Abstract
The identification of the non-random chromosome rearrangements between the EWS gene on chromosome 22q12 and members of the ETS gene family in Ewing’s sarcoma, peripheral primitive neuroectodermal tumour, Askin tumour, and neuroepithelioma has been a key advance in understanding their common histogenesis and defining the Ewing’s sarcoma family of tumours (ESFT). In addition to improvements in diagnosis and potentially the stratification of patients for risk, biological investigations of these gene fusions may define targets for much needed therapeutic strategies to eliminate minimal residual disease or metastatic disease. Insight into their relation with other oncogenic events in ESFT will advance risk group analysis and ultimately may improve clinical management and survival for patients with this disease.
Keywords: Ewing’s sarcoma, diagnosis, prognosis, treatment, translocations
Ewing’s sarcoma is the second most common malignant bone tumour occurring in children and young adults, and accounts for 10–15% of all primary bone tumours.1 The annual incidence is approximately 0.6/million total population, and it usually occurs between the ages of 10 and 20 years. It affects 13/million 0–24 year olds each year in the UK,2 and is slightly more common in males than females (ratio, 1.5 : 1). It has been described in siblings,3,4 although this is rare and the disease does not appear to be implicated in familial cancer syndromes. Genetic influences may play some role in its aetiology because black Afro-Caribbean and Chinese populations are less frequently affected than the white population.5,4
Ewing’s sarcoma can affect any bone but the most common sites are the lower extremity (45%), followed by the pelvis (20%), upper extremity (13%), axial skeleton and ribs (13%), and face (2%).6 The femur is the most frequently affected bone, with the tumour usually arising in the midshaft. Typically, by light microscopy, the tumour consists of small round cells with regular round nuclei containing finely dispersed chromatin and inconspicuous nucleoli, and a narrow rim of clear or pale cytoplasm. Ultrastructurally, the tumour contains primitive cells with a smooth nuclear surface, scanty organelles, and cytoplasmic glycogen in pools or aggregates.
“The femur is the most frequently affected bone, with the tumour usually arising in the midshaft”
Tumours with similar histology also arise in soft tissues. These include peripheral primitive neuroectodermal tumour (pPNET), neuroepithelioma, and Askin tumour. pPNET is the second most common soft tissue malignancy in childhood, accounting for 20% of sarcomas.7 It is frequently found in the chest wall (Askin tumour), paraspinal tissues, abdominal wall, head and neck, and extremities.7,8 However, soft tissue extension is common in osseous Ewing’s sarcoma and infiltration of adjacent bone is frequent in soft tissue pPNETs, which often makes it difficult to determine the primary site of tumour origin.
The identification of the non-random t(11;22)(q24;q12) chromosome rearrangement9,10 in these aggressive malignant tumours arising in bone and soft tissue is strong evidence for their common histogenesis, and provides a valuable characteristic for their differential diagnosis from other small round cell tumours of childhood. These tumours, including Ewing’s sarcoma, pPNET, Askin tumour, and neuroepithelioma, are now collectively recognised as the Ewing’s sarcoma family of tumours (ESFT). In this review, the diagnostic, prognostic, and therapeutic power of the t(11;22)(q24;q12) translocation and other molecular abnormalities in ESFT will be reviewed.
DIAGNOSIS
Accurate diagnosis of ESFT is crucial for the most appropriate clinical management of patients. Adequate clinical information and the recognition of the morphological, immunocytochemical, and sometimes ultrastructural features (table 1) of ESFT are all required for its differential diagnosis from other small round cell tumours of childhood, such as neuroblastoma, rhabdomyosarcoma, lymphoma (non-Hodgkin’s), other primitive neuroectodermal tumours, desmoplastic small round cell tumour, poorly differentiated synovial sarcoma, and small cell osteosarcoma.
Table 1.
EWS fusion types described in Ewing’s sarcoma family of tumours (ESFT) and other sarcomas
| Translocation | Gene fusion | Tumour type (% of tumours with this EWS gene rearrangement) |
| t(11;22)(q24;q12) | EWS–FLI1 | ESFT (85%) |
| t(21;22)(q22;q12) | EWS–ERG | ESFT (10%) |
| t(7;22)(p22;q12) | EWS–ETV1 | ESFT (rare) |
| t(17;22)(q12;q12) | EWS–E1AF | ESFT (rare) |
| t(2;22)(q33;q12) | EWS–FEV | ESFT (rare) |
| t(12;22)(q13;q12) | EWS–AFT1 | Clear cell sarcoma |
| t(11;22)(q13;q12) | EWS–WT1 | Desmoplastic small round cell tumour |
| t(9;22)(q22;q12) | EWS–CHN | Myxoid chondrosarcoma |
| t(12;22)(q13;q12) | EWS–CHOP | Myxoid liposarcoma |
The presence of EWS–ETS gene rearrangements is increasingly used to define ESFT. The involvement of the EWS gene on chromosome 22q is consistent; this can partner with several different ETS gene family members located on various chromosomes, but most frequently with FLI1 on chromosome 11 in ESFT. Rearrangements of the EWS gene on chromosome 22q12 with other chromosomes have also been described in other less common sarcoma types (shown in italics).
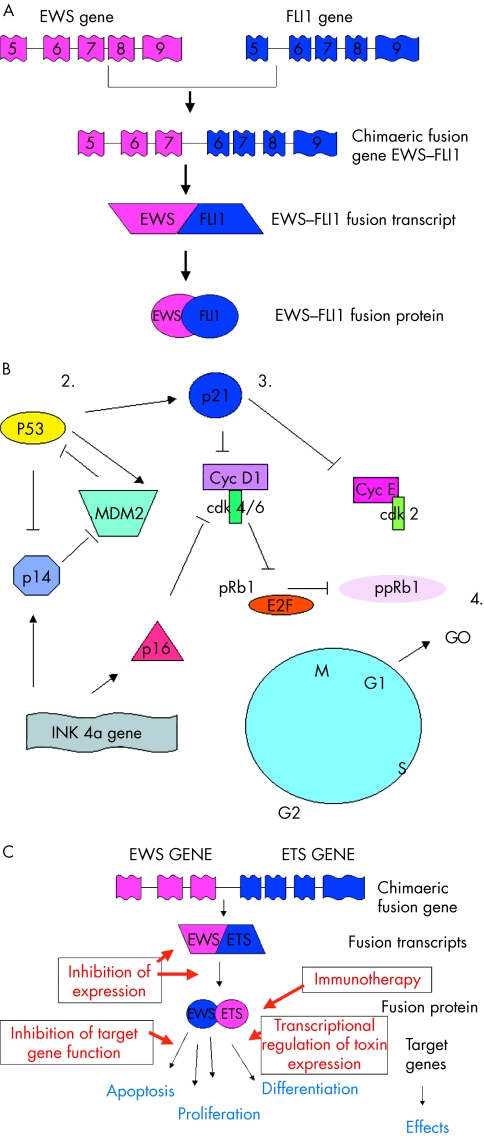
Increasingly, ESFT are being characterised by non-random gene rearrangements between the EWS gene on 22q12 and various members of the ETS gene family (table 1). The most frequent gene rearrangement is the t(11;22)(q24;q12) translocation, found in about 85% of these tumours (fig 1; table 1). Fusion of the EWS gene on 22q12 with the FLI1 gene on 11q24 results in a chimaeric fusion transcript EWS–FLI1 (fig 2A). Substitution of the EWS domain with a portion of the FLI1 transcriptional domain results in an EWS–FLI1 fusion transcript with increased transcriptional activity compared with that of normal FLI1. The chimaeric protein product is capable of transforming NIH3T3 cells in vitro,11–13 but neither EWS nor FLI1 alone have this capacity.
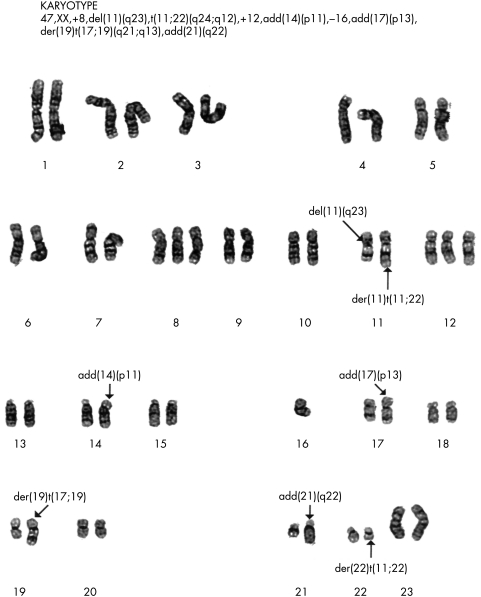
Figure 1.
G banding of chromosomes from a Ewing’s sarcoma showing the t(11.22)(q12.24) translocation. This tumour is also characterised by trisomy of chromosome 8 and 12 and loss of chromosome 16, frequently described secondary aberrations in the Ewing’s sarcoma family of tumours. Reproduced with the permission of P Roberts, Department of Cytogenetics, St James’s University Hospital, Leeds, UK.
Figure 2.
(A) Diagrammatical representation of the t(11;22) (q24;q12) translocation resulting in the generation of the EWS–FLI1 type 1 fusion transcript. (B) Cell cycle regulatory proteins implicated in regulation of the G1 checkpoint. 1. Despite the absence of cytogenetically detectable 9p21 chromosomal aberrations, p16 deletions are one of the most frequent secondary molecular aberrations identified to date in the Ewing’s sarcoma family of tumours (ESFT). 2. p53 is rarely mutated in primary ESFT, although mutation in this tumour suppressor gene has been described in up to 50% of ESFT cell lines. Mutation of p53 in ESFT is associated with a poor outcome for a small group of patients. 3. Mutation of the p21WAF gene and/or downregulation of its nuclear expression is a common aberration in ESFT. For most tumours, loss of p21WAF expression is independent of p53 status, suggesting gene silencing of WAF1 by mutation or hypermethylation. 4. Loss of pRb1 is rarely described in ESFT, but abrogation of the G1 checkpoint appears to be important in the progression and development of the ESFT clinical phenotype. Cdk, cyclin dependent kinase; Cyc, cyclin; pRb1, retinoblastoma protein 1; ppRb1, phosphorylated pRb1. (C) Potential strategies to exploit EWS–ETS gene rearrangements for therapeutic purposes.
Other EWS–ETS gene family rearrangements have been identified in the remaining 15% of tumours, with the t(21;22)(q22;q12) translocation resulting in fusion of EWS with the ERG gene on 21q22 being the second most common (table 1).14 The fusion protein derived from EWS–ERG is similar to that of the EWS–FLI1 gene product, with EWS replacing the transcriptional domain of ERG. Other variant translocations have been described where EWS is fused with ETV1 (t(7;22)(p22;q12)),14 E1AF (t(17;22)(q21;q12)),15 and FEV (t(2;22)(q33;q12)),16 in addition to more complex translocations involving multiple chromosomes such as t(11;14;22)(q24;q11;q12) and t(10;11;22)(p11.2;q24;q12). The biological importance of these more complex gene rearrangements is not yet clear, but they may be associated with more aggressive tumour behaviour (P Roberts, personal communication, 2002). There has also been at least one case of cryptic EWS–FLI1 fusion in a tumour with normal chromosomes 11 and 22.17
“Accurate diagnosis of the Ewing’s sarcoma family of tumours is crucial for the most appropriate clinical management of patients”
Therefore, cytogenetic evaluation of ESFT is an essential part of their diagnostic evaluation. Conventional G banding may identify EWS–ETS gene rearrangements in a proportion of tumours, although diagnostic accuracy can be enhanced using either the reverse transcriptase polymerase chain reaction (RT-PCR)18 and/or fluorescent in situ hybridisation (FISH).19–21 The presence of EWS–ETS gene fusion products detected by RT-PCR or FISH correlates well with conventional G banding studies, but perhaps more interestingly these techniques can be used to identify gene fusions in tumours where conventional G banding has been uninformative, either because of a very complex gene rearrangement or the lack of mitotic cells. Indeed, the ability of RT-PCR to detect fusion transcripts in small pieces of tumour, even in fine needle biopsies or bone marrow aspirations, makes this a very attractive tool to aid in the diagnosis of ESFT.18
The EWS gene is also rearranged in several other tumours, which are clinically and morphologically distinct from ESFT (table 1). A multiplex RT-PCR for these different gene rearrangements can be very useful to aid in the differential diagnosis of sarcomas.18 However, there are some reports of EWS–ETS gene arrangements in biphenotypic sarcomas with features of muscle and neural differentiation,22,23 and rarely in rhabdomyosarcomas,24 neuroblastomas,25 rhabdoid tumour,26 extraskeletal mesenchymal chondrosarcoma,27 and solid papillary tumour of the pancreas.28 Whether these examples represent extremes of the ESFT or rare pathological entities themselves is not clear. Consequently, the presence of an EWS–ETS gene rearrangement alone may not be sufficiently powerful to diagnose ESFT, but in conjunction with other clinical and pathological criteria it is pathognomonic of this tumour group.29
PROGNOSTIC VALUE
Improved outcome may be achieved by stratifying patients for treatment according to risk. A recent systematic review of prognostic tumour markers in ESFT has been undertaken (NHS health and technology assessment programme; grant number 97/15/03; for further details of these results see http://www.prw.le.ac.uk/epidemio/personal/rdr3/paed.html). However, very few prognostic markers are routinely used in ESFT, probably as a result of the small number of studies and patients evaluated in clinical outcome investigations. This identifies the need for multicentre, quality controlled, prospective clinical outcome studies in ESFT. Currently in ESFT the most useful prognostic indicators in clinical practice are clinical features, including the presence of metastatic disease at diagnosis (measured by imaging and histological examination of bone marrow), tumour volume, and primary tumour site (patients with a primary tumour of the pelvis have a worse prognosis).
Using RT-PCR to detect EWS–ETS fusion transcripts several alternative forms of EWS–FLI1 have been described, reflecting the different breakpoints in FLI1 and EWS. The most common type, designated EWS–FLI1 type 1, consists of the first seven exons of EWS joined to exons 6 to 9 of FLI1, and accounts for approximately 60% of cases (fig 2A). The type 2 EWS–FLI1 fusion also includes FLI1 exon 5 and is present in a further 25%.30 Recent studies have suggested that the fusion transcript type may be prognostically powerful,30,31 with the presence of EWS–FLI1 fusion transcript type 1 being associated with improved outcome compared with that in patients with other fusion transcript types.30,31 However, this may be limited to patients with localised disease.30 The EWS–FLI1 fusion transcript type 1 appears to encode a less active chimaeric transcription factor32 and to be associated with a lower proliferation index33 than that of tumours with other fusion transcript types; this might provide the biological basis for the correlation between fusion transcript type and clinical outcome. However, other studies have shown that the two gene fusion products EWS–FLI1 and EWS–ERG do not define distinct clinical phenotypes, suggesting that differences in the C-terminal partner of ESFT gene fusions are not associated with significant phenotypic differences.34 The clinical and biological relevance of different fusion transcript types is currently being evaluated through a European prospective, quality controlled, clinical outcome study, namely: the European Ewing tumour working initiative of national groups 99 (Euro EWING 99).
The presence of disseminated disease, detected by imaging and cytological and histological examination of bone marrow at diagnosis in ESFT, is one of the most powerful markers of poor prognosis. However, some patients with apparently localised disease at diagnosis rapidly develop metastases and aggressive disease, from which they subsequently die. This suggests that these patients have low level metastatic disease that is not detected by current routine methods, such as imaging and histological examination of bone marrow at diagnosis. Fusion products of specific EWS–ETS gene rearrangements have successfully been used as targets for RT-PCR to identify low numbers of circulating ESFT cells with improved sensitivity and specificity.35–37 Disease detected in the peripheral blood by RT-PCR appears not to be clinically informative, although in bone marrow it may predict a poor clinical outcome.35,36,38–41 This suggests that RT-PCR for the EWS–ETS fusion transcript type in bone marrow may be useful to improve the stratification of patients for treatment and could result in a redefinition of the term disease free. This hypothesis is currently being evaluated in Euro EWING 99.
Additional chromosomal abnormalities have also been described in ESFT, although these are less frequently described than rearrangements of the EWS–ETS genes and their clinical implication less well understood. They consist of both numerical and structural aberrations, including gains of chromosomes 842–46 and 12,43–46 the unbalanced translocation t(1;16),42,46–50 and deletions at the short arm of chromosome 1.46,50 Initial studies suggested that individuals with gain of chromosome 8 might have a poor clinical outcome,43,45 although this has recently been disputed.48 Deletion of chromosome 1 is reported to be associated with an unfavourable outcome in individuals with localised disease.48 Up to 15 different chromosomes, other than those relating to EWS–ETS gene rearrangements, have been implicated in ESFT, although much of this has been in the form of individual patient data (see http://www.prw.le.ac.uk/epidemio/personal/rdr3/paed.html). It is important to evaluate the frequency and clinical importance of these additional chromosomal aberrations in large clinical outcome studies, because a more complex karyotype with multiple chromosomal aberrations appears to be associated with poor outcome and may be prognostically powerful in ESFT.46,50
Loss of cell cycle control occurs in a variety of malignancies, often as secondary genetic changes during multistage progression. In ESFT, abrogation of the G1 checkpoint appears to be important in the progression and development of the clinical phenotype (fig 2B),50–52 consistent with the hypothesis of unchecked cell division. This hypothesis is supported by recent studies demonstrating changes in G1/S regulatory genes after downregulation and forced expression of the EWS–FLI1 fusion gene.53
Homozygous deletions and/or mutations of the INK4a gene on chromosome 9p21 have been reported in seven of 4154 and eight of 2750 primary ESFTs, although one study failed to identify mutations of this gene in all 70 primary ESFTs tested.55 Interestingly, loss of the INK4a gene product p16INK4a in four of 20 cases was associated with shortened survival and the presence of metastatic disease at diagnosis. Conversely, loss of the alternatively spliced INK4a variant p14ARF is rarely described even in tumours where p16INK4a is lost.52 This is consistent with the hypothesis that selective loss of p16INK4a function may save p14ARF,56 with epigenetic inactivation by promoter hypermethylation of the E1α exon resulting in the loss of p16INK4a expression but retaining expression of functional p14ARF.57
“Although the loss of p21WAF expression appears to be the most common aberration in primary Ewing’s sarcoma family of tumours, no correlation with clinical behaviour has yet been reported”
The loss of retinoblastoma gene expression is very rarely described in ESFT,50,52 and although mutations in the tumour suppressor gene p53 have been described in up to 50% of ESFT cell lines,58,59 this appears to be a rare event in primary tumours.51,60–62 However, in the small clinical subset of patients with ESFT in whom p53 is mutated this is associated with a very poor outcome,51,62 although the expression of p53 protein is reported by some to correlate with a poor prognosis.63 Amplification of the MDM2 gene, which inactivates the p53 protein, is also rare in ESFT,51,54 consistent with the hypothesis that p53 and regulators of its activity may not play a dominant role in ESFT transformation and progression.
One of the most frequent aberrations of a tumour suppressor in ESFT reported to date is loss of p21WAF expression (11 of 20,52 and 30 of 5062). Because p21WAF is transactivated by wild-type but not mutated p53, loss of its expression might reflect inactivation of p53. This is true for some ESFTs,64 but for most loss of p21WAF expression is independent of p53 status, suggesting that gene silencing of WAF1 by mutation or hypermethylation may be important in these tumours. Although the loss of p21WAF expression appears to be the most common aberration in primary ESFT, no correlation with clinical behaviour has yet been reported, although there are reports in other tumour types of its prognostic power.65–67
THERAPEUTIC VALUE
In human ESFT cells, knockout studies using anti-sense EWS–FLI1 oligonucleotides or cDNAs have shown reduced growth in in vitro68–70 and in vivo69,71 models. These observations imply that EWS–ETS gene rearrangements are important for the maintenance of the ESFT malignant phenotype. Coupled with the high frequency of these rearrangements in ESFT, this suggests that the products of these gene rearrangements play a role in the genesis of ESFT. As the genetics and biology of these gene fusions and their protein products are elucidated, their potential as targets for therapeutic intervention are being explored so they may be exploited for the benefit of adolescents and adults with these tumours (fig 2C).
In animal models, the chimaeric fusion transcripts are reported to act as dominant oncogenes, contributing to the development of ESFT by binding to the promoter region of specific genes, and altering their expression.11,12,72–74 Deletion analysis suggests that the EWS gene functions as the regulatory or modulating domain, with the ETS gene partners possibly acting to establish promoter activity and also to link with signal transduction pathways.75 Gene expression is a multistage process potentially providing several steps that may be targeted as a therapeutic strategy. Although general inhibitors of transcription and translation may effectively downregulate EWS–FLI1 protein expression and induce growth arrest in vitro,76 this is unlikely to be specific enough for therapeutic use. However, more specific targeting of oncogene expression, through the use of antisense reagents to interfere with ribosomal translation, degradation, or processing of pre-mRNA complexes may be more successful. Because EWS–ETS oncoproteins are thought to protect ESFT cells from apoptosis, antisense treatment may increase the efficacy of chemotherapeutic strategies,77 or conversely may have a direct effect by downregulating the role of EWS–FLI1 in maintaining cell proliferation.68,69,76 Recent studies have shown that the delivery of antisense oligonucleotides in polyalkylcyanoacrylate nanocapsules results in a greater inhibition of ESFT growth in nude mice than that seen with free antisense oligonucleotide injections,71 although the delivery of antisense reagents in vivo remains a challenge in therapeutic trials. Alternatively, the oncogenic function of the EWS–ETS proteins may be targeted by inhibition of the protein itself using—for example, a competitive dominant negative strategy for the selective disruption of gene translation and expression.68
“Recent studies have also shown that EWS–ETS may interfere with normal RNA processing, suggesting that the gene fusions may function both in transcriptional and post transcriptional processes to modulate growth”
The primary goal for much research that aims to exploit the therapeutic value of EWS–ETS gene fusions has been to identify the EWS–ETS fusion target genes responsible for its oncogenic activity. Because the transformed phenotype of these tumours is thought to be dependent on the deregulated expression of target genes, transcriptional repressors that downregulate the same transcriptional targets may be used to inhibit the function of these oncoproteins.78 This approach has been evaluated in alveolar rhabdomyosarcoma.79 Alternatively, fusion oncoprotein function may be inhibited by expressing antibody fragments intracellularly.80 Although the genes targeted by the EWS–ETS fusion proteins are not yet fully defined, analysis of RNAs differentially expressed by NIH3T3 cells expressing FLI1 or EWS–FLI1 have identified some interesting candidate genes including Manic Fringe (m-FNG),81 stromelysin,82 EAT-2,74 E2-C,83 transforming growth factor βII receptor (TGFβ2 receptor),84,85 and platelet derived growth factor C (PDGF-C).86 However, no single or group of gene(s) has yet been identified that reproduces the EWS–FLI1 phenotype, and thus far direct target genes have not been distinguished from indirect ones.
The function of these novel fusion proteins as potent transcription factors may also be exploited to increase the delivery of cytotoxin directed treatments. Because some of the fusion transcripts have been shown to be more potent transcriptional activators than their wild-type counterparts, the delivery of exogenous toxic genes under the control of these regulatory elements is a potentially specific mechanism to target these tumours. Although normal cells may express the wild-type gene, its reduced transcriptional activity is hypothesised to result in little or no activation of the exogenous toxin. This approach has yet to be evaluated in ESFT, but in alveolar rhabdomyosarcoma target expression of the diphtheria toxin A gene has resulted in expression and appropriately selective toxicity.87
Take home messages.
EWS–ETS gene rearrangements are important for the differential diagnosis of the Ewing’s sarcoma family of tumours
EWS–ETS fusion transcript type may predict clinical behaviour
Elucidating the biology of these gene fusions and their protein products may provide targets for novel therapeutic intervention
Insight into other molecular abnormalities in ESFT is essential
Recent studies have also shown that EWS–ETS may interfere with normal RNA processing,88,89 suggesting that the gene fusions may function both in transcriptional and post transcriptional processes to modulate growth. Further evidence that EWS–ETS fusion transcripts may operate through mechanisms other than promoters of specific target genes comes from the observations that EWS–FLI1 binds a nuclear ribonucleoprotein involved in RNA splicing, which can result in inhibition of EWS–FLI1 transactivation,90 and that an artificial EWS–FLI1 DNA binding domain mutant that has no in vitro binding activity still retains some transforming activity.91
An alternative therapeutic strategy in ESFT may be to exploit the EWS–ETS gene fusions as novel tumour antigens for immunotherapy. The intracellular expression of translocation protein products protects them from immune surveillance. If the fusion transcripts are to be exploited as novel tumour antigens to increase immune recognition of tumour cells then the peptides derived from these fusion proteins (in conjunction with major histocompatability complex class I molecules) must be expressed on the tumour cell surface. This strategy is being explored in alveolar rhabdomyosarcoma,92 synovial sarcoma, clear cell sarcoma, desmoplastic small round cell tumour,93 and a variety of other sarcomas.94,95
CONCLUSIONS
In summary, as in many cancers, improved diagnostic and staging methodologies for patients with ESFT will lead to better risk group analysis and ultimately may improve clinical management and survival for patients with this disease. The identification and characterisation of the EWS gene rearrangements in ESFT has been the most important advance made in these tumours in the past two decades, leading to improvements in diagnosis and potentially stratification of patients for risk. Current biological investigations of these gene fusions may define targets for much needed therapeutic strategies to eliminate minimal residual or minimal metastatic disease, although delivery of these novel agents remains a major challenge. Understanding the role of these tumour specific fusion transcripts and protein products is essential, but insight into their relation with other oncogenic events in ESFT is crucial so that molecular abnormalities may be exploited for the maximum benefit of individuals afflicted with this disease.
Abbreviations
ESFT, Ewing’s sarcoma family of tumours
Euro EWING 99, European Ewing tumour working initiative of national groups 99
FISH, fluorescent in situ hybridisation
pPNET, peripheral primitive neuroectodermal tumour
RT-PCR, reverse transcriptase polymerase chain reaction
REFERENCES
- 1.Huvos AG. Ewing’s sarcoma. In: Huvos AG, ed. Bone tumors: diagnosis, treatment and prognosis, 2nd ed. Philadelphia PA: Sanders, 1991:523–52.
- 2.Cotterill SJ, Parker L, Malcolm AJ, et al. Incidence and survival for cancer in children and young adults in the North of England, 1968–1995: a report from the Northern Region young persons’ malignant disease registry. Br J Cancer 2000;83:397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hutter RVP, Francis KC, Foote FW. Ewing’s sarcoma in siblings. Am J Surg 1964;107:598. [DOI] [PubMed] [Google Scholar]
- 4.Zamora P, Garcia de Paredes ML, Gonzalez Baron M, et al. Ewing’s tumor in brothers. An unusual observation. Am J Clin Oncol 1986;9:358–60. [DOI] [PubMed] [Google Scholar]
- 5.Joyce MJ, Harmon DC, Mankin HJ, et al. Ewing’s sarcoma in female siblings: a clinical report and review of the literature. Cancer 1984;53:1959–62. [DOI] [PubMed] [Google Scholar]
- 6.Grier HE. The Ewing family of tumors. Ewing’s sarcoma and primitive neuroectodermal tumors. Pediatr Clin North Am 1997;44:991–1104. [DOI] [PubMed] [Google Scholar]
- 7.Harms D. Soft tissue sarcoma in the Kiel pediatric registry. Curr Top Pathol 1995;89:31–45. [DOI] [PubMed] [Google Scholar]
- 8.Kimber C, Michalski A, Spitz L, et al. Primitive neuroectodermal tumours: anatomic location, extent of surgery, and outcome. J Pediatr Surg 1998;33:39–41. [DOI] [PubMed] [Google Scholar]
- 9.Aurias A, Rimbaut C, Buffe D, et al. Translocation involving chromosome 22 in Ewing’s sarcoma. A cytogenetic study of four fresh tumors. Cancer Genet Cytogenet 1984;12:21–5. [DOI] [PubMed] [Google Scholar]
- 10.Turc-Carel C, Philip I, Berger MP, et al. Chromosome study of Ewing’s sarcoma (ES) cell lines. Consistency of a reciprocal translocation t(11;22)(q24;q12). Cancer Genet Cytogenet 1984;12:1–19. [DOI] [PubMed] [Google Scholar]
- 11.May WA, Lessnick SL, Braun BS, et al. The Ewing’s sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol Cell Biol 1993;13:7393–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohno T, Rao VN, Reddy ES. EWS/Fli-1 chimeric protein is a transcriptional activator. Cancer Res 1993;53:5859–63. [PubMed] [Google Scholar]
- 13.Chan JHC. Molecular analysis of primitive neuroectodermal tumors: a new model for the study of solid tumors showing specific chromosomal translocations. Adv Anat Pathol 1994;1:87. [Google Scholar]
- 14.Sorensen PH, Triche TJ. Gene fusions encoding chimaeric transcription factors in solid tumours. Semin Cancer Biol 1996;7:3–14. [DOI] [PubMed] [Google Scholar]
- 15.Urano F, Umezawa A, Hong W, et al. A novel chimera gene between EWS and EIA-F, encoding the adenovirus enhancing-binding protein, in extraosseous Ewing’s sarcoma. Biochem Biophys Res Commun 1996;219:608–12. [DOI] [PubMed] [Google Scholar]
- 16.Peter M, Couturier J, Pacquement H, et al. A new member of the ETS family fused to EWS in Ewing’s tumors. Oncogene 1997;14:1159–64. [DOI] [PubMed] [Google Scholar]
- 17.Batanian JR, Bridge JA, Wickert R, et al. EWS/FLI-1 fusion signal inserted into chromosome 11 in one patient with morphologic features of Ewing sarcoma, but lacking t(11;22). Cancer Genet Cytogenet 2002;133:72–5. [DOI] [PubMed] [Google Scholar]
- 18.Peter M, Gilbert E, Delattre O. A multiplex real-time PCR assay for the detection of gene fusions observed in solid tumors. Lab Invest 2001;81:905–912. [DOI] [PubMed] [Google Scholar]
- 19.Desmaze C, Zucman J, Delattre O, et al. Unicolor and bicolour in situ hybridization in the diagnosis of peripheral neuroepithelioma and related tumors. Genes Chromosomes Cancer 1992;5:30–4. [DOI] [PubMed] [Google Scholar]
- 20.Taylor C, Patel K, Jones T, et al. Diagnosis of Ewing’s sarcoma and peripheral neuroectodermal tumour based on the detection of t(11;22) using fluorescence in situ hybridisation. Br J Cancer 1993;67:128–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McManus AP, Gusterson BA, Pinkerton CR, et al. Diagnosis of Ewing’s sarcoma and related tumours by detection of chromosome 22q12 translocations using fluorescence in situ hybridization on tumour touch imprints. J Pathol 1995;176:137–42. [DOI] [PubMed] [Google Scholar]
- 22.Sorensen PHB, Shimada H, Liu XF, et al. Biphenotypic sarcomas with myogenic and neural differentiation express the Ewing’s sarcoma EWS/FLI1 fusion gene. Cancer Res 1995;55:1385–92. [PubMed] [Google Scholar]
- 23.Tan S-Y, Burchill S, Brownhill SC, et al. Small round cell tumour with biphenotypic differentiation and variant of t(21;22)(q22;q12). Pediatr Dev Pathol 2001;4:391–6. [DOI] [PubMed] [Google Scholar]
- 24.Thorner P, Squire J, Chilton-MacNeil S, et al. Is the EWS–FLI-1 fusion transcript specific for Ewing sarcoma and peripheral primitive neuroectodermal tumor? A report of four cases showing this transcript in a wider range of tumor types. Am J Pathol 1996;148:1125–38. [PMC free article] [PubMed] [Google Scholar]
- 25.Burchill SA, Wheeldon J, Cullinane C, et al. EWS–FLI1 fusion transcripts identified in patients with typical neuroblastoma. Eur J Cancer 1997;33:239–43. [DOI] [PubMed] [Google Scholar]
- 26.Mastik MF, Molenaar WM, Boudewijn E, et al. Translocation (11;22)(q24;q12) in a small cell tumor of the thigh in a two-year-old boy: immunohistology, cytogenetics, molecular genetics, and review of the literature. Hum Pathol 1999;30:352–5. [DOI] [PubMed] [Google Scholar]
- 27.Sainati L, Scapinello A, Montaldi A, et al. A mesenchymal chondrosarcoma of a child with the reciprocal translocation (11; 22)(q24;q12). Cancer Genet Cytogenet 1993;71:144–7. [DOI] [PubMed] [Google Scholar]
- 28.Maitra A, Weinberg AG, Schneider N, et al. Detection of the t(11;22)(q24;q12) translocation and EWS–FLI1 fusion transcript in a case of solid pseudopapillary tumor of the pancreas. Pediatr Dev Pathol 2000;3:603–5. [DOI] [PubMed] [Google Scholar]
- 29.de Alava E. Transcripts, transcripts everywhere. Adv Anat Pathol 2001;8:264–72. [DOI] [PubMed] [Google Scholar]
- 30.Zoubek A, Dockhorn-Dworniczak B, Delattre O, et al. Does expression of different EWS chimeric transcripts define clinically distinct risk groups of Ewing tumor patients? J Clin Oncol 1996;14:1245–51. [DOI] [PubMed] [Google Scholar]
- 31.de Alava E, Kawai A, Healey JH, et al. EWS–FLI1 fusion transcript structure is an independent determinant of prognosis in Ewing’s sarcoma. J Clin Oncol 1998;16:1248–55. [DOI] [PubMed] [Google Scholar]
- 32.Lin PP, Brody RI, Hamelin AC, et al. Differential transactivation by alternative EWS–FLI1 fusion proteins correlates with clinical heterogeneity in Ewing’s sarcoma. Cancer Res 1999;59:1428–14. [PubMed] [Google Scholar]
- 33.de Alava E, Panizo A, Antonescu CR, et al. Association of EWS–FLI1 type 1 fusion with lower proliferative rate in Ewing’s sarcoma. Am J Pathol 2000;156:849–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ginsberg JP, de Alava E, Ladanyi M, et al. EWS–FLI1 and EWS–ERG gene fusions are associated with similar clinical phenotypes in Ewing’s sarcoma. J Clin Oncol 1999;17:1809–14. [DOI] [PubMed] [Google Scholar]
- 35.Peter M, Magdelenat H, Michon J, et al. Sensitive detection of occult Ewing’s cells by the reverse transcriptase-polymerase chain reaction. Br J Cancer 1995;72:96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.West DC, Grier HE, Swallow MM, et al. Detection of circulating tumor cells in patients with Ewing’s sarcoma and peripheral primitive neuroectodermal tumor. J Clin Oncol 1997;15:583–8. [DOI] [PubMed] [Google Scholar]
- 37.de Alava E, Lozano MD, Patino A, et al. Ewing family tumours: potential prognostic value of reverse-transcriptase polymerase chain reaction detection of minimal residual disease in peripheral blood samples. Diagn Mol Pathol 1998;7:152–7. [DOI] [PubMed] [Google Scholar]
- 38.Pfleiderer C, Zoubek A, Gruber B, et al. Detection of tumour cells in peripheral blood and bone marrow from Ewing tumour patients by RT-PCR. Int J Cancer 1995;64:135–9. [DOI] [PubMed] [Google Scholar]
- 39.Toretsky JA, Neckers L, Wexler LH. Detection of (11;22)(q24;q12) translocation-bearing cells in peripheral blood progenitor cells of patients with Ewing’s sarcoma family of tumors. J Natl Cancer Inst 1995;87:385–6. [DOI] [PubMed] [Google Scholar]
- 40.Zoubek A, Ladenstein R, Windhager R, et al. Predictive potential of testing for bone marrow involvement in Ewing tumor patients by RT-PCR: a preliminary evaluation. Int J Cancer 1998;79:56–60. [DOI] [PubMed] [Google Scholar]
- 41.Fagnou C, Michon J, Peter M, et al. Presence of tumor cells in bone marrow but not in blood is associated with adverse prognosis in patients with Ewing’s tumor. J Clin Oncol 1998;16:1707–11. [DOI] [PubMed] [Google Scholar]
- 42.Mugneret F, Lizard S, Aurias A, et al. Chromosomes in Ewing’s sarcoma. II. Nonrandom additional changes, trisomy 8 and der(16)t(1;16). Cancer Genet Cytogenet 1988;32:239–45. [DOI] [PubMed] [Google Scholar]
- 43.Armengol G, Tarkkanen M, Virolainen M, et al. Recurrent gains of 1q, 8 and 12 in the Ewing family of tumours by comparative genomic hybridisation. Br J Cancer 1997;75:1403–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maurici D, Perez-Atayde A, Grier HE, et al. Frequency and implications of chromosome 8 and 12 gains in Ewing sarcoma. Cancer Genet Cytogenet 1998;100:106–10. [DOI] [PubMed] [Google Scholar]
- 45.Tarkkanen M, Kiuru-Kuhlefelt S, Blomqvist C, et al. Clinical correlations of genetic changes by comparative genomic hybridization in Ewing sarcoma and related tumors. Cancer Genet Cytogenet 1999;114:35–41. [DOI] [PubMed] [Google Scholar]
- 46.Zielenska M, Zhang ZM, Ng K, et al. Acquisition of secondary structural chromosomal changes in pediatric Ewing sarcoma is a probable prognostic factor for tumor response and clinical outcome. Cancer 2001;91:2156–64. [DOI] [PubMed] [Google Scholar]
- 47.Douglass EC, Rowe ST, Valentine M, et al. A second nonrandom translocation, der(16)t(1;16)(q21;q13), in Ewing sarcoma and peripheral neuroectodermal tumor. Cytogenet Cell Genet 1990;53:87–90. [DOI] [PubMed] [Google Scholar]
- 48.Hattinger CM, Rumpler S, Strehl S, et al. Prognostic impact of deletions at 1p36 and numerical aberrations in Ewing tumors. Genes Chromosomes Cancer 1999;24:243–54. [DOI] [PubMed] [Google Scholar]
- 49.Kullendorff CM, Mertens F, Donner M, et al. Cytogenetic aberrations in Ewing sarcoma: are secondary changes associated with clinical outcome? Med Pediatr Oncol 1999;32:79–83. [DOI] [PubMed] [Google Scholar]
- 50.Kovar H, Jug G, Aryee DN, et al. Among genes involved in the RB dependent cell cycle regulatory cascade, the p16 tumor suppressor gene is frequently lost in the Ewing family of tumors. Oncogene 1997;15:2225–32. [DOI] [PubMed] [Google Scholar]
- 51.Lopez-Guerrero JA, Pellin A, Noguers R, et al. Molecular analysis of the 9p21 locus and p53 genes in Ewing family tumors. Lab Invest 2001;81:803–14. [DOI] [PubMed] [Google Scholar]
- 52.Maitra A, Roberts H, Weinberg AG, et al. Aberrant expression of tumor suppressor proteins in the Ewing family of tumors. Arch Pathol Lab Med 2001;125:1207–12. [DOI] [PubMed] [Google Scholar]
- 53.Matsumoto Y, Tanaka K, Nakatani F, et al. Downregulation and forced expression of EWS–FLI1 fusion gene results in changes in the expression of G(1) regulatory genes. Br J Cancer 2001;23:768–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei G, Antonescu CR, de Alava E, et al. Prognostic impact of INK4A deletion in Ewing sarcoma. Cancer 2000;89:793–9. [DOI] [PubMed] [Google Scholar]
- 55.Park YK, Chi SG, Kim YW, et al. Mutational alteration of the p16CDKN2a tumor suppressor gene is infrequent in Ewing’s sarcoma. Oncol Rep 1999;6:1261–6. [DOI] [PubMed] [Google Scholar]
- 56.Quelle DE, Cheng M, Ashmun RA, et al. Cancer-associated mutations at the INK4a locus cancel cell cycle arrest by p16INK4a but not by the alternative reading frame protein p19ARF. Proc Natl Acad Sci U S A 1997;94:669–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vonlanthen S, Heighway J, Tschan MP, et al. Expression of p16INK4a/p16alpha and p19ARF/p16beta is frequently altered in non-small cell lung cancer and correlates with p53 overexpression. Oncogene 1998;17:2779–85. [DOI] [PubMed] [Google Scholar]
- 58.Kovar H, Auinger A, Jug G, et al. Narrow spectrum of infrequent p53 mutations and absence of MDM2 amplification in Ewing tumours. Oncogene 1993;8:2683–90. [PubMed] [Google Scholar]
- 59.Hamelin R, Zucman J, Melot T, et al. p53 mutations in human tumors with chimeric EWS/FLI-1 genes. Int J Cancer 1994;57:336–40. [DOI] [PubMed] [Google Scholar]
- 60.Wadayama B, Toguchida J, Yamaguchi T, et al. P53 expression and its relationship to DNA alterations in bone and soft tissue sarcomas. Br J Cancer 1993;68:1134–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Radig K, Schneider-Stock R, Rose I, et al. p53 and ras mutations in Ewing’s sarcoma. Pathol Res Pract 1998;194:157–62. [DOI] [PubMed] [Google Scholar]
- 62.de Alava E, Antonescu CR, Panizo A, et al. Prognostic impact of p53 status in Ewing sarcoma. Cancer 2000;89:783–92. [PubMed] [Google Scholar]
- 63.Abudu A, Mangham DC, Reynolds GM, et al. Overexpression of the p53 protein in primary Ewing’s sarcoma of bone: relationship to tumour stage, response and prognosis. Br J Cancer 1999;79:1185–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maeda T, Nishimura N, Nakamura H, et al. p21 (WAF1/Cip1/Sdi1/Pic1) mRNA is expressed in neuroblastoma cell lines but not in Ewing’s sarcoma and primitive neuroectodermal tumor cell lines. Acta Paediatr Jpn 1997;39:590–4. [DOI] [PubMed] [Google Scholar]
- 65.Jiang M, Shao ZM, Wu J, et al. p21/waf1/cip1 and mdm-2 expression in breast carcinoma patients as related to prognosis. Int J Cancer 1997;74:529–34. [DOI] [PubMed] [Google Scholar]
- 66.Gomyo Y, Ideda M, Osaki M, et al. Expression of p21 (waf1/cip1/sdi1), but not p53 protein, is a factor in the survival of patients with advanced gastric carcinoma. Cancer 1997;79:2067–72. [DOI] [PubMed] [Google Scholar]
- 67.Stein JP, Ginsberg DA, Grossfeld GD, et al. Effect of p21WAF1/CIP1 expression on tumor progression in bladder cancer. J Natl Cancer Inst 1998;90:1072–9. [DOI] [PubMed] [Google Scholar]
- 68.Kovar H, Aryee DN, Jug G, et al. EWS/FLI-1 antagonists induce growth inhibition of Ewing tumor cells in vitro. Cell Growth Differ 1996;7:429–37. [PubMed] [Google Scholar]
- 69.Tanaka K, Iwakuma T, Harimaya K, et al. EWS–Fli1 antisense ligodeoxynucleotide inhibits proliferation of human Ewing’s sarcoma and primitive neuroectodermal tumor cells. J Clin Invest 1997;99:239–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dohjima T, Ohno T, Banno Y, et al. Preferential down-regulation of phospholipase C-beta in Ewing’s sarcoma cells transfected with antisense EWS–Fli-1. Br J Cancer 2000;82:16–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lambert G, Bertrand JR, Fattal E, et al. EWS fli-1 antisense nanocapsules inhibits Ewing sarcoma-related tumor in mice. Biochem Biophys Res Commun 2000;279:401–6. [DOI] [PubMed] [Google Scholar]
- 72.Bailly RA, Bosselut R, Zucman J, et al. DNA-binding and transcriptional activation properties of the EWS–FLI-1 fusion protein resulting from the t(11;22) translocation in Ewing sarcoma. Mol Cell Biol 1994;14:3230–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mao X, Miesfeldt S, Yang H, et al. The FLI-1 and chimeric EWS–FLI-1 oncoproteins display similar DNA binding specificities. J Biol Chem 1994;269:18216–22. [PubMed] [Google Scholar]
- 74.Thompson AD, Braun BS, Arvand A, et al. EAT-2 is a novel SH2 domain containing protein that is up regulated by Ewing’s sarcoma EWS/FLI1 fusion gene. Oncogene 1996;13:2649–58. [PubMed] [Google Scholar]
- 75.Arvand A, Denny CT. Biology of EWS/ETS fusions in Ewing’s family tumors. Oncogene 2001;20:5747–54. [DOI] [PubMed] [Google Scholar]
- 76.Wang M, Xie Y, Girnita L, et al. Regulatory role of mevalonate and N-linked glycosylation in proliferation and expression of the EWS/FLI-1 fusion protein in Ewing’s sarcoma cells. Exp Cell Res 1999;246:38–46. [DOI] [PubMed] [Google Scholar]
- 77.Yi H, Fujimura Y, Ouchida M, et al. Inhibition of apoptosis by normal and aberrant Fli-1 and erg proteins involved in human solid tumors and leukemias. Oncogene 1997;14:1259–68. [DOI] [PubMed] [Google Scholar]
- 78.Fredericks WJ, Ayyanathan K, Rauscher FJ, 3rd. Regulating the neoplastic phenotype using engineered transcriptional repressors. Cancer Lett 2001;162(suppl):S23–32. [DOI] [PubMed] [Google Scholar]
- 79.Fredericks WJ, Ayyanathan K, Herlyn M, et al. An engineered PAX3–KRAB transcriptional repressor inhibits the malignant phenotype of alveolar rhabdomyosarcoma cells harboring the endogenous PAX3–FKHR oncogene. Mol Cell Biol 2000;20:5019–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bosilevac JM, Olsen RJ, Bridge JA, et al. Tumor cell viability in clear cell sarcoma requires DNA binding activity of the EWS/ATF1 fusion protein. J Biol Chem 1999;274:34811–18. [DOI] [PubMed] [Google Scholar]
- 81.May WA, Arvand A, Thompson AD, et al. EWS/FLI1-induced manic fringe renders NIH 3T3 cells tumorigenic. Nat Genet 1997;17:495–7. [DOI] [PubMed] [Google Scholar]
- 82.Braun BS, Frieden R Lessnick SL, et al. Identification of target genes for the Ewing’s sarcoma EWS/FLI fusion protein by representational difference analysis. Mol Cell Biol 1995;15:4623–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Arvand A, Bastians H, Welford SM, et al. EWS/FLI1 up regulates mE2-C, a cyclin-selective ubiquitin conjugating enzyme involved in cyclin B destruction. Oncogene 1998;17:2039–45. [DOI] [PubMed] [Google Scholar]
- 84.Hahm KB, Cho K, Lee C. Repression of the gene encoding the TGF-beta type II of the EWS–FLI-1 oncoprotein. Nat Genet 1999;23:222–7. [DOI] [PubMed] [Google Scholar]
- 85.Im YH, Kim HT, Lee C, et al. EWS–FLI1, EWS–ERG, and EWS–ETV1 oncoproteins of Ewing tumor family all suppress transcription of transforming growth factor beta type II receptor gene. Cancer Res 2000;60:1536–40. [PubMed] [Google Scholar]
- 86.Zwerner JP, May WA. PDGF-C is an EWS/FLI induced transforming growth factor in Ewing family tumors. Oncogene 2001;20:626–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Massuda ES, Dunphy EJ, Redman RA, et al. Regulated expression of the diphtheria toxin A chain by a tumor-specific chimeric transcription factor results in selective toxicity for alveolar rhabdomyosarcoma cells. Proc Natl Acad Sci U S A 1997;94:14701–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang L, Chansky HA, Hickstein DD. EWS–Fli-1 fusion protein interacts with hyperphosphorylated RNA polymerase II and interferes with serine-arginine protein-mediated RNA splicing. J Biol Chem 2000;275:37612–18. [DOI] [PubMed] [Google Scholar]
- 89.Yang L, Embree LJ, Hickstein DD. TLS-ERG leukaemia fusion protein inhibits RNA splicing mediated by serine-arginine proteins. Mol Cell Biol 2000;20:3345–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Knoop LL, Baker SJ. EWS/FLI1 alters 5′-splice site selection. J Biol Chem 2001;276:22317–22. [DOI] [PubMed] [Google Scholar]
- 91.Jaishankar S, Zhang J, Roussel MF, Baker SJ. Transforming activity of EWS/FLI is not strictly dependent upon DNA-binding activity. Oncogene 1999;18:5592–7. [DOI] [PubMed] [Google Scholar]
- 92.Mackall C, Berzofsky J, Helman LJ. Targeting tumor specific translocations in sarcomas in pediatric patients for immunotherapy. Clin Orthop 2000;373:25–31. [DOI] [PubMed] [Google Scholar]
- 93.Worley BS, van den Broeke LT, Goletz TJ, et al. Antigenicity of fusion proteins from sarcoma-associated chromosomal translocations. Cancer Res 2001;61:6868–75. [PubMed] [Google Scholar]
- 94.Linehan DC, Bowne WB, Lewis JJ. Immunotherapeutic approaches to sarcoma. Semin Surg Oncol 1999;17:72–7. [DOI] [PubMed] [Google Scholar]
- 95.Maki RG. Soft tissue sarcoma as a model disease to examine cancer immunotherapy. Curr Opin Oncol 2001;13:270–4. [DOI] [PubMed] [Google Scholar]