expansions or clonal proliferations of T cells have been recognised. It will also describe various methods that are currently used to study T cell populations in body fluids and tissue, their diagnostic role, and current limitations of the methodology.
Keywords: T cells, T cell receptor gene rearrangement, clonality, polymerase chain reaction, flow cytometry, T cell receptor repertoire
The T cell receptor (TCR) is a membrane bound heterodimer composed of two polypeptide chains (αβ or γδ) linked by a disulfide bond and associated with a non-polymorphic cytoplasmic membrane bound complex of proteins collectively known as CD3.1–3 In the peripheral blood, most T cells express the αβ receptor and up to 15% express the γδ receptor.4–6 The extracellular domains of the TCR consist of a constant (C) domain and a variable (V) domain. Hypervariable regions or complementarity determining regions (CDR) encoded within the TCR V domains contact the peptide antigen and the major histocompatibility (MHC) molecules and therefore confer the specificity of the TCR. Furthermore, T cells are MHC restricted in that CD4 positive or helper T cells recognise peptides bound to MHC class II molecules and CD8 positive or suppressor T cells recognise peptides bound to MHC class I molecules.
THE TCR LOCI
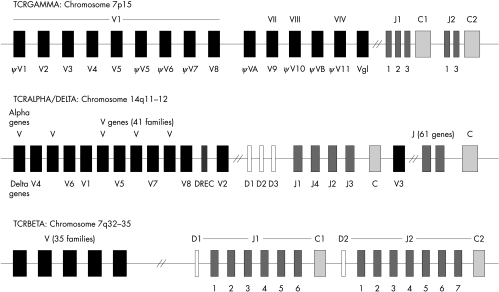
The TCR genes are located on chromosomes 7 and 14. The genes encoding the TCRα (TCRD) chain are located within the TCRα (TCRA) loci on chromosome 14q11–12, whereas the TCRβ (TCRB) and TCRγ (TCRG) genes are located at chromosomal positions 7q32–35 and 7p15, respectively (fig 1; table 1).8–14 The four genes are composed of C and V regions which are assembled together during thymic ontogeny by somatic recombination.15 Briefly, the V domains in the TCRA and TCRG chains are assembled from V and J gene segments whereas the TCRB and TCRD chains are assembled from V, diversity (D), and J segments. The multiple non-contiguous V, D, and J segments undergo rearrangement during T cell development to form complete V domain exons, which are responsible for antigen recognition. Diversity of the T cell repertoire is not only a consequence of different V, D, J recombinations but also results from nucleotide insertion and deletion at junctional sites (V–J, D–J, and V–D), thereby increasing diversity for recognition of a vast array of antigens.
Figure 1.
Germline organisation of the T cell receptor (TCR) genes. The TCRA, TCRB, and TCRG loci are located on the chromosomes at positions 14q11–12, 7q32–35, and 7p15, respectively. The TCRD loci are also situated on chromosome 14, within the TCRA loci. The gene segments are designated using the World Health Organisation standards for TCR nomenclature.7 The shaded boxes indicate the type of gene segment: V, variable (black); D, diversity (white); J, joining (dark grey); C, constant (light grey).
Table 1.
Genes encoding the T cell receptor (TCR) chains
| TCR gene | Chromosome | V segments | D segments | J segments | C segments |
| α (A) | 14q11–12 | 70 | 0 | 61 | 1 |
| β (B) | 7q32–35 | 67 | 2 | 13 | 2 |
| γ (G) | 7p15 | 14 | 0 | 5 | 2 |
| δ (D) | 14q11–12 | 8 | 3 | 4 | 1 |
C, constant; D, diversity; J, joining; V, variable.
“The multiple non-contiguous V, D, and J segments undergo rearrangement during T cell development to form complete V domain exons, which are responsible for antigen recognition”
The TCRA genes consist of 70 V gene segments, divided into 41 families16–19 and at least 61 J segments spread over 800 kb upstream from a single C gene. The 61 J gene segments, consisting of 50 functional J genes, are not clustered and are located at over 65 kb at various distances 5′ of the C gene.14,20
The TCRB locus comprises approximately 67 V genes of which 47 are functional and 19 are pseudogenes.21 The functional TCRBV families are grouped into 23 families based upon greater than 75% nucleotide sequence identity.22 Two C genes are associated with upstream clusters of J genes and a single D gene (D1 or D2) and are known as the D–J–C clusters. There are 13 J genes, designated J1S1 to J1S6 and J2S1 to J2S7.23
TCRG consists of up to 14 V segments and a duplicated J–C cluster consisting of three J genes (J1S1, J1S2, and J1S3) and one C gene, and two J genes (J2S1 and J2S3) and one C gene, respectively. The 14 V genes belong to four families. The TCRGVI family consists of nine members, five of which are functional and four of which are pseuodogenes, and the remaining three families (II, III, and IV) each consist of a single gene. The remaining two genes are VA, which lies 5′ to the TCRGVII, and VB, which lies 5′ to TCRGVIII.24,25
The TCRD locus is located within the TCRA locus between the AV and AJ gene segments. It is flanked by TCRD deleting elements (ιJA and δREC) such that rearrangements of the deleting elements to each other or rearrangement of AV to AJ gene segments causes deletion of the intervening TCRD gene locus. The germline encoded TCRD locus consists of eight V segments, three D segments, four J segments, and a single C segment.14 TCRDV2 is located immediately upstream of the D and J segments, whereas TCRDV3 is located downstream of the TCRDJ cluster in an inverted orientation. TCRDV1 is located among the TCRAV genes 360 kb upstream of the TCRDC gene cluster, whereas the remaining five TCRDV genes (described as TCRAV/DV) are also found among the TCRAV genes.14
V(D)J RECOMBINATION
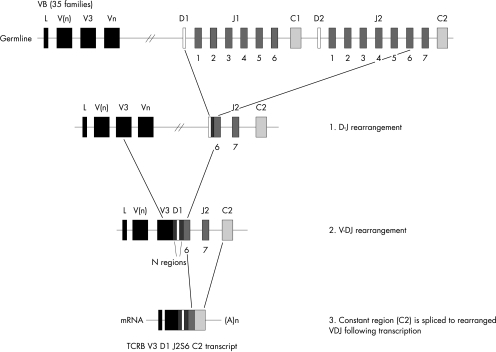
The T lymphocyte has a unique ability to recognise a vast array of non-self antigens and rearrangement of multiple V, D, and J gene segments accounts for the diversity and specificity necessary for this function. During TCR gene rearrangement the V, D (only in TCRB and TCRD chain rearrangement), and J segments are brought together into a continuous V–D–J coding block (fig 2).15 These events are mediated by recombination activating genes, RAG1 and RAG2,26,27 and the enzyme terminal deoxynucleotidyl transferase (TdT).28 The TCR gene segments, as with the immunoglobulin genes, are flanked by recombination signal sequences (RSS), which act as recognition sites for the VDJ recombinase and mark the points at which somatic recombination occurs. The RSS comprises two conserved sequences, a heptamer (CACA/TGTG) separated by 12 or 23 random nucleotides from a consensus nonamer sequence (ACAAAAACC or GGTTTTTGT). Recombination may only take place between gene segments flanked by different spacers,29,30 and this phenomenon is known as the “12–23” rule.31 Therefore, V gene segments flanked 3′ by a 23 bp spacer can recombine with D segments or directly with J segments, both of which are flanked 5′ by a 12 bp spacer. The rearrangement of TCRB and TCRD is a two step process, in which a D gene first recombines with a J gene and then a V gene recombines with the D–J block (fig 2). Because of the absence of D genes in TCRA and TCRG, only V–J recombination occurs in these two chains.
Figure 2.
T cell receptor β gene (TCRB) rearrangement. The RAG mediated recombination of the TCRB locus is a two step process in which a diversity (D) gene segment first recombines with a joining (J) segment, followed by the recombination of a variable (V) gene to the DJ block. The rearranged VDJ segment is then spliced post transcriptionally to the C transcript.
“Diversity of the T cell repertoire is not only a consequence of different V, D, J recombinations but also results from nucleotide insertion and deletion at junctional sites”
During these recombination events, diversity at junctional regions is further increased by the incorporation of template dependent palindromic “P” nucleotides,32,33 incorporation of template independent GC rich “N” nucleotides by TdT,34,35 deletion of gene encoded nucleotides by exonucleolytic activity, and imprecise joining of gene segments. The junctional region (V(D)J) encodes the hypervariable CDR3.36 The CDRs (CDR1 and CDR2 within the V genes and CDR3 at the V(D)J junction) are regions of greatest sequence variability, which constitute the binding site for antigen with the CDR3 positioned at the centre of the antigen binding site for direct contact with the MHC bound peptide. Therefore, diversity of the T cell repertoire is not only a consequence of different V, D, J recombinations but also results from nucleotide insertion and deletion at junctional sites, thereby increasing diversity for recognition of a vast array of antigens. Furthermore, diversity is also created by the combinatorial association of individual TCRA and TCRB chains, and TCRG and TCRD chains. It is estimated that 1015 unique TCRs can be produced in this way.31
THE HUMAN T CELL REPERTOIRE
There are several intrinsic and extrinsic factors that influence the expression of the TCR repertoire in humans. These include the human MHC (HLA) system37–39 and TCR gene polymorphisms,25,40–47 in addition to environmental factors, such as infection48–52 and superantigens.53–56 In normal individuals the TCR repertoire is stable and polyclonal. Whereas clonal populations are the hallmark of malignancy, clonal or oligoclonal populations of T and B cells may occur in non-malignant conditions including: human immunodeficiency virus (HIV) and Epstein-Barr virus (EBV) infections with specificity for virus,48,57–59 in the elderly,60,61 in autoimmunity,62–65 in common variable immunodeficiency (CVID),66,67 and in severe combined immunodeficiency (SC1D), where RAG1 or RAG2 gene defects allow only a limited number of successful antigen receptor gene rearrangements, as occurs in patients with the Omenn’s SCID variant.68
ANALYSIS OF TCRVB REPERTOIRE
Phenotype: flow cytometric analyses
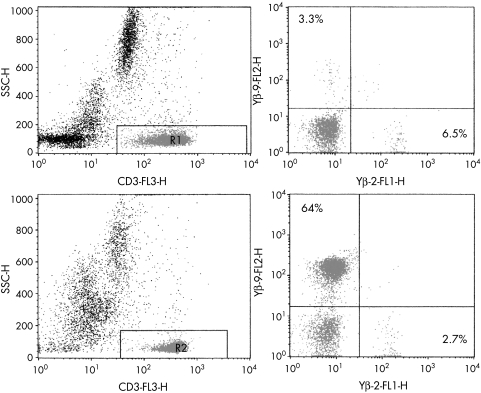
Flow cytometry offers a quick, relatively inexpensive method of analysing the TCR repertoire, which is reproducible between laboratories and allows a quantitative evaluation of the expression of different V genes within different T cell subsets—that is, CD4+ and CD8+ cells. Whole blood can be analysed by three/four colour flow cytometry using fluorescently labelled monoclonal antibodies against TCRV family gene segments, together with anti-CD3 and anti-CD4 or anti-CD8 monoclonal antibodies (fig 3). Monoclonal antibodies are available to all the TCR chains for detection of TCRαβ and γδ positive populations. In particular, monoclonal antibodies now cover > 80% of the whole TCRVβ repertoire and reference values of the Vβ repertoire in healthy controls from different age groups are available.69 In contrast, only a few monoclonal antibodies recognise the Vα genes because of the vast number of TCRA V and J recombinations and the polymorphic nature of the α genes. TCRVβ antibody studies can be used as an initial screening method for the detection of expanded families and/or skewing of the T cell repertoire in various disease states, including suspect T cell malignancies and proliferations, and immunodeficiency (fig 3).70
Figure 3.
Representative flow cytometric T cell receptor β variable gene (TCRVB) profiles of a healthy control compared with a patient with T cell lymphoma. TCRVβ repertoire analysis was performed using triple colour analysis with CD3 in combination with two different TCRVβ antibodies for each tube. Images on the left show CD3 versus side scatter (SSC) gating to identify the T cells. Images on the right show staining using Vβ2 and Vβ9 labelled antibodies. Only small percentages of Vβ positive T cells can be identified in the healthy control (upper panel), whereas a large T cell population expressing Vβ9 (64%) was identified in the T cell lymphoma (lower panel).
Genotype: PCR methods
The advent of the polymerase chain reaction (PCR) technique has largely replaced Southern blot analysis for clonality studies because PCR offers several advantages, with the principal being that it is quicker to perform and poor quality DNA can be used. The type of PCR approach will depend on whether DNA or RNA based methods are applicable. Paraffin wax embedded material is often the only material available in routine diagnostic pathology laboratories, so protocols are restricted to DNA based methods and the amplification of small (< 250 bp) PCR fragments.71 Both DNA and RNA methods are suitable for fresh material.
Reverse transcription PCR (RT-PCR) using RNA can be used for the analysis of the expressed TCR repertoire. Suitable PCR methods include: (1) amplification of all V segments using family specific primers complementary to all TCRV genes and a constant region primer70,72 (although easy and reliable, this method at best can only provide a semiquantitative profile of TCRV usage); (2) modification of all TCR transcripts so that they can be amplified by one oligonucleotide (anchor PCR)73; and (3) the production of circular transcripts by ligation (inverse PCR).74 These last two methods are technically demanding but allow quantitative analysis of the T cell population. Such analyses have been applied to clonality studies and the evaluation of the TCR repertoire in immunodeficiency, autoimmune disease, and alloreactive T cells in allografts.75
Methods for routine clonality assessment should be simple and robust with good detection rates and sensitivity rates of ∼ 5%. Although clonality assessment can be completed using RNA and RT-PCR, analysis of DNA is more common and applicable to poor quality DNA extracted from paraffin wax embedded, formalin fixed tissue. Many methods applicable to DNA rely on family consensus primers and therefore suffer from false negativity as a result of improper annealing of PCR primers to all gene segments because of the lack of homology of the primers to template DNA. Furthermore, some approaches have used nested PCR, which may suffer from contamination problems. Presently, a European project (BIOMED-2 concerted action (PL-96–3936)) is trying to deal with the problem of false negatives common to all PCR protocols. The project has involved the design of new and specific primers for all TCR and immunoglobulin (Ig) loci and the standardised testing of all cases. This approach has already increased detection rates and sensitivity.
PCR analysis of the TCRG genes is the protocol mostly widely applied to clonality studies in clinical pathology laboratories because of the restricted germline repertoire of the TCRG genes, which limits the number of required PCR primers.76 However, the limited repertoire also results in high background amplification of similar rearrangements in normal T cells. Furthermore, TCRG gene rearrangements can be used as markers of clonality, not only in TCRγδ positive malignancies (where the rearrangements are functional) but also in TCRαβ positive malignancies, where non-functional TCRG gene rearrangements are seen as a consequence of the ordered hierarchy of TCR loci gene rearrangements.
Analysis of TCRD genes is mainly restricted to TCRγδ positive malignancies, which are fairly rare, because the TCRD genes are deleted in most TCRαβ positive T cell malignancies. However, the TCRD locus is extensively used as a target in minimal residual disease (MRD) detection in both T and B lineage acute lymphoblastic leukaemia (ALL), particularly because cross lineage TCRD gene rearrangements occur extensively in precursor B cell ALL (B-ALL).77 The TCRD is a useful target as a result of the combination of three factors, namely: (1) limited germline repertoires, (2) specific recombination patterns in different subtypes of ALL, and (3) extensive junctional diversity.78
“PCR analysis of the TCRG genes is the protocol mostly widely applied to clonality studies in clinical pathology laboratories because of the restricted germline repertoire of the TCRG genes, which limits the number of required PCR primers”
The TCRB locus is the locus of choice for establishing clonality of TCRαβ expressing tumours, which comprise more than 90% of lymphoid malignancies. DNA based methods have exploited the extensive TCRB germline repertoire with varied success.71,79 However, recently family specific BV primers have been applied to DNA as part of the BIOMED-2 programme and have achieved improved detection rates for TCRB clonality. The TCRA locus has proved too complex for suitable DNA clonal analyses.
Detection systems
TCR PCR fragments may be detected in a variety of ways to discriminate the junctional diversity between polyclonal and clonal populations. These methods include fluorescent gene scanning,80,81 heteroduplex analysis,82–84 single strand conformation polymorphism (SSCP),85 denaturing gradient gel electrophoresis (DGGE),86,87 cloning, and sequencing.88
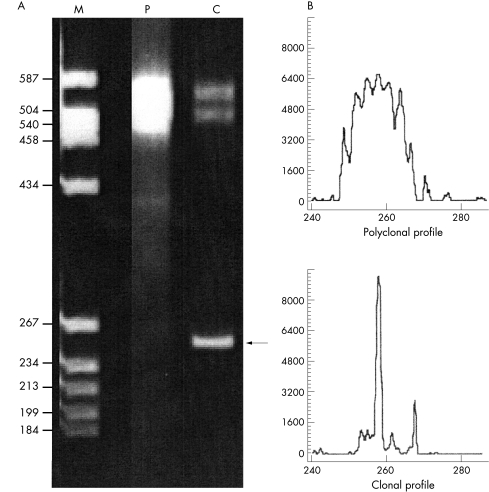
Fluorescent gene scanning involves the amplification of specific TCR genes using fluorescently labelled primers. Such analysis is used in routine clonality assessment of TCRG and TCRB genes, TCRBV family usage, and MRD studies. The resulting PCR products are run on a gene sequencer and analysed using GeneScan software (Applied Biosystems, Warrington, Cheshire, UK). Polyclonal populations with varying PCR fragment lengths demonstrate a normal Gaussian distribution visualised as a series of peaks. Clonal populations are seen as distinct peaks (fig 4).
Figure 4.
Heteroduplex and fluorescent gene scanning analyses. The same T cell receptor β gene (TCRB) polymerase chain reaction (PCR) products, amplified using the BIOMED TCR VB–JB primers (JJM van Dongen et al, unpublished data) are shown by (A) heteroduplex analysis and (B) fluorescent gene scanning. (A) M, molecular weight size marker, with sizes indicated in base pairs; P, polyclonal peripheral blood control; C, clonal T cell lymphoma control. In the polyclonal control, only heteroduplexes are visible as a smear at ∼ 500–550 bp, which results from the re-annealing of the polyclonal PCR fragments of different sizes and sequences. The clonal homoduplex PCR product is indicated by an arrow at about ∼ 259 bp, whereas the heteroduplex PCR products in the sample run behind at about 500–550 bp. (B) Relative fluorescent intensities (ordinate) are plotted as a function of PCR fragment size (abscissa). The polyclonal control in the upper panel shows a normal Gaussian distribution of PCR products of different sizes within the appropriate size range (250–290 bp). The clonal T cell lymphoma control in the lower panel shows a dominant fluorescent peak indicative of a clonal population with identical PCR fragment sizes at ∼ 259 bp.
Heteroduplex analysis (and the less frequently used SSCP analysis), rely on conformational changes caused by mismatch at the junctional region visualised by retardation on a gel matrix. True clonal PCR products give rise to homoduplexes, whereas polyclonal PCR products form heteroduplexes upon renaturation. Because of differences in conformation, the clonal homoduplexes (PCR products with identical junctional regions from clonal populations) migrate more rapidly through the gel and are seen as intense clonal bands, whereas heteroduplexes with less perfectly matched sequences (PCR products with heterogeneous junctional regions) appear as ill defined smears running behind the homoduplex (fig 4). DGGE allows rapid screening for single base changes at the CDR3 region. This technique is based on the migration of double stranded DNA through a polyacrylamide gel containing increasing concentrations of a denaturing agent.89
Cloning and sequencing provides perhaps the best method of assessing clonality within T cell populations, allowing a detailed analysis of the CDR3 region, including V, D, and J gene identification (using internet databases such as www.ncbi.nim.nih.gov/BLAST and http://imgt.cines.fr.8104), in addition to the identification of non-template encoded nucleotides; however, this technique is both costly and labour intensive.
Cross lineage rearrangements
It was originally hoped that clonal gene rearrangements of TCR and Ig loci would accurately assign T or B cell lineages to clonal lymphoid proliferations, respectively; however, it was soon evident that cross lineage rearrangements—that is, TCR and IgH gene rearrangements in B and T cell lymphoid neoplasms, respectively—occurred.90 Both Ig and TCR genes are flanked by RSS and therefore inappropriate gene rearrangements reflect gene segments that are accessible for recombination, but which do not reflect the final lineage commitment of the tumour cells. Approximately 25–30% of high grade lymphoid tumours and 5–10% of low grade tumours have cross lineage rearrangements: partial rearrangements (D–J recombinations) occur more frequently than complete (VDJ or VJ) rearrangements. Furthermore, cross lineage rearrangements are more common among immature lymphoid neoplasms (ALL) than mature neoplasms.77,90 Cross lineage Ig gene rearrangements are usually restricted to the Ig heavy chain gene in T cell neoplasia; Ig light chain genes have rarely been found.
THE DIAGNOSTIC ROLE OF TCR GENE REARRANGEMENT STUDIES
T cell lymphomas and leukaemias
The molecular diagnosis of non-Hodgkins lymphoma and leukaemia is based on the fact that all cells have a common clonal origin and > 98% of cases exhibit clonal rearrangements of TCR and Ig genes, reflecting their cellular origins from T or B cells, respectively.77 Molecular analyses are extremely helpful for diagnosis in cases with unusual morphological features, confusing immunophenotypic characteristics, or where a phenotypic clonal marker is lacking. Nevertheless, it should be remembered that the presence of clonal gene rearrangements does not necessarily imply malignancy because several benign or reactive inflammatory conditions may show such a pattern. Some examples include benign inflammatory disorders of skin, such as lymphomatoid papulosis and pityriasis lichenoides et varioliformis acuta, where clonal T cell expansions are evident.91–93 Conversely, the absence of a clonal gene rearrangement does not rule out malignancy because the population of malignant cells may be at a level below the sensitivity of the technique. The current consensus view is that lymphomas can only be defined by a combination of clinical, histological, and immunological data.94
Table 2 shows the incidence of clonal rearrangements in various lymphoproliferative disorders. At our centre, 60% of all lymphoproliferations referred for clonal investigations are suspect T cell lymphoproliferations, and more than 65% of these specimens are referred as formalin fixed, paraffin wax embedded biopsies. Two problematical areas of diagnosis include lymphoproliferations in skin and peripheral T cell lymphoma (PTCL).
Table 2.
Incidence of clonal gene rearrangements in T and B cell lymphomas and leukaemias
| T cell receptor | Immunoglobulin | |||||
| Disease | TCRB | TCRG | TCRD | IgH | IgK | IgL |
| Lymphoblastic T cell malignancies | ||||||
| CD3− T ALL | 84 | 85 | 76 | 16 | 0 | 0 |
| CD3+ T ALL | 96 | 100 | 58 | 6 | 0 | 0 |
| Lymphoblastic T NHL | 79 | 100 | 10 | 33 | 0 | |
| Mature T cell malignancies | ||||||
| T CLL | 84 | 97 | 3 | 0 | 0 | |
| T PLL | 100 | >90 | 25 | 7 | ||
| ATLL | 100 | 100 | 0 | 3 | 0 | 0 |
| Peripheral T NHL | 80 | 80 | 20 | 4 | 12 | 0 |
| CTLL/MF | 100 | 100 | 7 | 0 | 0 | |
| Lymphoblastic B cell malignancies | ||||||
| Null ALL | 9 | 22 | 0 | 95 | 13 | 9 |
| Common ALL | 41 | 66 | 54 | 99 | 32 | 2 |
| Pre B ALL | 23 | 44 | 20 | 100 | 8 | 20 |
| B ALL | 0 | 0 | 100 | |||
| Mature B cell malignancies | ||||||
| B CLL | 7 | 0 | 0 | 100 | 100 | 30 |
| HCL | 0 | 0 | 0 | 100 | 100 | 70 |
| B NHL | 6 | 6 | 100 | 100 | 30 | |
| Multiple myeloma | 5 | 0 | 0 | 100 | 100 | 30 |
Table adapted from van Dongen and Wolvers-Tettero77 and Gill and Gulley95.
ALL, acute lymphoblastic leukaemia; ATLL, HTLV1 positive adult T cell leukaemia/lymphoma; CLL, chronic lymphocytic leukaemia; CTLL, cutaneous T cell lymphoma; HCL, hairy cell leukaemia; MF, mycosis fungoides; NHL, non-Hodgkin’s lymphoma; PLL, prolymphocytic leukaemia.
More than 25% of referrals to our laboratory are skin lymphoproliferations. The diagnosis of lymphoproliferations in skin is a difficult task, because lymphoid infiltrates with similar histological features may represent widely disparate clinical diseases. Conversely, the histological picture associated with certain diagnoses such as mycosis fungoides can vary greatly between individuals and at different times in the clinical course of the disease. TCR and clonality studies have proved to be of immense help in the diagnosis and follow up of patients with cutaneous T cell lymphomas.91
The interpretation of clonality in PTCL can also be difficult, especially those of the angioimmunoblastic type (AILD), where the interpretation of clonality is complicated by the heterogeneity of TCR and IgH rearrangements. In addition, clonal rearrangements have been shown to regress or appear during the course of disease, suggesting that these cases harbour multiple clones similar to those found in cases with acquired immunosuppression. Although most cases exhibit TCR rearrangements, we have recently shown that functional T and B cell oligoclones can be found in up to 30% of cases.96 Our data do not support the concept of AILD as a clearly defined PTCL, but offer experimental evidence in support of the observation of Lipford et al,97 who concluded that AILD is a disease of proliferating lymphoid clones in which supervening malignant lymphomas may develop by a process of clonal selection. Figure 4 shows a malignant T cell proliferation as detected by heteroduplex analysis and fluorescent gene scanning.
“The presence of clonal gene rearrangements does not necessarily imply malignancy because several benign or reactive inflammatory conditions may show such a pattern”
During the treatment of many haematological malignancies, tumour cells may become undetectable in blood, bone marrow, etc by conventional means. However, many cases may still relapse, indicating that biological remission has not been achieved. MRD describes the presence of tumour cells at a level below that detectable by conventional methodology and provides accurate information about the efficacy of treatment regimens.77,98 Furthermore, MRD analysis can predict outcome by determining malignant cell burden during initial treatment. Most MRD assays use “real time” technology to quantitate residual disease. Such assays exploit the 5′→3′ nuclease activity of Taq polymerase to detect and quantify specific PCR products.99,100 Using this technique, the PCR product is measured at each cycle by cleavage of a fluorogenic intrinsic probe by the emission of a fluorescent signal that accumulates during the reaction. Because post-PCR processing is not necessary, quantitative data can be accumulated quickly. Real time primers and probes have been applied to patient specific junctional regions in ALL, in addition to BCR–ABL fusion transcripts in chronic myeloid leukaemia, to provide accurate and reliable data for MRD detection.101,102
Large granular lymphocytoses
The syndrome of large granular lymphocytosis (LGL) with chronic neutropenia was recognised as a distinct clinical entity in 1977. Two types of LGLs have been described on the basis of their lineages: CD3+ LGL and CD3− LGL. Whereas CD3+ LGLs express and rearrange TCRs, CD3− LGLs are composed of natural killer cells which do not rearrange their TCR or Ig genes.103,104 Although clonality can be easily demonstrated in the more common CD3+ LGL by the detection of TCR gene rearrangement, clonality is less easy to demonstrate in CD3− LGL, although clonal analysis using the lineage independent method of X inactivation has been applied to this group.105 Furthermore, the detection of a single episomal form of EBV has been used as evidence that the LGL has arisen from a single EBV infected cell.106
One of the key questions in this disorder has been whether LGL represents a “reactive” or “neoplastic” proliferation. It should also be noted that oligoclonal/clonal T cell expansions are frequently seen in the peripheral blood of normal healthy individuals. Such expansions are most prominent among CD8+ T cells and are increasingly present with age, being detectable in adults (> 35 years of age) and becoming very frequent in the elderly (> 70 years of age).60,107–109 However, such populations are not unknown in the young. We have recently described two paediatric cases presenting in the 2nd decade of life with neutropenia and sepsis. Both cases had expanded clonal populations of TCRBV13 CD8+ T cells. Both patients received cyclosporin and showed pronounced clinical improvement. In contrast, the CD4+ T cell repertoire remains largely polyclonal throughout life, although expanded CD4+/CD45RO+ populations have been detected in elderly individuals.61
Although some reports have highlighted that clonality studies and CD8 TCRVB family profiling are useful in monitoring disease progression and response to treatment in LGL, it has been suggested that the morbidity and mortality in LGL is primarily related to neutropenia rather than lymphocytosis.104 Most of our current knowledge regarding the management of these patients comes from anecdotal case reports and with the uncertainty in the diagnosis, the indication for beginning aggressive immunosuppressive treatment remains unclear.
Infection
Infections, particularly viral (EBV, cytomegalovirus (CMV), human T cell lymphotropic virus (HTLV), and HIV) and staphylococcal toxic shock syndrome, are known to induce lymphocytosis, usually polyclonal, but occasionally clonal.110–112 The lymphocytoses in CMV, EBV, and HTLV infection are usually associated with an expansion of LGLs. After EBV infection, memory clonotypic CD8+ cells persist at relatively high circulating frequencies and are necessary for maintaining a balanced host–virus relationship in normal individuals. More recently, oligoclonality has also been seen in hepatitis B virus specific CD8+ cells using HLA-A2 peptide tetramers.113
EBV is commonly associated with post transplant lymphoproliferative disease of oligoclonal B cell origin. T cell proliferations after transplantation are uncommon but such populations have been reported.114 Reactive oligoclonal populations can also occur following transplantation that represent re-population of the bone marrow, and which cannot be interpreted as re-occurrence of malignancy without reference to the original clone.
TCR family expansions have also been reported in HIV positive patients with low CD4+ cell counts, and it has been suggested that the virus per se may have a superantigen effect on the TCR.115,116 Recent studies have also shown that in the early phases of HIV infection there is a transient expansion of CD8+ T cells, which in some patients may persist for six to 30 months.117 CD8+ T cells play an important role in protection and control of HIV-1 by direct cytolysis of infected cells and by suppression of viral replication by secreting cytokines, chemokines, and CD8+ T cell antiviral factors. Although clonality and oligoclonality can be demonstrated in a proportion of these cells they represent a reactive expansion of CD8+ T cells rather than malignancy. A decline in reactive clonal CD8 cells has been linked to progression to AIDS, with a strong inverse correlation between the in vivo frequency of HIV specific clones and levels of viraemia.118
Immunodeficiency
In many primary immunodeficiency disorders a high degree of clonality/oligoclonality is present within lymphocyte populations. Restricted heterogeneity of T cell subpopulations has been described in Omenn’s syndrome, with skewing of the TCRAB repertoire and the presence of oligoclonal T cell populations.68,119 Omenn’s syndrome was first described in 1965 as a severe form of SCID, characterised by erythroderma, failure to thrive, and lymphadenopathy. High numbers of activated, anergic T cells are usually present and infiltrate skin and gut epithelium. Recent reports have described mutations in RAG1 and RAG2 genes in this syndrome.120,121 However, clinically defined Omenn’s syndrome may be a more complex entity than is currently recognised.120 We have recently studied eight children with clinically defined Omenn’s syndrome and two of the eight demonstrated RAG gene mutations. Alterations of the TCRVB repertoire were seen with five of the eight children demonstrating oligoclonal TCRV gene rearrangements. Although failure to demonstrate TCRVB clonality may arise from the lack of homology of V primers to all known V genes, these results suggest that TCRVB skewing and clonality may not be seen in all cases and may, in part, be related to the type of RAG gene mutation.123 Furthermore, oligoclonal B cell populations found in one child in the absence of clonal T cells were confirmed by gene sequencing (J L Smith et al, unpublished data, 2002). Such data suggest that Omenn’s syndrome is a more heterogeneous disease than previously recognised.
The differentiation of materno–fetal graft versus host disease from Omenn’s syndrome in patients with SCID and similar presenting features can be resolved by chimaerism studies. Although materno–foetal grafts have not been reported to show the alterations in the T cell repertoire commonly seen in patients with Omenn’s syndrome, studies for TCRG/TCRD clonality analysis may be useful. We have identified three cases of materno–fetal engraftment with clonal TCRGD populations in the blood (J L Smith et al, unpublished data, 2002).
Take home messages.
T cell receptor (TCR) gene rearrangement is an important event in T cell ontogeny that enables T cells to recognise antigens specifically, and any dysregulation in this process may result in disease
Techniques such as Southern blot hybridisation, polymerase chain reaction (PCR), and flow cytometry have enabled T cell proliferations to be characterised in malignancy and in diseases where T cells have been implicated in the pathogenesis, such as immunodeficiency, autoimmunity, and atopy
One of the most important of these applications is the diagnosis of lymphocytic disease and its differentiation from reactive lymphoproliferations
These applications are less well defined for patient monitoring but are useful for monitoring minimal residual disease (MRD) and in patients with CD8+ lymphocytoses
Methods based on real time PCR are also available for monitoring MRD after treatment or bone marrow transplantation
TCR gene studies have been diagnostically useful for the definition of immunodeficiency states, most notably, Omenn’s syndrome, and for the characterisation of T cells at disease sites in patients with autoimmune disease or allergy
However, PCR technology has a high incidence of false negativity, resulting from the use of consensus primers, so that several groups are working towards the design of better primers
Clonal or oligoclonal lymphocyte populations can also be found in other children with primary immunodeficiency. In a study of genomic DNA, extracted from blood lymphocytes from 86 children with defined and undefined immunodeficiency, and using heteroduplex analysis we have found clonal or oligoclonal populations of T and/or B cells in 37 cases. Of these 37 children, 84% had clonal TCR gene rearrangements and 35% had clonal IgH rearrangements. In a control study of 26 children matched for age and sex we did not find clonal/oligoclonal populations (J L Smith, et al unpublished data, 2002). The origin of these clonal populations is uncertain; however, from the diversity of the patient population studied it is clear that intrinsic factors alone cannot explain these results and that extrinsic factors may have an important role.121
“Although clonal rearrangements are the hallmark of lymphoid tumours, monoclonal proliferations have been reported in patients with CVID without progression”
The most common type of primary immunodeficiency in clinical practice is CVID.124 Significant differences in TCRBV gene usage has been reported in patients with CVID, and in particular those with an associated CD8+ lymphocytosis.124,125 Up to 15% of patients with CVID have lymphoadenopathy, with granulomatous lymphadenopathy occurring in 75% of all biopsies.126 The remainder, including reactive and atypical lymphoid hyperplasia, lack distinctive features and in these patients prediction of clinical outcome is highly unsatisfactory.66 Lymphomas occurring in patients with CVID are usually diffuse and are of intermediate or high grade in the working formulation. The diagnosis is complicated by the fact that lymphoid tissue from all patients with CVID can be histologically abnormal because germinal centres are frequently absent or poorly developed, so that stimulation by infection can result in histologies that are difficult to differentiate from lymphoma.127
Lymphomas of B cell type predominate in CVID, although T cell types have been reported128 The interpretation of clonality in this group of patients is complex. Although clonal rearrangements are the hallmark of lymphoid tumours, monoclonal proliferations have been reported in patients without progression. In a recent UK study, we have reviewed clonality in biopsies of reactive and lymphoma histologies. Although clearly present in lymphoma tissue, clonal populations of B cells were also found in reactive biopsies and biopsies of equivocal lymphoma histology. Apparent clonality may result from small numbers of B cells present in some biopsies, but this was not true in all biopsies of reactive histology. In CVID, clonal populations cannot be definitive for lymphoma and these findings must be interpreted with the immunohistology of the biopsy and the clinical presentation of the patient.
Clonality in autoimmunity and atopy
In many autoimmune and atopic disorders localised T cell expansions have been described in the affected tissue and subsequent clonality studies have identified increased oligoclonality within T cells at these sites. In both rheumatoid arthritis and multiple sclerosis, various different TCRAV and TCRBV segments have been found to be expanded, and oligoclonal/clonal populations of CD4+ and CD8+ T cells have been identified at the disease sites.62,129–133; however, no consistent usage of TCRBV families has been identified between studies. Some of these studies have implications for immunotherapy.134
Clonal and oligoclonal lymphocyte expansions can be found in the lungs of atopic and non-atopic patients with asthma and normal individuals.135 Therefore, it is difficult to comment on the clinical relevance of these findings; however, clonal populations can also be found after allergen challenge, suggesting a specific T cell response in some cases. Although the lung T cell repertoire is broadly representative of blood T cells, there does appear to be selection and amplification of specific T cell families in the lung in response to local antigenic stimulation.136 In a recent study, we examined the clonality of the T cell response to peanut in MHC class II identical peanut allergy discordant siblings. In vitro stimulation of peripheral blood mononuclear cells with crude peanut extract resulted in stimulation of clonal/oligoclonal T cell responses, together with increased usage of TCRVB11 in peanut allergic individuals.137
Analysis of clonality is a further means of characterising T cells in clinical hypersensitivity reactions; however, apparent specific T cell responses to antigen in some atopic individuals, characterised by oligoclonality of T cell populations at the affected site, do not always provide unique clonality profiles that could be used as diagnostic markers for these diseases.
CONCLUSION
Advances in molecular biological techniques have provided a variety of methods for the investigation of disease. Methods are applicable to RNA and DNA, including poor quality DNA isolated from paraffin wax embedded biopsy material, thus enabling retrospective analyses to be carried out. Foremost among these applications is the diagnosis of lymphocytic disease and differentiation from reactive lymphoproliferations. These applications are less well defined for patient monitoring but are useful for monitoring MRD and in patients with CD8+ lymphocytoses. Methods based on real time PCR are also available for monitoring residual disease after treatment or bone marrow transplantation. The study of the T cell repertoire has proved diagnostically useful for the definition of immunodeficiency states, most notably, Omenn’s syndrome, and for the characterisation of T cells at disease sites in patients with autoimmune disease or allergy. Nevertheless, one of the limitations of PCR technology is the high incidence of false negativity, resulting from the use of consensus primers. Currently, there are several groups working towards the design of better primers,38 which we hope will be available in forthcoming years.
Abbreviations
B-ALL, B cell acute lymphoblastic leukaemia
C, constant
CDR, complementarity determining regions
CMV, cytomegalovirus
CVID, common variable immunodeficiency
D, diversity
DGGE, denaturing gradient gel electrophoresis
EBV, Epstein-Barr virus
HIV, human immunodeficiency virus
HLA, human major histocompatibility complex
HTLV, human T cell lymphotropic virus
Ig, immunoglobulin
LGL, large granular lymphocytosis
J, joining
MHC, major histocompatibility complex
MRD, minimal residual disease
PCR, polymerase chain reaction
PTCL, peripheral T cell lymphoma
RSS, recombination signal sequences
RT, reverse transcription
SCID, severe combined immunodeficiency
SSCP, single strand conformation polymorphism
TCR, T cell receptor
TdT, terminal deoxynucleotidyl transferase
V, variable
REFERENCES
- 1.Clevers H, Alarcon B, Wileman T, et al. The T cell receptor/CD3 complex: a dynamic protein ensemble. Annu Rev Immunol 1988;6:629–62. [DOI] [PubMed] [Google Scholar]
- 2.Rudd CE, Janssen O, Cai YC, et al. Two-step TCR zeta/CD3-CD4 and CD28 signaling in T cells: SH2/SH3 domains, protein-tyrosine and lipid kinases. Immunol Today 1994;15:225–34. [DOI] [PubMed] [Google Scholar]
- 3.Weiss A, Iwashima M, Irving B, et al. Molecular and genetic insights into T cell antigen receptor signal transduction. Adv Exp Med Biol 1994;365:53–62. [DOI] [PubMed] [Google Scholar]
- 4.Esin S, Shigematsu M, Nagai S, et al. Different percentages of peripheral blood gamma delta + T cells in healthy individuals from different areas of the world. Scand J Immunol 1996;43:593–6. [DOI] [PubMed] [Google Scholar]
- 5.Ciccone E, Ferrini S, Bottino C, et al. A monoclonal antibody specific for a common determinant of the human T cell receptor gamma/delta directly activates CD3+. J Exp Med 1988;168:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dechanet J, Merville P, Lim A, et al. Implication of gammadelta T cells in the human immune response to cytomegalovirus. J Clin Invest 1999;103:1437–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO-IUIS Nomenclature Sub-committee on TCR Designation. Nomenclature for T-cell receptor (TCR) gene segments of the immune system. Immunogenetics 1995;42:451–3. [DOI] [PubMed] [Google Scholar]
- 8.Caccia N, Kronenberg M, Saxe D, et al. The T cell receptor beta chain genes are located on chromosome 6 in mice and chromosome 7 in humans. Cell 1984;37:1091–9. [DOI] [PubMed] [Google Scholar]
- 9.Morton CC, Duby AD, Eddy RL, et al. Genes for beta chain of human T-cell antigen receptor map to regions of chromosomal rearrangement in T cells. Science 1985;228:582–5. [DOI] [PubMed] [Google Scholar]
- 10.Isobe M, Erikson J, Emanuel BS, et al. Location of gene for beta subunit of human T-cell receptor at band 7q35, a region prone to rearrangements in T cells. Science 1985;228:580–2. [DOI] [PubMed] [Google Scholar]
- 11.Griesser H, Champagne E, Tkachuk D, et al. The human T cell receptor alpha-delta locus: a physical map of the variable, joining and constant region genes. Eur J Immunol 1988;4:641–4. [DOI] [PubMed] [Google Scholar]
- 12.Wilson RK, Lai E, Concannon P, et al. Structure, organization and polymorphism of murine and human T-cell receptor alpha and beta chain gene families. Immunol Rev 1988;101:149–72. [DOI] [PubMed] [Google Scholar]
- 13.Rabbitts TH, Lefranc MP, Stinson MA, et al. The chromosomal location of T-cell receptor genes and a T cell rearranging gene: possible correlation with specific translocations in human T cell leukaemia. EMBO J 1985;4:1461–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lefranc M-P. Locus maps and genomic repertoire of the human T cell repertoire. The Immunologist 2000;8:72–8. [Google Scholar]
- 15.Alt FW, Oltz EM, Young F, et al. VDJ recombination. Immunol Today 1992;13:306–14. [DOI] [PubMed] [Google Scholar]
- 16.Roman-Roman S, Ferradini L, Azocar J, et al. Studies on the human T cell receptor alpha/beta variable region genes. I. Identification of 7 additional V alpha subfamilies and 14 J alpha gene segments. Eur J Immunol 1991;21:927–33. [DOI] [PubMed] [Google Scholar]
- 17.Yoshikai Y, Clark SP, Taylor S, et al. Organization and sequences of the variable, joining and constant region genes of the human T-cell receptor alpha-chain. Nature 1985;316:837–40. [DOI] [PubMed] [Google Scholar]
- 18.Kimura N, Toyonaga B, Yoshikai Y, et al. Sequences and repertoire of the human T cell receptor alpha and beta chain variable region genes in thymocytes. Eur J Immunol 1987;17:375–83. [DOI] [PubMed] [Google Scholar]
- 19.Klein MH, Concannon P, Everett M, et al. Diversity and structure of human T-cell receptor alpha-chain variable region genes. Proc Natl Acad Sci U S A 1987;84:6884–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koop BF, Hood L. Striking sequence similarity over almost 100 kilobases of human and mouse T-cell receptor DNA. Nat Genet 1994;7:48–53. [DOI] [PubMed] [Google Scholar]
- 21.Rowen L, Koop BF, Hood L. The complete 685-kilobase DNA sequence of the human beta T cell receptor locus. Science 1996;272:1755–62. [DOI] [PubMed] [Google Scholar]
- 22.Toyonaga B, Mak TW. Genes of the T-cell antigen receptor in normal and malignant T cells. Annu Rev Immunol 1987;5:585–620. [DOI] [PubMed] [Google Scholar]
- 23.Arden B, Clarke SP, Kabelitz D. Human T-cell receptor variable gene segment families. Immunogenetics 1995;42:455–500. [DOI] [PubMed] [Google Scholar]
- 24.Lefranc MP, Forster A, Baer R, et al. Diversity and rearrangement of the human T cell rearranging gamma genes: nine germ-line variable genes belonging to two subgroups. Cell 1986;45:237–46. [DOI] [PubMed] [Google Scholar]
- 25.Forster A, Huck S, Ghanem N, et al. New subgroups in the human T cell rearranging V gamma gene locus. EMBO J 1987;6:1945–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oettinger MA, Schatz DG, Gorka C, et al. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science 1990;248:1517–23. [DOI] [PubMed] [Google Scholar]
- 27.Notarangelo LD, Santagata S, Villa A. Recombinase activating gene enzymes of lymphocytes. Curr Opin Haematol 2001;8:41–6. [DOI] [PubMed] [Google Scholar]
- 28.Landau NR, Schatz DG, Rosa M, et al. Increased frequency of N-region insertion in a murine pre-B-cell line infected with a terminal deoxynucleotidyl transferase retroviral expression vector. Mol Cell Biol 1987;7:3237–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Early P, Huang H, Davis M, et al. An immunoglobulin heavy chain variable region gene is generated from three segments of DNA: VH, D and JH. Cell 1980;19:981–2. [DOI] [PubMed] [Google Scholar]
- 30.Tonegawa S. Somatic generation of antibody diversity. Nature 1983;302:575–81. [DOI] [PubMed] [Google Scholar]
- 31.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature 1988;334:395–402. [DOI] [PubMed] [Google Scholar]
- 32.Lewis SM. P nucleotides, hairpin DNA and V(D)J joining: making the connection. Semin Immunol 1994;6:131–41. [DOI] [PubMed] [Google Scholar]
- 33.Nadel B, Feeney AJ. Nucleotide deletion and P addition in V(D)J recombination: a determinant role of the coding-end sequence. Mol Cell Biol 1997;17:3768–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alt FW, Baltimore D. Joining of immunoglobulin heavy chain gene segments: implications from a chromosome with evidence of three D–JH fusions. Proc Natl Acad Sci U S A 1982;79:4118–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilfillan S, Dierich A, Lemeur M, et al. Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science 1993;261:1175–8. [DOI] [PubMed] [Google Scholar]
- 36.Jorgensen JL, Esser U, Fazekas de St Groth B, et al. Mapping T-cell receptor–peptide contacts by variant peptide immunization of single-chain transgenics. Nature 1992;355:224–30. [DOI] [PubMed] [Google Scholar]
- 37.Akolkar PN, Gulwani-Akolkar B, Pergolizzi R, et al. Influence of HLA genes on T cell receptor V segment frequencies and expression levels in peripheral blood lymphocytes. J Immunol 1993;150:2761–73. [PubMed] [Google Scholar]
- 38.Gulwani-Akolkar B, Shi B, Akolkar PN, et al. Do HLA genes play a prominent role in determining T cell receptor V alpha segment usage in humans? J Immunol 1995;154:3843–51. [PubMed] [Google Scholar]
- 39.Walser-Kuntz DR, Weyand CM, Fulbright JW, et al. HLA-DRB1 molecules and antigenic experience shape the repertoire of CD4 T cells. Hum Immunol 1995;44:203–9. [DOI] [PubMed] [Google Scholar]
- 40.Kay RA, Snowden N, Hajeer A, et al. Genetic control of the human V beta 13.2 T cell repertoire: importance of allelic variation outside the coding regions of the TCRBV13S2 gene. Eur J Immunol 1994;24:2863–7. [DOI] [PubMed] [Google Scholar]
- 41.Vissinga CS, Charmley P, Concannon P. Influence of coding region polymorphism on the peripheral expression of a human TCR V beta gene. J Immunol 1994;152:1222–7. [PubMed] [Google Scholar]
- 42.Reyburn H, Cornelis F, Russell V, et al. Allelic polymorphism of human T-cell receptor V alpha gene segments. Immunogenetics 1993;38:287–91. [DOI] [PubMed] [Google Scholar]
- 43.Robinson MA. Allelic sequence variations in the hypervariable region of a T-cell receptor beta chain: correlation with restriction fragment length polymorphism in human families and populations. Proc Natl Acad Sci U S A 1989;86:9422–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Szabo P, Robinson MA, et al. Allelic variations in the human T cell receptor V beta 6.7 gene products. J Exp Med 1990;171:221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pile K, Wordsworth P, Liote F, et al. Analysis of a T-cell receptor V beta segment implicated in susceptibility to rheumatoid arthritis: V beta 2 germline polymorphism does not encode susceptibility. Ann Rheum Dis 1993;52:891–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomolka M, Epplen C, Buitkamp J, et al. Novel members and germline polymorphisms in the human T-cell receptor Vb6 family. Immunogenetics 1993;37:257–65. [DOI] [PubMed] [Google Scholar]
- 47.Posnett DN, Vissinga CS, Pambuccian C, et al. Level of human TCRBV3S1 (V beta 3) expression correlates with allelic polymorphism in the spacer region of the recombination signal sequence. J Exp Med 1994;179:1707–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishihara S, Okada S, Wakiguchi H, et al. Clonal lymphoproliferation following chronic active Epstein-Barr virus infection and hypersensitivity to mosquito bites. Am J Hematol 1997;54:276–81. [DOI] [PubMed] [Google Scholar]
- 49.Soudeyns H, Campi G, Rizzardi GP, et al. Initiation of antiretroviral therapy during primary HIV-1 infection induces rapid stabilization of the T-cell receptor beta chain repertoire and reduces the level of T-cell oligoclonality. Blood 2000;95:1743–51. [PubMed] [Google Scholar]
- 50.Pantaleo G, Demarest JF, Soudeyns H, et al. Major expansion of CD8+ T cells with a predominant V beta usage during the primary immune response to HIV. Nature 1994;370:463–7. [DOI] [PubMed] [Google Scholar]
- 51.Wang EC, Moss PA, Frodsham P, et al. CD8highCD57+ T lymphocytes in normal, healthy individuals are oligoclonal and respond to human cytomegalovirus. J Immunol 1995;155:5046–56. [PubMed] [Google Scholar]
- 52.Lehner PJ, Wang EC, Moss PA, et al. Human HLA-A0201-restricted cytotoxic T lymphocyte recognition of influenza A is dominated by T cells bearing the V beta 17 gene segment. J Exp Med 1995;181:79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi YW, Kotzin B, Herron L, et al. Interaction of Staphylococcus aureus toxin “superantigens” with human T cells. Proc Natl Acad Sci U S A 1989;86:8941–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ulrich RG. Evolving superantigens of Staphylococcus aureus. FEMS Immunol Med Microbiol 2000;27:1–7. [DOI] [PubMed] [Google Scholar]
- 55.McCormick JK, Pragman AA, Stolpa JC, et al. Functional characterization of streptococcal pyrogenic exotoxin J, a novel superantigen. Infect Immun 2001;69:1381–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bernal A, Proft T, Fraser JD, et al. Superantigens in human disease. J Clin Immunol 1999;19:149–57. [DOI] [PubMed] [Google Scholar]
- 57.Ishii E, Kimura N, Kato K, et al. Clonal change of infiltrating T-cells in children with familial hemophagocytic lymphohistiocytosis: possible association with Epstein-Barr virus infection. Cancer 1999;85:1636–43. [DOI] [PubMed] [Google Scholar]
- 58.Islam SA, Hay CM, Hartman KE, et al. Persistence of human immunodeficiency virus type 1-specific cytotoxic T-lymphocyte clones in a subject with rapid disease progression. J Virol 2001;75:4907–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Than S, Kharbanda M, Chitnis V, et al. Clonal dominance patterns of CD8 T cells in relation to disease progression in HIV-infected children. J Immunol 1999;162:3680–6. [PubMed] [Google Scholar]
- 60.Posnett DN, Sinha R, Kabak S, et al. Clonal populations of T cells in normal elderly humans: the T cell equivalent to “benign monoclonal gammapathy”. J Exp Med 1994;179:609–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwab R, Szabo P, Manavalan JS, et al. Expanded CD4+ and CD8+ T cell clones in elderly humans. J Immunol 1997;158:4493–9. [PubMed] [Google Scholar]
- 62.Davey MP, Burgoine GA, Woody CN. TCRB clonotypes are present in CD4+ T cell populations prepared directly from rheumatoid synovium. Hum Immunol 1997;55:11–21. [DOI] [PubMed] [Google Scholar]
- 63.Michalowska-Wender G. The early stage of multiple sclerosis in the light of molecular and immunological studies. Folia Neuropathol 1999;37:273–6. [PubMed] [Google Scholar]
- 64.Oksenberg JR, Panzara MA, Begovich AB, et al. Selection for T-cell receptor V beta–D beta–J beta gene rearrangements with specificity for a myelin basic protein peptide in brain lesions of multiple sclerosis. Nature 1993;362:68–70. [DOI] [PubMed] [Google Scholar]
- 65.Ben Nun A, Liblau RS, Cohen L, et al. Restricted T-cell receptor V beta gene usage by myelin basic protein-specific T-cell clones in multiple sclerosis: predominant genes vary in individuals. Proc Natl Acad Sci U S A 1991;88:2466–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sander CA, Medeiros LJ, Weiss LM, et al. Lymphoproliferative lesions in patients with common variable immunodeficiency syndrome. Am J Surg Pathol 1992;16:1170–82. [DOI] [PubMed] [Google Scholar]
- 67.Elenitoba-Johnson KS, Jaffe ES. Lymphoproliferative disorders associated with congenital immunodeficiencies. Semin Diagn Pathol 1997;14:35–47. [PubMed] [Google Scholar]
- 68.Brooks EG, Filipovich AH, Padgett JW, et al. T-cell receptor analysis in Omenn’s syndrome: evidence for defects in gene rearrangement and assembly. Blood 1999;93:242–50. [PubMed] [Google Scholar]
- 69.van den Beemd R, Boor PP, van Lochem EG, et al. Flow cytometric analysis of the Vbeta repertoire in healthy controls. Cytometry 2000;40:336–45. [DOI] [PubMed] [Google Scholar]
- 70.Langerak AW, van den Beemd R, Wolvers-Tettero IL, et al. Molecular and flow cytometric analysis of the Vbeta repertoire for clonality assessment in mature TCRalphabeta T-cell proliferations. Blood 2001;98:165–73. [DOI] [PubMed] [Google Scholar]
- 71.Slack DN, McCarthy KP, Wiedemanm LM, et al. Evaluation of sensitivity, specificity, and reproducibility of an optimised method for detecting clonal rearrangements of immunoglobulin and T cell receptor genes in formalin fixed, paraffin embedded sections. Diagn Mol Pathol 1993;2:223–32. [PubMed] [Google Scholar]
- 72.Pannetier C, Even J, Kourilisky P. T-cell repertoire diversity and clonal expansions in normal and clinical samples. Immunol Today 1995:16:176–81. [DOI] [PubMed] [Google Scholar]
- 73.Rosenberg WM, Moss PA, Bell JI. Variation in human T cell receptor V beta and J beta repertoire analysis using anchor polymerase chain reaction. Eur J Immunol 1992;22:541–9. [DOI] [PubMed] [Google Scholar]
- 74.Uematsu Y, Wege H, Straus A, et al. The T-cell receptor repertoire in the synovial fluid of a patient with rheumatoid arthritis is polyclonal. Proc Natl Acad Sci U S A 1991;19:8534–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gagne K, Brouard S, Giral M, et al. Highly altered V beta repertoire of T cells infiltrating long-term rejected kidney allografts. J Immunol 2000;164:1553–63. [DOI] [PubMed] [Google Scholar]
- 76.McCarthy KP, Sloan JP, Kabarowski JHS, et al. A simplified method of detection of clonal rearrangements of the T-cell receptor γ chain gene. Diagn Mol Pathol 1992;1:173–9. [PubMed] [Google Scholar]
- 77.van Dongen JJM, Wolvers-Tettero LM. Analysis of immunoglobulin and T cell receptor genes. Part II: possibilities and limitations in the diagnosis and management of lymphoproliferative diseases and related disorders. Clin Chim Acta 1991;198:93–174. [DOI] [PubMed] [Google Scholar]
- 78.Bartram CR. Molecular genetic techniques for detection of minimal residual disease in acute lymphoblastic leukemia: possibilities and limitations. Recent Results Cancer Res 1993;131:149–55. [DOI] [PubMed] [Google Scholar]
- 79.Kneba M, Boilz I, Linke B, et al. Analysis of rearranged T-cell receptor beta-chains by polymerase chain reaction (PCR) DNA sequencing and automated high resolution PCR fragment analysis. Blood 1995;86:3930–7. [PubMed] [Google Scholar]
- 80.Assaf C, Hummel M, Dippel E, et al. High detection rate of T-cell receptor beta chain rearrangements in T-cell lymphoproliferation by family specific polymerase chain reaction in combination with the GeneScan technique and DNA sequencing. Blood 2000;95:640–6. [PubMed] [Google Scholar]
- 81.Even J, Lim A, Puisieux I, et al. T-cell repertoires in healthy and diseased human tissues analysed by T-cell receptor beta-chain CDR3 size determination: evidence for oligoclonal expansions in tumours and inflammatory diseases. Res Immunol 1995;146:65–80. [DOI] [PubMed] [Google Scholar]
- 82.Marguerie C, Lunardi C, So A. PCR-based analysis of the TCR repertoire in human autoimmune diseases. Immunol Today 1992;13:336–8. [DOI] [PubMed] [Google Scholar]
- 83.Martinelli G, Farabegoli P, Testoni N, et al. Detection of clonality by heteroduplex analysis of amplified junctional region of T-cell receptor gamma in patients with T-cell acute lymphoblastic leukemias. Haematologica 1997;82:161–5. [PubMed] [Google Scholar]
- 84.Langerak AW, Szczepanski T, van der Burg M, et al. Heteroduplex PCR analysis of rearranged T cell receptor genes for clonality assessment in suspect T cell proliferations. Leukemia 1997;11:2192–9. [DOI] [PubMed] [Google Scholar]
- 85.Yamamoto K, Masuko-Hongo K, Tanaka A, et al. Establishment and application of a novel T cell clonality analysis using single-strand conformation polymorphism of T cell receptor messenger signals. Hum Immunol 1996;48:23–31. [DOI] [PubMed] [Google Scholar]
- 86.Offermans MT, Sonneveld RD, Bakker E, et al. Denaturing and non-denaturing gel electrophoresis as methods for the detection of junctional diversity in rearranged T cell receptor sequences. J Immunol Methods 1995;181:101–14. [DOI] [PubMed] [Google Scholar]
- 87.Wood GS, Tung RM, Haeffner AC, et al. Detection of clonal T-cell receptor gamma gene rearrangements in early mycosis fungoides/Sezary syndrome by polymerase chain reaction and denaturing gradient gel electrophoresis (PCR/DGGE). J Invest Dermatol 1994;103:34–41. [DOI] [PubMed] [Google Scholar]
- 88.Wilson KB, Quayle AJ, Suleyman S, et al. Heterogeneity of the TCR repertoire in synovial fluid T lymphocytes responding to BCG in a patient with early rheumatoid arthritis. Scand J Immunol 1993;38:102–12. [DOI] [PubMed] [Google Scholar]
- 89.Fodde R, Losekoot M. Mutation detection by denaturing gradient gel electrophoresis (DGGE). Hum Mutat 1994;3:83–94. [DOI] [PubMed] [Google Scholar]
- 90.Griesser H. Gene rearrangements and chromosomal translocations in T cell lymphoma—diagnostic applications and their limits. Virchows Arch 1995;426:323–38. [DOI] [PubMed] [Google Scholar]
- 91.Terhune MH, Cooper KD. Gene rearrangements and T-cell lymphomas. Arch Dermatol 1993;129:1484–90. [PubMed] [Google Scholar]
- 92.Weiss LM, Wood GS, Ellisen LW, et al. Clonal T-cell populations in pityriasis lichenoides et varioliformis acuta (Mucha-Habermann disease). Am J Pathol 1987;126:417–21. [PMC free article] [PubMed] [Google Scholar]
- 93.Wood GS, Strickler JG, Abel EA, et al. Immunohistology of pityriasis lichenoides et varioliformis acuta and pityriasis lichenoides chronica. Evidence for their interrelationship with lymphomatoid papulosis. J Am Acad Dermatol 1987;16:559–70. [DOI] [PubMed] [Google Scholar]
- 94.Harris NL, Jaffe ES, Stein H, et al. A revised European–American classification of lymphoid neoplasms: a proposal from the international lymphoma study group. Blood 1994;84:1361–92. [PubMed] [Google Scholar]
- 95.Gill JI, Gulley MD. Immunoglobulin and T cell receptor gene rearrangement. Diagn Haematol 1994;8:751–70. [PubMed] [Google Scholar]
- 96.Smith JL, Hodges E, Quin CT, et al. Frequent T and B cell oligoclones in histologically and immunophenotypically characterized angioimmunoblastic lymphadenopathy. Am J Pathol 2000;156:661–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lipford EH, Smith HR, Pittaluga S, et al. Clonality of angioimmunoblastic lymphadenopathy and implications for its evolution to malignant lymphoma. J Clin Invest 1987;79:637–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Campana D, Pui CH. Detection of minimal residual disease in acute leukemia: methodologic advances and clinical significance. Blood 1995;85:1416–34. [PubMed] [Google Scholar]
- 99.Heid CA, Stevens J, Livak KJ, et al. Real time quantitative PCR. Genome Res 1996;6:986–94. [DOI] [PubMed] [Google Scholar]
- 100.Holland PM, Abramson RD, Watson R, et al. Detection of specific polymerase chain reaction product by utilizing the 5′→3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci U S A 1991;88:7276–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pongers-Willemse M J, Verhagen OJ, Tibbe GJ, et al. Real-time quantitative PCR for the detection of minimal residual disease in acute lymphoblastic leukemia using junctional region specific Taqman probes. Leukemia 1998;12:2006–14. [DOI] [PubMed] [Google Scholar]
- 102.Bolufer P, Sanz GF, Barragan E, et al. Rapid quantitative detection of BCR–ABL transcripts in chronic myeloid leukemia patients by real-time reverse transcriptase polymerase-chain reaction using fluorescently labeled probes. Haematologica 2000;85:1248–54. [PubMed] [Google Scholar]
- 103.Loughran TP, Jr. Clonal diseases of large granular lymphocytes. Blood 1993;82:1–14. [PubMed] [Google Scholar]
- 104.Loughran TP. Large granular lymphocytic leukemia: an overview. Hosp Pract (Off Ed) 1998;33:133–8. [DOI] [PubMed] [Google Scholar]
- 105.Kelly A, Richards SJ, Sivakumaran M, et al. Clonality of CD3 negative large granular lymphocyte proliferations determined by PCR based X-inactivation studies. J Clin Pathol 1994;47:399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Akashi K, Mizuno S. Epstein-Barr virus infected natural killer cell leukemia. Leuk Lymphoma 2000;40:57–66. [DOI] [PubMed] [Google Scholar]
- 107.Wack A, Cossarizza A, Heltai S, et al. Age-related modifications of the human alphabeta T cell repertoire due to different clonal expansions in the CD4+ and CD8+ subsets. Int Immunol 1998;10:1281–8. [DOI] [PubMed] [Google Scholar]
- 108.Fitzgerald JE, Ricalton NS, Meyer AC, et al. Analysis of clonal CD8+ T cell expansions in normal individuals and patients with rheumatoid arthritis. J Immunol 1995;154:3538–47. [PubMed] [Google Scholar]
- 109.Hingorani R, Choi IH, Akolkar P, et al. Clonal predominance of T cell receptors within the CD8+ CD45RO+ subset in normal human subjects. J Immunol 1993;151:5762–9. [PubMed] [Google Scholar]
- 110.Lamy T, Loughran TP. Current concepts: large granular lymphocytic leukaemia. Blood Rev 1999:13:230–40. [DOI] [PubMed] [Google Scholar]
- 111.Martin MP, Biggar RJ, Hamlin-Green G, et al. Large granular lymphocytosis in a patient infected with HTLV-II. AIDS Res Hum Retroviruses 1993;9:715–19. [DOI] [PubMed] [Google Scholar]
- 112.Choi YW, Herman A, DiGiusto D, et al. Residues of the variable region of the T-cell-receptor beta-chain that interact with S. aureus toxin superantigens. Nature 1990;346:471–3. [DOI] [PubMed] [Google Scholar]
- 113.Lechner F, Cuero AL, Kantzanou M, et al. Studies of human antiviral CD8+ lymphocytes using class I peptide tetramers. Rev Med Virol 2001;11:11–22. [DOI] [PubMed] [Google Scholar]
- 114.Wang LC, Lu MY, Yu Y, et al. T cell lymphoproliferative disorder following bone marrow transplantation for sever aplastic anaemia. Bone Marrow Transplant 2000;26:893–7. [DOI] [PubMed] [Google Scholar]
- 115.Hodara VL, Jeddi-Tehrani M, Grunewald J, et al. HIV infection leads to differential expression of T-cell receptor V beta genes in CD4+ and CD8+ T cells. AIDS 1993;7:633–8. [DOI] [PubMed] [Google Scholar]
- 116.Bansal AS, Green LM, Khoo SH, et al. HIV induces deletion of T cell receptor variable gene product-specific T cells. Clin Exp Immunol 1993;94:17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Smith PR, Cavenagh JD, Milne T, et al. Benign monoclonal expansion of CD8+ lymphocytes in HIV infection. J Clin Pathol 2000;53:177–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gorochov G, Neumann AU, Parizot C, et al. Down-regulation of CD8+ T-cell expansions in patients with human immunodeficiency virus infection receiving highly active combination therapy. Blood 2001;97:1787–95. [DOI] [PubMed] [Google Scholar]
- 119.Signorini S, Imberti L, Pirovano S, et al. Intrathymic restriction and peripheral expansion of the T-cell repertoire in Omenn syndrome. Blood 1999;94:3468–78. [PubMed] [Google Scholar]
- 120.Santagata S, Gomez CA, Sobacchi C, et al. N-terminal RAG1 frameshift mutations in Omenn’s syndrome: internal methionine usage leads to partial V(D)J recombination activity and reveals a fundamental role in vivo for the N-terminal domains. Proc Natl Acad Sci U S A 2000;97:14572–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Villa A, Santagata S, Bozzi F, et al. Partial V(D)J recombination activity leads to Omenn syndrome. Cell 1998;93:885–96. [DOI] [PubMed] [Google Scholar]
- 122.Wade T, Takei K, Kudo M, et al. Characterisation of immune function and analysis of RAG gene mutations in Omenn syndrome and related disorders. Clin Exp Immunol 2000;119:148–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Villa A, Sobacchi C, Notarangelo LD, et al. V(D)J recombination defects in lymphocytes due to RAG mutations: severe immunodeficiency with a spectrum of clinical presentations. Blood 2001;97:81–8. [DOI] [PubMed] [Google Scholar]
- 124.Katial RK, Lieberman MM, Muehlbauer SL, et al. Gamma delta T lymphocytosis associated with common variable immunodeficiency. J Clin Immunol 1997;17:34–42. [DOI] [PubMed] [Google Scholar]
- 125.Duchmann R, Jaffe J, Ehrhardt R, et al. Differential usage of T-cell receptor V beta gene families by CD4+ and CD8+ T cells in patients with CD8hi common variable immunodeficiency: evidence of a post-thymic effect. Immunology 1996;87:99–107. [PMC free article] [PubMed] [Google Scholar]
- 126.Mechanic LJ, Dikman S, Cunningham-Rundles C. Granulomatous disease in common variable immunodeficiency. Ann Intern Med 1997;127:613–17. [DOI] [PubMed] [Google Scholar]
- 127.Cottier H, Kraft R, Meister F. Primary immunodeficiency syndromes and their manifestations in lymph nodes. Curr Top Pathol 1991;84:81–155. [DOI] [PubMed] [Google Scholar]
- 128.Gottesman SR, Haas D, Ladanyi M, et al. Peripheral T cell lymphoma in a patient with common variable immunodeficiency disease: case report and literature review. Leuk Lymphoma 1999;32:589–95. [DOI] [PubMed] [Google Scholar]
- 129.Schmidt D, Martens PB, Weyand CM, et al. The repertoire of CD4+. Mol Med 1996;2:608–18. [PMC free article] [PubMed] [Google Scholar]
- 130.Goronzy JJ, Bartz-Bazzanella P, Hu W, et al. Dominant clonotypes in the repertoire of peripheral CD4+ T cells in rheumatoid arthritis. J Clin Invest 1994;94:2068–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wucherpfennig KW, Hafler DA. A review of T-cell receptors in multiple sclerosis: clonal expansion and persistence of human T-cells specific for an immunodominant myelin basic protein peptide. Ann N Y Acad Sci 1995;756:241–58. [DOI] [PubMed] [Google Scholar]
- 132.Babbe H, Roers A, Waisman A, et al. Clonal expansions of CD8(+) T cells dominate the T cell infiltrate in active multiple sclerosis lesions as shown by micromanipulation and single cell polymerase chain reaction. J Exp Med 2000;192:393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shimonkevitz R, Colburn C, Burnham JA, et al. Clonal expansions of activated gamma/delta T cells in recent-onset multiple sclerosis. Proc Natl Acad Sci U S A 1993;90:923–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Moreland JW, Morgan EE, Adamson TC, 3rd. et al. T cell receptor peptide vaccination in rheumatoid arthritis: a placebo-controlled trial using a combination of Vbeta3, Vbeta14, and Vbeta17 peptides. Arthritis Rheum 1998;41:1919–29. [DOI] [PubMed] [Google Scholar]
- 135.Umibe T, Kita Y, Nakao A, et al. Clonal expansion of T cells infiltrating the airways of non-atopic asthmatics. Clin Exp Immunol 2000;119:390–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hodges E, Dasmahapatra J, Smith JL, et al. T cell receptor (TCR) Vbeta gene usage in bronchoalveolar lavage and peripheral blood T cells from asthmatic and normal subjects. Clin Exp Immunol 1998;112:363–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bakakos P, Smith JL, Warner JO, et al. Modification of T-cell receptor Vbeta repertoire in response to allergen stimulation in peanut allergy. J Allergy Clin Immunol 2001;107:1089–94. system. Immunogenetics 1995;42: 451–3. [DOI] [PubMed] [Google Scholar]
- 138.Van Dongen JJM, Langerak AW, San Miguel JF, et al. PCR-based clonality studies for early diagnosis of lymphoproliferative disorders: report of the BIOMED-2 concerted action [abstract]. Blood 2001;98:129a. [Google Scholar]