Abstract
Background: Alterations in the methylation patterns of promoter CpG islands have been associated with the transcriptional inhibition of genes in many human cancers. These epigenetic alterations could be used as molecular markers for the early detection of cancer—that is, while potentially curable according to current therapeutic strategies. In prostate cancer, GSTP1 hypermethylation is the most common epigenetic alteration, and can be detected in up to 90% of cases. Thus, screening for methylation of other loci would probably increase the number of primary tumours amenable to screening. Moreover, previous studies have shown that the endothelin B receptor (EDNRB) gene is abnormally methylated in a high proportion of prostate tumours (∼70%).
Aims: To investigate the potential use of EDNRB gene hypermethylation as a prostate cancer specific marker.
Methods: Methylation specific polymerase chain reaction (MSP) for the promoter region of EDNRB was performed on prospectively collected tissue samples from 48 patients harbouring clinically localised prostate cancer, and in a group of 23 patients with benign prostatic hyperplasia (BPH). Genomic DNA was isolated from the samples and the methylation status was examined in a blinded manner.
Results: EDNRB methylation was found in 40 of 48 of the adenocarcinomas. However, the same alteration was found in the paired normal tissue, and 21 of 23 of the BPH samples were found to harbour EDNRB hypermethylation.
Conclusions: EDNRB hypermethylation at CpG sites upstream of the transcription start site can be detected in a high proportion of prostate adenocarcinomas. However, because this same alteration is also present in normal and hyperplastic tissue, it does not distinguish normal from neoplastic prostate cells, thus precluding its use as a prostate cancer marker.
Keywords: benign prostate hyperplasia, EDNRB, methylation specific polymerase chain reaction, hypermethylation, prostate cancer
Prostate adenocarcinoma is the second most common cause of cancer related death in men from North America and Western Europe.1 Indeed, at current rates of diagnosis, a man in the USA has a one in five chance of developing invasive prostate cancer during his lifetime.1 Whereas organ confined prostate adenocarcinoma can be cured in most patients, the treatment of more extensive tumours has met with limited success. Thus, the development of new and reliable methods for the early detection of localised tumours increases the likelihood of cure after radical treatment, and may have strong implications for patient outcome.2
CpG islands are 1 kb length regions often associated with promoters or transcribed exons of genes.3 These islands normally remain unmethylated in the germ line and in normal adult tissue,4 and rarely become methylated in somatic cells.5 Moreover, methylation of cytosines at CpG islands has been recently recognised as an important epigenetic alteration, which may play a decisive role in the control of gene expression, namely during mammalian development.6 Alterations in the methylation patterns of promoter CpG islands has been associated with the transcriptional inhibition of genes in many human cancers and stands as an alternative mechanism of gene inactivation.7–9 Examples of genes that are frequent targets for de novo methylation include p16, p15, RB1, GSTP1, the oestrogen receptor gene (ESR1), and DNA repair genes such as MLH1 and MGMT.10–17 Furthermore, these epigenetic alterations have been proposed as molecular markers for the detection of several tumours, most notably in prostate cancer.18–21 Indeed, p16 methylation was reported in three of five prostate cancer cell lines analysed, although this alteration was found to be less common in prostate primary tumours (13%).22 Studies on E-cadherin and CD44 also yielded a low frequency of promoter hypermethylation in prostate cancer.23,24 On the contrary, GSTP1 was found to be methylated frequently in this cancer (∼90% of cases), but additional molecular markers should be sought to increase the detection rate.20,21,25,26
“The EDNRB gene joins a growing number of genes that are of importance in normal development and may become deregulated in cancer”
The endothelin B receptor (EDNRB) gene is located on chromosome 13, and its role in carcinogenesis is still largely unknown, although recent findings suggest that EDNRB signalling is necessary during embryogenesis.27,28 Therefore, the EDNRB gene joins a growing number of genes that are of importance in normal development and may become deregulated in cancer.29 Previous studies have shown that the EDNRB gene is abnormally methylated in a high proportion of prostate tumours,27,28 and that no methylation was found in normal tissues.30 Thus, a potential use of this marker in the molecular detection of prostate cancer could be envisaged.
Hence, to test the usefulness of the detection of EDNRB somatic methylation as a prostate cancer marker, we analysed the methylation status of the promoter region of this gene in a series of 48 adenocarcinomas and paired morphologically normal prostate tissues. For control purposes, tissue from patients with benign prostatic hyperplasia (BPH) was also analysed.
METHODS
Patients and sample collection
Forty eight patients with clinically localised prostate adenocarcinoma, consecutively diagnosed and primarily treated with radical prostatectomy at the Portuguese Cancer Institute, Porto, were selected for our study. All cases were identified by raised serum prostate specific antigen (PSA) in routine analysis and confirmed by sextant prostate biopsy (stage T1c31). In addition, 23 patients with BPH, submitted to transurethral resection of the prostate (TURP), were included for control purposes. All histological slides were reviewed by two pathologists, and each adenocarcinoma was staged and graded according to the TNM staging system,31 and the Gleason grading system.32 Fresh prostatic tissue was collected from each surgical specimen, snapped frozen in isopentane and stored at −80°C. Sections were cut for the identification of areas of morphologically normal tissue from the peripheral zone and adenocarcinoma (radical prostatectomy specimens) and BPH (TURP specimens). These areas were then carefully microdissected from 12 μm thick sections for cell enrichment. DNA was extracted from either hyperplastic, normal, or tumour tissue collected from each patient, according to the method described by Ahrendt et al. Briefly, DNA was digested overnight at 48°C with proteinase K (0.5 mg/ml) in 1% sodium dodecyl sulfate, Tris (1M, pH 8.8), EDTA (0.5M, pH 8.0), and NaCl (5M), followed by phenol/chloroform extraction and ethanol precipitation.33
Bisulfite treatment
Sodium bisulfite conversion of 2 μg of genomic DNA was performed by a modification of a previously described method.34 In brief, DNA was denatured in 0.2M NaOH for 20 minutes at 50°C. A volume of 500 μl freshly made solution containing 2.5M sodium bisulfite (Sigma, Steinheim, Germany) and 125mM hydroquinone (Sigma), at pH 5.0, was added to each sample and incubation was continued at 50°C in the dark. After three hours of incubation, the modified DNA was desalted through a column (Wizard DNA purification resin; Promega Corp, Madison, Wisconsin, USA), according to the manufacturer’s instructions. After treatment with NaOH (final concentration, 0.3M) for 10 minutes at 37°C, isolation was continued with 75 μl 7.5M ammonium acetate, followed by an incubation step of five minutes at room temperature. Finally, the modified DNA was precipitated with 2.5 volumes of 100% ethanol and 2 μl glycogen (5 mg/ml). The pellet was washed with 70% ethanol, dried, and eluted in 30 μl 5mM Tris (pH 8.0).
Methylation specific polymerase chain reaction (MSP) analysis
For PCR amplification, 2 μl of bisulfite modified DNA was added to a final volume of 25 μl PCR mix containing 1× PCR buffer (16.6mM ammonium sulfate/67mM Tris, pH 8.8/6.7mM MgCl2/10mM 2 mercaptoethanol), dNTPs (each at 1.25mM), and primers (300 ng each/reaction). The primer sequences were: 5′-TGGTGAAGAGGTTGTGGGTGGTATT AGTG-3′ (sense) and 5′-ACCTACTCCAAAAACA TCCAATAACCA-3′ (antisense) for unmethylated DNA and 5′-cgaagaggttgcgggcggtattagcg-3′ (sense) and 5′-TACTCCAAAAACGTCCGATAACCG-3′ (antisense) for methylated DNA. Because the nucleotide positions are numbered relative to the transcription start site (+1), the PCR amplified region for the methylated alleles spanned from −139 to −9, and for unmethylated alleles it spanned from −141 to −7. This region contains nine CpG dinucleotides, including six CpGs at the primer annealing sites. PCR was performed using the following conditions: one cycle at 95°C for one minute; 35 cycles of one minute at 95°C, one minute at 62°C, and one minute at 72°C, and a final extension step for five minutes at 70°C. In each PCR performed, treated DNA extracted from a prostate cancer cell line (PC3) and from normal lymphocytes was used for positive and negative control purposes, respectively. The PCR products were loaded directly on to a non-denaturing 6% polyacrylamide gel, stained with ethidium bromide, and visualised under ultraviolet illumination.
RESULTS
We prospectively studied 48 patients with clinically localised prostate adenocarcinoma with a median age of 63 years (range, 48–74). As a control group, 23 patients with BPH were included (median age, 67 years; range, 58–81). No significant difference was found between the age distribution of these two groups of patients (p = 0.33). The median value of the preoperative serum PSA was 9.9 ng/ml (range, 5.1–28.5) and 4.73 ng/ml (range, 1.8–9) for patients with cancer and BPH, respectively, and this difference is significant (p < 0.0001).
The median Gleason score of the prostate adenocarcinomas was 6 (range, 5–9). In these same radical prostatectomy specimens, nine cases were staged as pT2a, 21 cases as pT2b, and 18 cases as pT3a, according to the TNM staging system.31
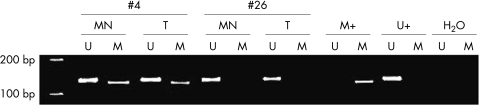
We determined the promotor methylation status of the EDNRB gene in the tissue samples, both for patients with prostate cancer and for controls. By MSP analysis of the 5′ region of the EDNRB gene located at the fringe of the CpG island, 40 of 48 adenocarcinomas were found to be methylated (fig 1). The paired normal tissue of these 40 patients was also methylated. In the remaining eight patients, both the tumour and the normal tissue samples were unmethylated. Moreover, we found that only two cases of BPH were not methylated at the same CpG sites. The primer sets used in our study included two CpG sites (−130 and −8) analysed in a previous report.35
Figure 1.
Illustrative example of methylation specific polymerase chain reaction for the EDNRB promoter region: morphologically normal (MN) and tumour (T) tissue of patients number 4 and 26. Lanes U and M correspond to unmethylated (134 bp) and methylated (130 bp) reactions, respectively. In each case, normal lymphocyte DNA was used as a negative control for methylation (U+), DNA from the PC3 cell line was used as a positive control for methylation (M+), and water was used as negative PCR control (H2O). On the right hand side the HiLo marker is depicted.
DISCUSSION
DNA hypermethylation in neoplastic tissue (when compared with the normal tissue) has been described in many instances, and it has been suggested that these changes could be useful markers for the early detection of cancer cells.18–21 A promising marker has been found for prostate cancer—namely, GSTP1 hypermethylation—which can be detected both in tissue and body fluids.14,20,21,25,26 Because GSTP1 hypermethylation is present in ∼90% of prostate adenocarcinomas, additional molecular markers should be sought to increase the detection rate.20,21,25,26 A previous study by Nelson and co-workers showed that the EDNRB gene was abnormally methylated in ∼70% of prostate tumours, and no methylation was found in normal tissues.30 Thus, we hypothesised that EDNRB hypermethylation could potentially be used as an additional molecular marker for prostate cancer. Our results are in accordance with that previous study, which showed CpG −130 methylation in prostate adenocarcinoma and normal adjacent tissue, although in the normal tissue the degree of methylation was generally lower, as would be expected for normal tissues.35 However, because conventional MSP was used in our study, no conclusions can be drawn regarding the degree of methylation.
“These frequently methylated sites may play an important role as starting points for methylation in more downstream CpG sites, which are frequently methylated in prostate tumours but not in normal tissue”
The region of the EDNRB gene promotor analysed in our study was chosen because of a previous study indicating that the most 3′ CpG dinucleotide was more heavily methylated than the 5′ end.30 Our results confirm this finding concerning the methylation status of adenocarcinoma samples. However, because of the high sensitivity (1/1000) of the MSP method used in our present study,36 we were also able to detect DNA hypermethylation in paired normal tissue and hyperplastic tissue from the control group (BPH). Moreover, the increased sensitivity of the method may explain the larger proportion of methylated tumours found in our present study. These findings are consistent with the results reported by Pao et al, who found that EDNRB methylation levels varied from 11% to 25% in all the five normal samples analysed, and varying from 11% to > 50% in the paired prostate tumours.35 Indeed, the primer sets used in our study included two CpG sites (−130 and −8) analysed in the aforementioned report.35 Thus, EDNRB methylation at these CpG sites does seem to be a useful marker for the detection of prostate cancer. Indeed, Pao et al suggested that selected CpG sites located more downstream in the CpG island of the EDNRB gene could be more reliable markers of malignancy.35
The finding that the EDNRB methylation status at these CpG sites in prostate adenocarcinoma cases parallels the respective normal tissue does not seem to support an important role for this epigenetic alteration in prostate carcinogenesis, as previously anticipated.30 Alternatively, EDNRB hypermethylation could be envisaged as a preneoplastic alteration, with no corresponding change in the morphological appearance of the prostate epithelium. Moreover, these frequently methylated sites may play an important role as starting points for methylation in more downstream CpG sites, which are frequently methylated in prostate tumours but not in normal tissue.35 In this regard, the analysis of these more downstream sites in the cases found to be unmethylated in our study might help test this hypothesis.
In conclusion, the detection of EDNRB gene hypermethylation at CpG sites upstream to the transcription start site does not allow for the distinction between normal and neoplastic prostate cells, thus preventing its use as a prostate cancer marker. However, further analyses of more downstream sites in a large series of patients may reveal a role for EDNRB gene methylation in prostate cancer detection.
Take home messages
A high proportion of prostate adenocarcinomas show EDNRB hypermethylation at CpG sites upstream of the transcription start site
However, this same alteration is also present in normal and hyperplastic tissue, and does not distinguish normal from neoplastic prostate cells, thus precluding its use as a prostate cancer marker
Additional analyses of sites further downstream may yet reveal a role for EDNRB gene methylation in prostate cancer detection
Acknowledgments
CJ is supported by a grant from the Fundação para a Ciência e Tecnologia, Portugal (Program PRAXIS XXI - BD 13398/97). The support of the Liga Portuguesa Contra o Cancro – Núcleo Regional do Norte is acknowledged.
Abbreviations
BPH, benign prostatic hyperplasia
MSP, methylation specific polymerase chain reaction
PSA, prostate specific antigen
TURP, transurethral resection of the prostate
REFERENCES
- 1.Landis SH, Murray T, Bolden S, et al. Cancer statistics. CA Cancer J Clin 1999;49:8–31. [DOI] [PubMed] [Google Scholar]
- 2.Andriole GL, Catalona WJ. The case for aggressive diagnosis and therapy of localized prostate cancer. In: Raghavan D, Scher HI, Leibel AS, et al. Principles and practice of genitourinary oncology. Philadelphia: Lippincott-Raven, 1996:457–64.
- 3.Cooper DN, Krawczak M. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum Genet 1989;83:181–8. [DOI] [PubMed] [Google Scholar]
- 4.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet 1999;21:163–7. [DOI] [PubMed] [Google Scholar]
- 5.Antequera F, Boyes J, Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell 1990;62:503–14. [DOI] [PubMed] [Google Scholar]
- 6.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 1992;69:915–26. [DOI] [PubMed] [Google Scholar]
- 7.Baylin SB, Herman JG, Graff JR, et al. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res 1998;72:141–6. [PubMed] [Google Scholar]
- 8.Jones PA. DNA methylation errors and cancer. Cancer Res 1996;56:2463–7. [PubMed] [Google Scholar]
- 9.Liang G, Salem CE, Yu MC, et al. DNA methylation differences associated with tumor tissues identified by genome scanning analysis. Genomics 1998;53:260–8. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez-Zulueta M, Bender CM, Yang AS, et al. Methylation of the 5′CpG island of the p16/CDKN2 tumor suppressor gene in normal and transformed human tissues correlates with gene silencing. Cancer Res 1995;55:4531–5. [PubMed] [Google Scholar]
- 11.Merlo A, Herman JG, Mao L, et al. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med 1995;1:686–92. [DOI] [PubMed] [Google Scholar]
- 12.Herman JG, Jen J, Merlo A, et al. Hypermethylation-associated inactivation indicates a tumor suppressor role for p15INK4B. Cancer Res 1996;56:722–7. [PubMed] [Google Scholar]
- 13.Stirzaker C, Millar DS, Paul CL, et al. Extensive DNA methylation spanning the Rb promoter in retinoblastoma tumors. Cancer Res 1997;57:2229–37. [PubMed] [Google Scholar]
- 14.Esteller M, Corn PG, Urena JM, et al. Inactivation of glutathione S-transferase P1 gene by promoter hypermethylation in human neoplasia. Cancer Res 1998;58:4515–18. [PubMed] [Google Scholar]
- 15.Issa JP, Ottaviano YL, Celano P, et al. Methylation of the oestrogen receptor CpG island links aging and neoplasia in human colon. Nat Genet 1994;7:536–40. [DOI] [PubMed] [Google Scholar]
- 16.Herman JG, Umar A, Polyak K, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A 1998;95:6870–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esteller M, Hamilton SR, Burger PC, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res 1999;59:793–7. [PubMed] [Google Scholar]
- 18.Esteller M, Sanchez-Cespedes M, Rosell R, et al. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res 1999;59:67–70. [PubMed] [Google Scholar]
- 19.Sanchez-Cespedes M, Esteller M, Wu L, et al. Gene promoter hypermethylation in tumors and serum of head and neck patients. Cancer Res 2000;60:892–5. [PubMed] [Google Scholar]
- 20.Cairns P, Esteller M, Herman JG, et al. Detection of prostate cancer in urine by GSTP1 hypermethylation. Clin Cancer Res 2001;7:2727–0. [PubMed] [Google Scholar]
- 21.Jerónimo C, Usadel H, Henrique R, et al. Quantitation of GSTP1 methylation in non-neoplastic prostatic tissue and organ confined prostate adenocarcinoma. J Natl Cancer Inst 2001;93:1747–52. [DOI] [PubMed] [Google Scholar]
- 22.Jarrard DF, Bova GS, Ewing CM, et al. Deletional, mutational, and methylation analyses of CDKN2 (p16/MST1) in primary and metastatic prostate cancer. Genes Chromosomes Cancer 1997;19:90–6. [PubMed] [Google Scholar]
- 23.Graff JR, Herman JG, Lapidus RG, et al. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res 1995;55:5195–9. [PubMed] [Google Scholar]
- 24.Lou W, Krill D, Dhir R, et al. Methylation of the CD44 metastasis suppressor gene in human prostate cancer. Cancer Res 1999;59:2329–31. [PubMed] [Google Scholar]
- 25.Lee W-H, Morton RA, Epstein JI, et al. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc Natl Acad Sci U S A 1994;91:11733–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee W-H, Isaacs WB, Bova GS, et al. CG island methylation changes near the GSTP1 gene in prostatic carcinoma cells detected using the polymerase chain reaction: a new prostate cancer biomarker. Cancer Epidemiol Biomark Prev 1997;6:443–50. [PubMed] [Google Scholar]
- 27.Arai H, Nakao K, Takaya K, et al. The human endothelin-B receptor gene, structural organization and chromosomal assignment. J Biol Chem 1993;268:3463–70. [PubMed] [Google Scholar]
- 28.Shin MK, Levorse JM, Ingram RS, et al. The temporal requirement for endothelin receptor-B signalling during neural crest development. Nature 1999;402:496–501. [DOI] [PubMed] [Google Scholar]
- 29.Ford HL. Homeobox genes: a link between development, cell cycle, and cancer? Cell Biol Int 1998;22:397–400. [DOI] [PubMed] [Google Scholar]
- 30.Nelson JB, Lee WH, Nguyen SH, et al. Methylation of the 5′ CpG island of the endothelin B receptor gene is common in human prostate cancer. Cancer Res 1997;57:35–7. [PubMed] [Google Scholar]
- 31.Hermanek P, Hunter RVP, Sobin LH, et al. Prostate. In: Hermanek P, Hutter RVP, Sobin LH, et al. Illustrated guide to the TNM/pTNM classification of malignant tumors, 4th ed. Heidelberg: Springer-Verlag, 1997:272–80.
- 32.Gleason DF, Mellinger GT. Prediction of prognosis for prostatic adenocarcinoma by combined histologic grading and clinical staging. J Urol 1974;111:58–64. [DOI] [PubMed] [Google Scholar]
- 33.Ahrendt SA, Chow JT, Xu L-H, et al. Molecular detection of tumor cells in bronchoalveolar lavage fluid from patients with early stage lung cancer. J Natl Cancer Inst 1999;91:332–339. [DOI] [PubMed] [Google Scholar]
- 34.Olek A, Oswald J, Walter JA. A modified and improved method of bisulfite based cytosine methylation analysis. Nucleic Acids Res 1996;24:5064–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pao MM, Tsutsumi M, Liang G, et al. The endothelin B receptor (EDNRB) promoter displays heterogeneous, site specific methylation patterns in normal and tumor cells. Hum Mol Genet 2001;10:903–10. [DOI] [PubMed] [Google Scholar]
- 36.Herman JG, Graff JR, Myohanen S, et al. Methylation-specific PCR. A novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 1996;93:9821–6. [DOI] [PMC free article] [PubMed] [Google Scholar]