Abstract
Aims: To investigate the genetic differences among the strains of adenovirus type 8 (Ad8) circulating in Hiroshima city, Japan, and to study their circulation pattern.
Methods: One hundred and twenty nine strains of adenovirus type 8 (Ad8) were isolated in Hiroshima City over a 15 year period (1983–97) from patients with keratoconjunctivitis, and analysed with six restriction enzymes—BamHI, HindIII, PstI, SacI, SalI, and SmaI—to investigate possible relations among the isolates and their genetic variability. Seven hypervariable regions of the hexon gene that carry the type specific epitope were also sequenced to investigate the variation among the genome types.
Results: Restriction endonuclease analyses yielded three known genome types (Ad8A, 13 samples; Ad8B, seven samples; and Ad8E, 35 samples) and a novel genome type (Ad8I, 74 samples). Ad8A, Ad8B, and Ad8E were closely related, with 96% homology, whereas Ad8I had only 71% homology. Ad8A, Ad8B, and Ad8E shared 91.8% and 96.4% homology with regard to their amino acid and nucleotide sequences, respectively, with the isolate 1127 (accession no X74663). However, when compared with Ad8A, Ad8B, Ad8E, and isolate 1127, Ad8I shared only 62.7% and 69.9% homology with regard to amino acid and nucleotide sequences, respectively. Ad8A, Ad8B, and Ad8E had a unique 31 amino acid deletion in the hypervariable region 1 of the hexon gene, whereas Ad8I had a 33 residue deletion. The Ad8E strain that circulated from 1984 to 1995 was stable among the study population. Ad8I was isolated from an outbreak of epidemic keratoconjunctivitis in 1995 and was also isolated from sporadic cases until 1997.
Conclusions: These results confirmed that genetic variability occurs in Ad8 in the microenvironment and revealed the emergence of a new genome type (Ad8I).
Keywords: adenovirus, type 8, genetic characterisation, Hiroshima
The 51 serotypes of human adenovirus are classified into six subgenera, A to F, according to their biological, immunological, and biochemical properties.1,2 Adenovirus type 8 (Ad8) is the most common agent of epidemic keratoconjunctivitis (EKC). EKC is a highly contagious ocular infection and is often transmitted through ocular facilities, thus causing community epidemics.3–6 Type specific neutralisation, which is used for the identification of adenoviruses, cannot distinguish genetic variation among the isolates. Ad8 genomic variants (genome types) have been classified as Ad8A to Ad8H7–11 and Ad8/D1 to Ad8/D1212–14 by means of restriction enzyme analysis of prototype and field isolates. This genome typing has provided a powerful and practical tool for the study of the epidemiological and clinical patterns of adenoviruses. Although adenoviral ocular infection is common in Japan and other East Asian countries,15,16 the distribution of the various genome types has not been investigated extensively. So far, only the circulation of genome types Ad8A and Ad8B in the north part of Japan (Sapporo) has been reported.
“Epidemic keratoconjunctivitis is a highly contagious ocular infection and is often transmitted through ocular facilities, thus causing community epidemics”
In our study, Ad8 strains isolated in Hiroshima city over a 15 year period were analysed with six restriction enzymes (BamHI, HindIII, PstI, SacI, SalI, and SmaI) to investigate the genetic differences among the isolates, and their circulation pattern. Hypervariable regions (HVRs) of the hexon gene that carry type specific epitopes were also sequenced to document the degree of variation among the genome types, which may have some relation to the disease.
MATERIALS AND METHODS
Samples
In total, 129 strains were isolated from epidemic and sporadic cases of keratoconjunctivitis. The clinical diagnosis was recorded as EKC, acute haemorrhagic conjunctivitis, or acute conjunctivitis. All samples were isolated in Hep2 cells.
DNA extraction
DNA was extracted from the infected cells by a modified Hirt’s procedure. Briefly, confluent monolayers of Hep2 cells in 25 cm2 flasks were inoculated with virus stock and incubated at 35°C. When a 75–100% cytopathic effect was seen, cells were dislodged with a cell scraper and pelleted by low speed centrifugation. Cells were washed twice with phosphate buffered saline and resuspended in 1 ml of lysis buffer (10mM Tris/HCl (pH 7.4), 10mM EDTA, 1% sodium dodecyl sulfate (SDS)) for 15 minutes at room temperature. The suspension was incubated with 200 μg/ml of proteinase K (Sigma Chemical, St Louis, Missouri, USA) at 37°C for one hour. After incubation, 5M NaCl was added to a final concentration of 1M, and further incubated at 4°C overnight to precipitate cellular DNA. The suspension was then centrifuged at 15 000 ×g for 30 minutes. The supernatant was incubated with 30 μg of RNase A (Sigma Chemical) for one hour and extracted twice in phenol/chloroform. Next, the supernatant was precipitated in two volumes of 100% ethanol. After drying, the DNA was suspended in 50 μl of Tris/EDTA buffer (10mM Tris/HCl (pH 7.4), 10mM EDTA) and measured spectrophotometrically.
DNA restriction enzyme analysis
Restriction enzyme analyses were performed with BamHI, HindIII, PstI, SacI, SalI, and SmaI (Boehringer Mannheim, Mannheim, Germany). Briefly, a 2 μg aliquot of DNA was incubated with 10 units of restriction endonucleases in 20 μl of reaction mixture at an appropriate temperature (that recommended for each restriction endonuclease) for three hours. After digestion, all products were electrophoresed on a 1.2% horizontal submerged agarose gel at 90 V for three hours in 50 mM Tris acetate EDTA buffer (pH 8.0). The gel was stained with ethidium bromide (1 μg/ml) and photographed under ultraviolet light with a polaroid camera (Funakoshi, Tokyo, Japan). HindIII digests of λ DNA (Boehringer Mannheim) were used as molecular weight markers. Genomic homology between the two strains was calculated using the percentage of pairwise comigrating restriction fragments (PCRF) of a pair divided by the total number of bands in the pair. Genome type identifications were conducted by comparison of the resulting patterns with the published restriction patterns of the prototype and genome types.7,8,12
PCR, cycle sequencing, and sequence analysis
HVRs were sequenced by generating overlapping polymerase chain reaction (PCR) products and direct cycle sequencing. A set of six primers (forward primers: AdHD1N, 5‘-TGG ACC GCG GTC CCA GCT TCA A -3‘ (19 to 41); AdHD2F, 5‘-ATG AAA CCA TGC TAT GGC TC-3‘ (439 to 459); and AdHD3F, 5‘-TGG TCG ACT TGC AAG ACA G-3‘ (824–842); reverse primers: AdHD2R, 5‘-TAG GTT GAC CAT CTT CAG TGG T-3‘ (526–505); AdHD3R, 5‘-CTG TCC ACC GCA GAG TTC CA-3‘ (929–911); and AdHd4, 5‘-GCC ACG TTC GAG TAC AGA AAA C-3‘ (1187–1166)) were selected based on the alignment of hexon gene sequences available from GeneBank (Ad8 (X74663), Ad19 (X98539), Ad37 (X98360), Ad9 (X74664), and Ad15 (X74666)) from human adenovirus serotypes Ad8, Ad19, Ad37, Ad9, and Ad15, respectively. All products were sequenced in both directions with internal and template primers. Full length adenoviral DNA, extracted by Hirt’s method, was used as a template for PCR. The PCR amplification was carried out in 50 μl reaction mixtures containing 1 μl aliquots of DNA, 5 μl of 10× concentrated buffer, 0.5 μM each of the primer pair, 200 μM of each dNTP, and 1.25 U of Taq polymerase (Boehringer Mannheim). The assays were performed in a programmable heat block (model 9600-R; Perkin Elmer, Foster City, California, USA). Thermal cycling consisted of preliminary denaturation for three minutes at 94°C, followed by 35 cycles of denaturation at 94°C for one minute, annealing at 47°C for one minute and at 72°C for two minutes, and a final extension at 72°C for seven minutes. The amplification products were analysed on a 1.5% agarose gel. Next, the PCR products were purified using a DNA fragment purification kit (Mag Extractor-PCR and Gel Cleanup; Toyobo, Osaka, Japan) according to the manufacturer’s instructions. The cycle sequence reaction was carried out with an ABI prism dye terminator cycle sequencing ready reaction kit (Applied Biosystems, Chiba, Japan). The sequences were determined by a genetic analyser 310 (Applied Biosystems). The nucleotide sequences of four genome types (Ad8A, Ad8B, Ad8E, and Ad8I) were compared with the available sequence of the Ad8 isolate, 1127 (accession number, X74663). DNASIS software (Hitachi Software Ltd, Tokyo, Japan) was used for sequence alignment and analysis.
Nucleotide sequence accession numbers
Sequence data from this article have been deposited in GeneBank/DDBJ under the accession numbers: hexon gene Ad8A (AB090341), Ad8B (AB090342), Ad8E (AB090343), and Ad8I (AB090344). The amino acid sequences of the residues were deduced.
RESULTS
Prevalence among the age groups
Patients were divided into three age groups: 0–9 years, 10–19 years, and > 20 years. Thirty two (24.8%) of the stains isolated came from the 0–9 year old group, whereas only 9 (6.9%) were in the 10–19 year old group. Most isolates (88 (68.2%)) came from the > 20 years old group.
Cleavage patterns with the restriction endonucleases
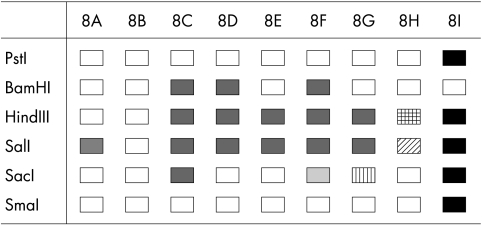
Restriction endonuclease cleavage patterns with SacI, PstI, and SmaI divided the isolates into two groups, whereas HindIII and SalI divided them into three groups (fig 1).
Figure 1.
Identification of adenovirus type 8 (Ad8) genome types in Hiroshima. The open boxes indicate cleavage patterns identical to those of Ad8B. The various shaded boxes indicate cleavage patterns different from those of Ad8B and also distinct from each other.
Cleavage pattern with HindIII
Upon digestion with HindIII, 20 isolates showed identical restriction patterns shared by Ad8A and Ad8B. Thirty five isolates, classified as Ad8E, showed a distinct restriction pattern. However, 74 isolates showed a different restriction pattern. These isolates are a novel genome type, designated Ad8I (fig 2C).
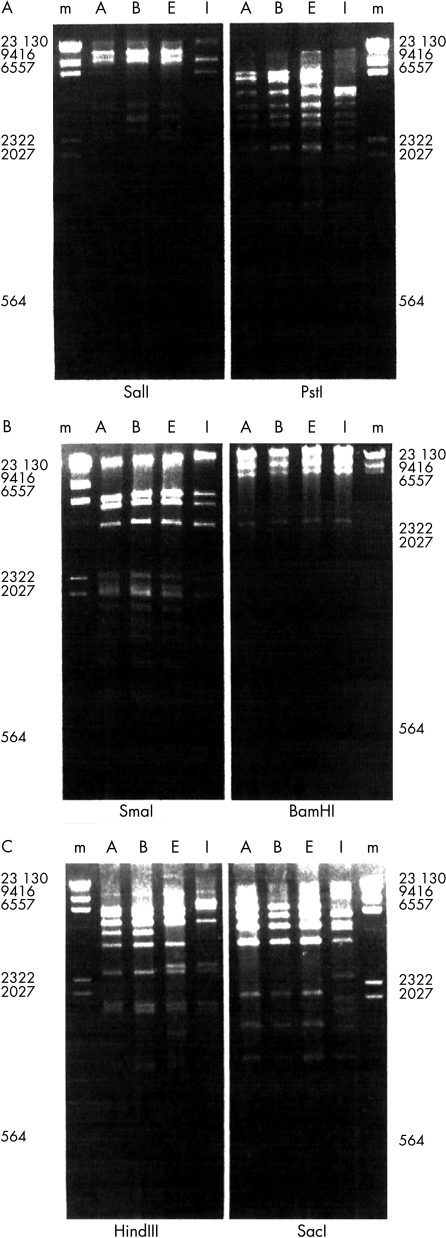
Figure 2.
Restriction fragments of adenovirus type 8 (Ad8) were electrophoresed on a 1.2% agarose gel with a HindIII digest of λ DNA (lane m) as molecular weight standard. Lanes A, B, E, and I contain genome types Ad8A (isolates from 1983), Ad8B (from 1984), Ad8E (from 1994), and Ad8I (from 1995), respectively. Ad8 digested with (A) SalI and PstI; (B) SmaI and BamHI; (C) HindIII and SacI.
Cleavage pattern with SalI
Upon digestion with SalI, 48 isolates showed a similar restriction pattern to that of Ad8A and Ad8E. The pattern of seven isolates was identical to that of Ad8B. However, 74 isolates showed a new restriction pattern (fig 2A).
Cleavage pattern with PstI, SmaI, and SacI
Fifty five isolates showed restriction patterns that correspond with Ad8A, Ad8B, and Ad8E. However, 23 isolates showed a new restriction pattern (fig 2A–C).
Cleavage pattern with BamHI
BamHI was not useful for distinguishing between the different isolates because they all showed an identical restriction pattern (fig 2B).
Genome type circulation
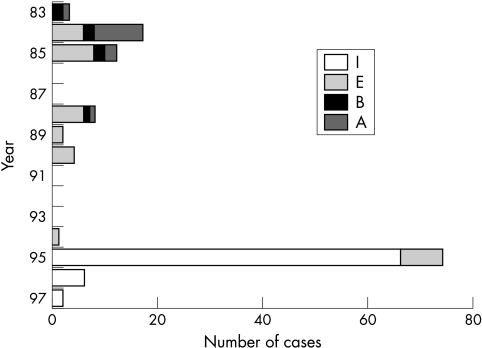
The analysis of 129 isolates from Hiroshima using six restriction enzymes (Bam HI, HindIII, PstI, SacI, SalI, and SmaI) yielded three known genome types, namely: Ad8A (13 isolates), Ad8B (seven isolates), and Ad8E (35 isolates) and a new genome type, designated Ad8I (74 isolates) (fig 2). Ad8A and Ad8B circulated between 1983 and 1988, and Ad8E between 1984 and 1995. Ad8I was first isolated from epidemic cases in 1995, and then from sporadic cases of EKC until 1997 (fig 3).
Figure 3.
Annual trends of adenovirus type 8 (Ad8) genome types in Hiroshima. The distribution of Ad8 genome types in Hiroshima on a yearly basis. Ad8A (A) and Ad8B (B) were co-circulating from 1983 to 1988. Ad8E (E) was co-circulating with Ad8A and Ad8B from 1984 to 1988 and circulated as a single genome type from 1989 to 1995. A new genome type Ad8I (I) was isolated from the outbreak of epidemic keratoconjunctivitis in 1995. All 66 cases from the outbreak were Ad8I. Ad8I was subsequently recovered from sporadic cases in 1996 and 1997.
Homology among the genome types
DNA homology studies of serotype 8 are often difficult, owing to its growth properties. Some strains grow well but others replicate slowly in the laboratory, as reported previously.12,14 In our study, there was not enough DNA for the analysis of small fragments with low molecular weight.
Pairwise comparison of PCRF showed that Ad8A has a total of 48 fragments for the six restriction enzymes. Ad8B and Ad8E shared 46 (96%) fragments with Ad8A, whereas Ad8I shared 34 (71%) fragments with Ad8A (table 1).
Table 1.
Pairwise comparison of comigrating restriction endonuclease cleavage fragments from the DNA of Ad8 genome type
| Number of comigrating fragments | |||||
| Enzyme | Ad8A | Ad8B | Ad8E | Ad81 | |
| BamHI | Ad8A | 4 | 4 | 4 | 4 |
| Ad8B | 4 | 4 | 4 | ||
| Ad8E | 4 | 4 | |||
| Ad81 | 4 | ||||
| HindIII | Ad8A | 9 | 9 | 7 | 6 |
| Ad8B | 9 | 7 | 6 | ||
| Ad8E | 9 | 7 | |||
| Ad81 | 9 | ||||
| PstI | Ad8A | 9 | 9 | 9 | 8 |
| Ad8B | 9 | 9 | 8 | ||
| Ad8E | 9 | 8 | |||
| Ad81 | 11 | ||||
| SacI | Ad8A | 7 | 7 | 7 | 4 |
| Ad8B | 7 | 7 | 4 | ||
| Ad8E | 7 | 4 | |||
| Ad81 | 9 | ||||
| SalI | Ad8A | 8 | 6 | 8 | 3 |
| Ad8B | 8 | 6 | 3 | ||
| Ad8E | 8 | 3 | |||
| Ad81 | 3 | ||||
| SmaI | Ad8A | 11 | 11 | 11 | 9 |
| Ad8B | 11 | 11 | 9 | ||
| Ad8E | 11 | 9 | |||
| Ad81 | 9 | ||||
| Total | Ad8A | 48 | 46 | 46 | 34 |
| Ad8B | 48 | 44 | 34 | ||
| Ad8E | 48 | 35 | |||
| Ad81 | 45 | ||||
Ad8, adenovirus type 8.
Nucleotide sequence analysis
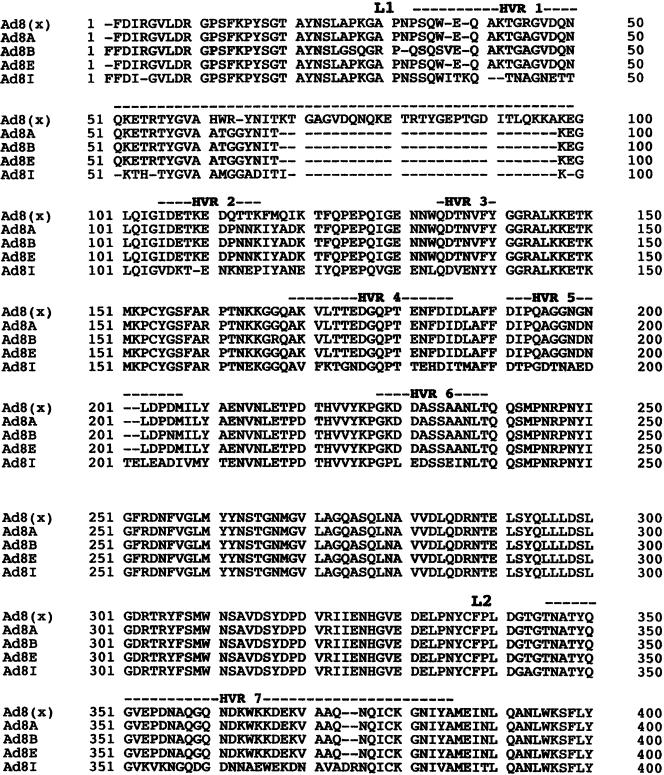
Ad8A, Ad8B, and Ad8E share 91.8% and 96.4% homology in their amino acid and nucleotide sequences, respectively, with the isolate 1127 (accession number, X74663) at the HVRs of the hexon gene. However, when compared with Ad8A, Ad8B, Ad8E, and isolate 1127, Ad8I shared only 62.7%. and 69.9% homology in amino acid and nucleotide sequences, respectively. Ad8A, Ad8B, and Ad8E showed a unique deletion of 31 amino acids in HVR 1, whereas Ad8I showed a 33 residue deletion (fig 4).
Figure 4.
Comparison of predicted amino acid sequences of seven hypervariable regions (HVRs) among the genome types of adenovirus type 8 (Ad8). The sequences of loop 1 (L1) and loop 2 (L2) have been aligned to obtain maximal homology. Deduced amino acid sequences of the Ad8 isolate, 1127, was obtained from Gene Bank (accession number, X74663). Ad8 (x) indicates isolate 1127.
DISCUSSION
Ad8 has a much higher tropism for conjunctival cells and produces more severe clinical manifestations and pathological alterations in EKC than do the Ad19 and Ad37 types. Thus, Ad8 has been the target of extensive studies, including those at the molecular level. This serotype persists in the population and causes sporadic epidemics, whereas Ad19 and Ad37 seemed to pass through the population in individual waves.9 Ad8 also has more genomic variants than Ad19 and Ad37.7,8,12,15 Genome typing by restriction endonucleases had been used successfully to establish separate identities at the molecular level of otherwise serologically identical strains. This typing offers a chance to follow the epidemiological distribution of the virus in different geographical regions and time periods. Ad8 is currently endemic in Japan and other East Asian countries.7,8,15 Ad8 isolates from the Asian Pacific and the USA are classified into genome types Ad8A to Ad8H (table 2). The interdomestic circulation of some genome types (for example, Ad8E in Taiwan, Korea, and Japan; Ad8D in Taiwan and the USA) has been documented. In Japan, molecular studies of Ad8 were conducted only in the northern part of the country (Sapporo) and only two genome types, Ad8A and Ad8B, were reported between 1975 and 1986.
Table 2.
Distribution of Ad8 genome types in Asia/Pacific
| Date | Country | Genome type |
| 1975–1997 | Japan | Ad8A, Ad8B, Ad8E, Ad8I |
| 1980–1994 | Taiwan | Ad8C, Ad8D, Ad8E, Ad8F, Ad8G, Ad8H |
| 1983 | Korea | Ad8E |
| 1983–1984 | Philippines | Ad8P |
| 1984–1986 | Australia | Ad8P |
Ad8A–Ad8I, genome types; Ad8P, prototype strain (Trim).
Among four genome types detected in Hiroshima, Ad8A and Ad8B circulated for a short period of time, from 1983 to 1988. However. Ad8E was genetically stable among the population of Hiroshima for a longer period of time, from 1984 to early 1995. In comparison, Ad8E circulated in Taiwan (Kaoshiung) and Korea (Pushan) in 1981–1987 and in 1983, respectively.
“The isolation of a new genome type is medically and epidemiologically important because the appearance of new genome types can result in more severe attacks of conjunctivitis”
The novel genome type Ad8I, which replaced Ad8E, circulated for a short period of time from 1995 to 1997. Ad8I was isolated from an outbreak of EKC in 1995, which affected mainly the older age group. The same genome type was isolated from sporadic cases until 1997. It is unclear whether this new genome type was imported from elsewhere or evolved indigenously. A large number of tourists visit Hiroshima every year. There is a strong possibility that some travellers may have unwittingly imported the new strain. This type of situation occurred in a Vietnamese refugee camp in Florida, USA in 1975, where Vietnamese children suffered from EKC as a result of Ad8D infection after they were transferred from Thailand to Florida. It is unlikely that Ad8I evolved from indigenous Ad8A, Ad8B, or Ad8E strains because Ad8B and Ad8E share 96% PCRF with Ad8A, whereas Ad8I shares only 71% restriction fragments. Therefore Ad8I is genetically distant from Ad8A, Ad8B, or Ad8E.
Ad8A, Ad8B, and Ad8E share 91.8% and 96.4% homology in their amino acid and nucleotide sequences, respectively, with the isolate 1127 (accession no, X74663). When compared with Ad8A, Ad8B, Ad8E, and isolate 1127, Ad8I shared only 62.7%. and 69.9% homology in amino acid and nucleotide sequences, respectively. Ad8A, Ad8B, and Ad8E all had a unique 31 amino acid deletion in HVR 1, whereas Ad8I had a 33 residue deletion. Changes in the hypervariable regions of the Ad8I strain provided it with its type specificity, as detected by the neutralisation assay and haemoagglutination inhibition test. Sequence variation in the hexon gene of Ad8I may have played an important role in the outbreak of EKC.17,18 An association was seen between the epidemic and the appearance of the new genome type (Ad8I), reminiscent of the isolation of a new Ad7 genotype in the Netherlands from epidemic cases.19
Take home messages.
Three known adenovirus type 8 (Ad8) genome types (Ad8A, Ad8B, and Ad8E) and a novel genome type (Ad8I) were detected in Hiroshima between 1983 and 1997
Ad8A, Ad8B, and Ad8E were closely related, with 96% homology, whereas Ad8I had only 71% homology
Ad8A, Ad8B, and Ad8E were closely related to the isolate 1127 (accession no X74663), whereas Ad8I was more distantly related
Ad8I was isolated from an outbreak of epidemic keratoconjunctivitis in 1995 and was also isolated from sporadic cases until 1997
The Ad8E strain that circulated from 1984 to 1995 was stable among the study population
Ad8 did not circulate as a single genome type in Hiroshima, but displayed a series of genetic changes, although the presence of Ad8E in Hiroshima over a 12 year period indicated that this genome type was very stable. The isolation of a new genome type is medically and epidemiologically important because the appearance of new genome types can result in more severe attacks of conjunctivitis.14 Continued investigations into the genome types of adenoviruses will help to define the unique evolutionary tendencies of these viruses.
Acknowledgments
Supported in part by grant in aid for Scientific Research (C) number 10470175 from the Ministry of Education, Science and Culture, Japan.
Abbreviations
Ad8, adenovirus type 8
EKC, epidemic keratoconjunctivitis
HVR, hypervariable region
PCR, polymerase chain reaction
PCRF, pairwise comigrating restriction fragments
SDS, sodium dodecyl sulfate
TE buffer, Tris/EDTA buffer
REFERENCES
- 1.Shenk T. Adenoviridae: the viruses and their replication. In: Fields BN, Knipe DM, Howley PM, eds. Fields’ virology. Philadelphia: Lippincort-Raven Publishers, 1996:2111–148.
- 2.De jong JC, Wermwnbol AG, Verweiji-Uijterwall MW, et al. Adenovirus from human immunodeficiency virus-infected individuals, including two strains that represent new candidate serotypes Ad50 and Ad51 of species B1 and D, respectively. J Clin Microbiol 1999;37:3940–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paval-Langston D. Viral diseases of the cornea and external eye. In: Albert DM, Jakobiec FA, Robinson N, eds. Ophthalmology. New York: WB Saunders Company, 1994:149–52.
- 4.Sprague JB, Hierholzer JC, Currier RW, et al. Epidemic keratoconjunctivitis: a severe industrial outbreak due to adenovirus type 8. N Engl J Med 1973;289:1341–6. [DOI] [PubMed] [Google Scholar]
- 5.Dawson C, Darnell R. Infection due to adenovirus type 8 in the United States I. An outbreak of epidemic keratoconjunctivitis originating in a physician’s office. N Engl J Med 1963;268:1301–4. [DOI] [PubMed] [Google Scholar]
- 6.Adhikary AK, Numaga J, Kaburaki T, et al. Rapid detection and typing of oculopathogenic strain of subgenus D adenoviruses by fiber based polymerase chain reaction and restriction enzyme analysis. Invest Ophthalmol Vis Sci 2001;42:201–15. [PubMed] [Google Scholar]
- 7.Fuji S, Nakazono N, Sawada H, et al. Restriction endonuclease cleavage analysis of adenovirus type 8: two new subtypes from patients with epidemic keratoconjunctivitis in Sapporo. Jpn J Med Sci Biol 1983;36:307–13. [DOI] [PubMed] [Google Scholar]
- 8.Fuji S, Nakazono N, Ishi K, et al. Molecular epidemiology of adenovirus type 8 (Ad8) in Taiwan: four subtypes recovered during the period of 1980–1981 from patients with epidemic keratoconjuctivitis. Jpn J Med Sci Biol 1984;37:161–9. [DOI] [PubMed] [Google Scholar]
- 9.Kemp MC, Hierholzer JC. Three adenovirus type 8 genome types defined by restriction enzyme analysis: prototype stability in geographically separated populations. J Clin Microbiol 1986;23:469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheu MM, Lin KH, Chen CW, et al. Changes of adenovirus type 8 isolated from viral conjunctivitis in the Kaoshiung area during 1980–1987: molecular epidemiological study by DNA endonuclease cleavage analysis. In: Shimizu K, ed. Current aspects in ophthalmology. Proceedings of the XIII Congress of the Asia-Pacific Academy of Ophthalmology, Kyoto. Amsterdam: Elsevier Science, 1992:228–33.
- 11.Chang CH, Sheu MM, Chern CL, et al. Epidemic keratoconjunctivitis caused by a new genotype of adenovirus type 8 (Ad8)—a chronological review of Ad8 in Southern Taiwan. Jpn J Ophthalmol 2001;45:160—6. [DOI] [PubMed] [Google Scholar]
- 12.Adrian T, Lauer HJ, Wigand R. Restriction site mapping of adenovirus type 8. Res Virol 1994;141:611–24. [DOI] [PubMed] [Google Scholar]
- 13.de Jong JC, Demazare M, L-Quillien MC, et al. New development in the molecular epidemiology of adenovirus 8 keratoconjuctivitis. J Med Virol 1992;38:102–7. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka K, Itoh N, Saitoh-Inagawa W, et al. Genetic characterization of an adenovirus strain isolated from a patient with acute conjunctivitis in the city of Sao Paulo, Brazil. J Med Virol 2000;61:143–9. [PubMed] [Google Scholar]
- 15.Ishii K, Nakazono N, Fujinaga K, et al. Comparative studies on viral conjunctivitis in three countries of east Asia–Japan, Taiwan and South Korea. Int J Epidemiol 1987;16:98–103. [DOI] [PubMed] [Google Scholar]
- 16.Yamadera S, Yamashita K, Akatsuka M, et al. Adenovirus surveillance, 1982–1995. Jpn J Med Sci Biol 1995;48:199–21. [PubMed] [Google Scholar]
- 17.Gall JG, Crystal RG, Falk-Pederson E. Construction and characterization of hexon-chimeric adenovirus: specification of adenovirus serotype. J Virol 1998;72:10260–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy S, Shirley PS, McClelland A, et al. Circumvention of immunity to the adenovirus major coat protein hexon. J Virol 1998;72:6875–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wadell G, de Jong JC, Wolonitis S. Molecular epidemiology of adenoviruses: alternating appearance of two different genome types of adenovirus 7 during epidemic outbreaks in Europe from 1958 to 1980. Infect Immun 1981;34:368–72. [DOI] [PMC free article] [PubMed] [Google Scholar]