Abstract
Background: Chronic neutrophilic leukaemia (CNL) is a rare myeloproliferative disorder of elderly patients characterised by sustained neutrophilia and splenomegaly. The diagnosis of CNL requires the exclusion of BCR/ABL positive chronic myelogenous leukaemia (CML) and of leukaemoid reactions (LRs). The differentiation between CNL and LR is problematic because both conditions share similar morphological features; it is also important because patients with CNL generally have a poor prognosis.
Aims: To determine whether CNL and LR could be distinguished on the basis of different clonality patterns.
Methods: Blood samples from 52 women were studied using the human androgen receptor gene assay (HUMARA).
Results: Monoclonality was found in the neutrophils in all 17 patients with different myeloproliferative syndromes (MPSs), including those with CNL. In four of the patients with CNL, autologous T cells were also monoclonal, suggesting that they belonged to the neoplastic clone. This finding was in contrast to other MPSs in which T cells were almost always polyclonal. Of nine patients with clinically suspected LR, the neutrophils of five were polyclonal, whereas three patients had monoclonal neutrophils, suggesting that they might be in the process of developing an MPS. Among 26 healthy blood donors, 20 had polyclonal neutrophils and five showed skewed clonality patterns. One case of LR and one normal blood donor were scored “not informative” at the HUMARA locus.
Conclusions: Clonality studies of blood neutrophils using HUMARA aid in distinguishing female patients with monoclonal CNL from those with LR. For the diagnosis of CNL, monoclonality of the neutrophils should be demonstrated whenever possible.
Keywords: chronic neutrophilic leukaemia, bone marrow, clonality, human androgen receptor gene assay
Chronic neutrophilic leukaemia (CNL) is a rare myeloproliferative syndrome (MPS) of elderly patients showing sustained neutrophilia and splenomegaly.1–4 To date, only 143 cases of CNL have been reported in the literature, including 14 of our own cases reported recently.5 The diagnosis of CNL is based on the exclusion of chronic myelogenous leukaemia (CML) and of leukaemoid reactions (LRs).2 In contrast to CML, which is characterised by a BCR/ABL translocation, no definite molecular marker is known in CNL.1,2 The differential diagnosis between CNL and LR may be difficult or even impossible because both conditions share identical morphological features, including a raised neutrophil alkaline phosphatase (NAP) score. The spectrum of disorders capable of causing LR is so wide that clinicians may not be able to exclude all possible causes of LR. It is important to differentiate CNL from LR because the prognosis of patients with CNL is poor, even worse than that of those with CML.6
“The diagnosis of chronic neutrophilic leukaemia is based on the exclusion of chronic myelogenous leukaemia and of leukaemoid reactions”
The human androgen receptor gene assay (HUMARA) for the analysis of clonality in tissues from female patients examines the inactivation patterns of the human androgen receptor gene on the X chromosome.7 This method relies on the length polymorphism of a human androgen receptor gene exon, which has restriction sites for methylase sensitive enzymes. Ideally, in polyclonal conditions such as LR, 50% of the cells show methylation of the maternal allele and 50% of the cells show methylation of the paternal allele. In contrast, neoplastic tissues such as leukaemic CML cells typically show complete methylation of one allele and demethylation of the other allele. In this study, we investigated blood samples of patients using HUMARA to determine whether CNL and LR could be distinguished on the basis of X chromosomal inactivation patterns.
MATERIALS AND METHODS
Selection of patients
In the archives of our department, which receives more than 10 000 bone marrow biopsies annually, we found five living female patients fulfilling the morphological and clinical criteria of CNL. These five patients belonged to a group of 14 CNL cases that we have reported previously.5 Clinically, these patients showed chronic neutrophilia, a variable degree of splenomegaly, and no thrombocytosis. The bone marrow was strongly hypercellular with expansion of the neutrophilic granulopoiesis, which was not left shifted. In the blood, moderate leucocytosis was present, with an excess of mature neutrophils and bands. In four of the cases the blood also contained myelocytes, but there were no blasts (table 1). In all our CNL cases, the NAP score was increased and the BCR/ABL translocation was excluded by reverse transcription polymerase chain reaction (PCR) and, in addition, by fluorescence in situ hybridisation (FISH).8 At the time of diagnosis, four of the patients with CNL had normal cytogenetics, but patient 4 showed an abnormal clone with trisomy 9 (20 of 21 metaphases). We performed clonality studies in blood samples using HUMARA in our five cases of CNL and compared the results with 12 patients who had untreated, newly diagnosed MPS, nine patients with clinically suspected LR, and 26 healthy blood donors (table 1). All the investigations were done according to the guidelines of our institute.
Table 1.
Haematological data of patients and results of the human androgen receptor gene assay (HUMARA) clonality studies
| Patient | Diagnosis | Age | BCR/ ABL | Hb | WBC | Differential blood count | Plt | N-clon | L-clon |
| 1 | CNL | 52 | Absent | 140 | 30.0 | Seg 70, Bn 3, My 9, Bas 1, Eo 2, Ly 10, Mo 5 | 182 | M | M |
| 2 | CNL | 81 | Absent | 108 | 35.0 | Seg 90, Bn 0, My 0, Bas 0, Eo 0, Ly 9, Mo 1 | 189 | M | P |
| 3 | CNL | 37 | Absent | 143 | 36.2 | Seg 64, Bn 15, My 13, Bas 0, Eo 0, Ly 7, Mo 1 | 273 | M | M |
| 4 | CNL | 72 | Absent | 89 | 38.0 | Seg 41, Bn 13, My 37, Bas 0, Eo 0, Ly 6, Mo 3 | 47 | M | M |
| 5 | CNL | 63 | Absent | 123 | 24.6 | Seg 60, Bn 6, My 17, Bas 5, Ery 2, Ly 9, Mo 1 | 210 | M | M |
| 6 | CML | 60 | b2a2 | 98 | 83.5 | Seg 28, Bn 14, My 39, Bas 1, Eo 1, Ly 10, Mo 7 | 718 | M | P |
| 7 | CML | 53 | b3a2 | 101 | 46.1 | Seg 63, Bn 4, My 12, Bas 0, Eo 1, Ly 13, Mo 7 | 365 | M | P |
| 8 | CML | 41 | b3a2 | 98 | 239.0 | Seg 45, Bn 5, My 39, Bas 3, Eo 0, Ly 2, Mo 6 | 619 | M | P |
| 9 | aCML | 48 | Absent | 142 | 32.7 | Seg 27, Bn 4, My 46, Bas 3, Eo 1, Ly 15, Mo 4 | 460 | M | P |
| 10 | CML | 64 | b2a2 | 116 | 39.2 | Seg 80, Bn 7, My 0, Bas 0, Eo 1, Ly 7, Mo 5 | 486 | M | P |
| 11 | CML | 58 | b3a2 | 136 | 24.1 | Seg 63, Bn 7, My 0, Bas 2, Eo 0, Ly 22, Mo 6 | 439 | M | M |
| 12 | CML | 65 | b3a2 | 130 | 33.9 | Seg 59, Bn 4, My 22, Ery 4, Bas 5, Eo 2, Ly 6, Mo 1 | 246 | M | P |
| 13 | CMML | 83 | Absent | 100 | 37.2 | Seg 63, Bn 0, My 0, Bas 0, Eo 0, Ly 9, Mo 28 | 191 | M | P |
| 14 | CMML | 74 | Absent | 114 | 8.2 | Seg 20, Bn 6, My 0, Bas 0, Eo 0, Ly 51, Mo 23 | 95 | M | P |
| 15 | PCV | 61 | Absent | 91 | 15.4 | Seg 80, Bn 4, Ery 4, Bas 0, Eo 0, Ly 10, Mo 2 | 44 | M | ND |
| 16 | CIMF | 73 | Absent | 105 | 12.2 | Seg 78, Bn 0, My 0, Bas 1, Eo 4, Ly 16, Mo 1 | 124 | M | P |
| 17 | CIMF | 83 | Absent | 116 | 30.4 | Seg 38, Bn 4, My 19, Bas 5, Eo 7, Ly 20, Mo 7 | 403 | M | P |
| 18 | LR (smoker) | 39 | Absent | Normal | 14.0 | Seg 63, Bn 2, My 2, Bas 1, Eo 4, Ly 25, Mo 3 | Normal | M | P |
| 19 | LR (diabetes) | 50 | Absent | 88 | 10.4 | Seg 61, Bn 0, My 0, Bas 0, Eo 3, Ly 32, Mo 4 | 348 | P | P |
| 20 | LR (fever) | 63 | ND | 99 | 18.1 | Seg 87, Bn 1, Eo 2, Ly 8, Mo2 | 518 | P | P |
| 21 | LR | 67 | ND | ND | 21.5 | ND | ND | P | P |
| 22 | LR (diabetes) | 93 | ND | ND | 20.1 | ND | ND | M | ND |
| 23 | LR (smoker) | 37 | ND | 134 | 13.6 | Seg 69, Bn 1, Ly24, others 6 | 204 | P | P |
| 24 | LR (Sharp sy.) | 50 | ND | 90 | 7.0 | Seg 80, Bn 1, Bas 1, Eo 5, Ly 7, Mo 6 | 307 | M | P |
| 25 | LR | 53 | Absent | 142 | 13.8 | Seg 56, Bn 2, Ly35, Mo 7 | 354 | NI | NI |
| 26 | LR (smoker) | 45 | Absent | 129 | 12.5 | Seg 66, Bn 4, Eo 6, Ly 20, Mo 4 | 403 | P | P |
| 27–52 | Blood donors | 20–77 | ND | ND | Normal | * | ND | See text | |
Diagnosis: aCML, atypical chronic myeloid leukaemia; CIMF, chronic idiopathic myelofibrosis; CML, chronic myelogenous leukaemia (chronic phase); CMML, chronic myelomonocytic leukaemia; CNL, chronic neutrophilic leukaemia; LR, leukaemoid reaction; PCV, polycythaemia vera; Sharp sy., Sharp syndrome (a mixed connective tissue disease). Age, age at time of diagnosis; BCR/ABL, status of BCR/ABL translocation; Hb, haemoglobin concentration (g/l); WBC, white blood cell count (×109/l); differential blood count: Seg, segmented neutrophils; Bn, bands; My, myelocytes; Bas(ophils), Eo(sinophils), Ly(mphocytes), Mo(nocytes), Ery(throblasts); Plt, platelet count (×109/l); N-clon/L-clon, clonal status of neutrophils and phytohaemagglutinin expanded lymphocytes using HUMARA with the patterns (M) monoclonal and (P) polyclonal; NI, not informative; ND, not determined.
Blood sample clonality assay
The neutrophils of all 52 patients were enriched on a buffy coat by centrifuging 5 ml of blood at 800 ×g rpm for 15 minutes, followed by the lysis of red blood cells. Lymphocytes were isolated on a Ficoll gradient from 5 ml of blood (Nycomed Pharma AS, Oslo, Norway) and T cells were expanded in vitro using phytohaemagglutinin (PHA) and interleukin 2. Thus, in each patient, at least 3 million cultured T cells were available for investigation. DNA samples from the neutrophils and the PHA expanded T cells were isolated for each patient (PNA Blood Minikit; Qiagen, Hilden, Germany). These DNA samples were digested overnight at 37°C with Hpa II (Roche Diagnostics, Mannheim, Germany) and Hha I (Promega Corporation, Madison, Wisconsin, USA). A third DNA sample remained undigested. All samples were heated to 95°C for 10 minutes to stop the digestion and to denature the DNA. HUMARA sequences were amplified from the undigested DNA, the Hpa II and Hha I digested DNA samples obtained from the neutrophils, and the T cells for each patient by PCR using AmpliTaq Gold (Perkin-Elmer, Rodgau, Germany) with the following primers: 5′-GCTGTGAAGGTTGCTGTTCCTCAT-3′ (sense) and 5′-TCCAGAATCTGTTCCAGAGCGTGC-3′ (antisense).7 The sense primer was 5′-fluorescence labelled (6-FAM). The PCR conditions were 96°C for 10 minutes (first cycle), then 95°C for 30 seconds, 65°C for 45 seconds and 72°C for 90 seconds for 30 cycles, followed by 72°C for seven minutes. The PCR products were analysed by capillary electrophoresis on an ABI-310 sequencer (Perkin-Elmer, Weiterstadt, Germany).
RESULTS AND DISCUSSION
We studied the clonality patterns of leucocytes, which consisted mainly of neutrophils and PHA expanded T cells, from patients with MPS and LR. The T cells were used as an internal control cell population because both neutrophils and T cells are derived from haemopoietic progenitor cells.9–11 The results of the HUMARA study are summarised in table 1, and representative findings in two patients with CNL and one normal blood donor are illustrated in fig 1. In all five cases of CNL, the leukaemic neutrophils displayed a monoclonal HUMARA pattern. Similarly, all seven patients with CML or atypical CML and all five patients with other MPS had monoclonal leucocytes. The PHA expanded T cells of four of five patients with CNL showed a monoclonal HUMARA pattern. This finding suggests that in CNL the T cells are frequently derived from the neoplastic clone. However, we cannot completely exclude the possibility that the finding of monoclonal T cells in four of the five CNL cases could represent an extreme form of “skewing”, meaning an unbalanced, “skewed” pattern of X chromosomal inactivation in which cells with the inactivated maternal or paternal allele are predominant. Even though it is not always possible to differentiate clearly between true monoclonality and extreme skewing in the individual case, we think that such a concentration of cases showing extreme skewing in a small group of patients with CNL would be very unlikely. In contrast, in five of six patients with CML, in the patient with atypical CML, and in all five patients with other MPSs the T cells did not show a monoclonal HUMARA pattern, suggesting that they may be derived from residual normal progenitor cells. Thus, the different clonality patterns of the T cells may indicate that in CNL the neoplastic transformation occurs at an earlier stage of progenitor cell differentiation than in CML.
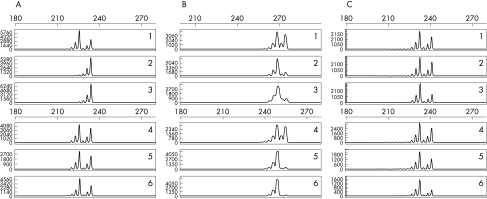
Figure 1.
Clonality analysis using human androgen receptor gene assay. (A) Chronic neutrophilic leukaemia (CNL) case 2: after predigestion of the DNA from the leukaemic neutrophils with the methylase sensitive enzymes only one of the human androgen receptor gene microsatellites can be amplified by PCR, whereas both X chromosomal alleles are amplified without predigestion (monoclonal pattern; rows 1–3). In contrast, PCR amplification of the human androgen receptor gene microsatellites from the DNA of this patient’s T cells is not affected by predigestion (polyclonal pattern; rows 4–6). A monoclonal pattern in the gene scan analysis is suggested if one allele peak completely disappears after enzymatic digestion, whereas a polyclonal profile is characterised by the persistence of two distinct allele peaks. The additional smaller peaks are caused by slippage of the Taq polymerase.12 (B) CNL case 1: monoclonal patterns in both neutrophils and T cells. (C) Normal blood donor: polyclonal patterns in both neutrophils and T cells. Rows 1 and 4, PCR from undigested DNA; rows 2 and 5, PCR from DNA predigested with HpaII; rows 3 and 6, PCR from DNA predigested with HhaI.
Most patients with the clinical diagnosis of LR showed polyclonal patterns for neutrophils and T cells, as expected. Three patients with clinically suspected LR had monoclonal neutrophils and polyclonal T cells, displaying a clonality pattern similar to the patients with MPS. Thus, these patients with clinically suspected LR might be in the initial phase of an MPS. At present, one year after the investigations were done, these patients are free of disease. The clonality analysis of 26 blood donors demonstrated 20 cases with polyclonal patterns of neutrophils and T cells. In five blood donors we found a skewed pattern of X chromosomal inactivation. This was seen in both the neutrophils and the T cells of these blood donors. The skewing has been interpreted as a natural phenomenon, detectable in some women upon aging.9,11,13–15 By definition, a cell fraction is considered as monoclonal or “skewed” if the expression of the dominant allele exceeds 75%.14 Thus, the results of HUMARA clonality studies should only be interpreted together with the clinical, blood, and bone marrow findings. Among the patients with LR and blood donors there were two “non-informative” cases, where the analysis of clonality was impossible because the two different PCR amplified X chromosomal microsatellites were of approximately equal size.
“The different clonality patterns of the T cells may indicate that in chronic neutrophilic leukaemia the neoplastic transformation occurs at an earlier stage of progenitor cell differentiation than in chronic myelogenous leukaemia”
To our knowledge this is the first report of HUMARA clonality studies in CNL. This technique is naturally restricted to female patients and gives meaningful results in 80% to 90% of cases, owing to the high rate of heterozygosity at the HUMARA locus.7,16–19 However, the results may be blurred by the excessive skewing that is seen in some normal blood donors.9,11,13–15 Thus far, the clonal nature of blood neutrophils in CNL has been documented in two cases. Froberg et al reported monosomy for a 11q23 probe using FISH in a 67 year old woman with CNL evolving from a myelodysplastic syndrome.20 In a 60 year old female patient, Kwong and Cheng21 found a monoclonal methylation pattern of the X linked hypoxanthine phosphoribosyl transferase (HPRT) gene. However, these authors did not provide data about the T cell clonality pattern, so that extreme skewing in that case cannot be excluded. Some of the cases that were reported in the literature as being “CNL” occurred in association with plasma cell dyscrasias like myeloma.2 However, when studied, a polyclonal pattern of the neutrophils at the HPRT gene of these suspected CNL cases was found,22,23 indicating that neutrophilia in these patients might have represented LR triggered by cytokines.2 In our five patients with CNL, plasma cell dyscrasias were absent. Significant dysplasia of the haemopoietic cells in the bone marrow has been described in some cases of “CNL”,4,20,24,25 but is not a diagnostic feature of CNL,1,2 and was not seen in our five patients with CNL. Thus, on the basis of the CNL cases published up to 2001, in his review, Reilly26 concluded that only 33 cases sufficiently fulfilled the criteria of “true” CNL, including the case of Kwong and Cheng,21 but excluding the case of Froberg et al.20 As was seen in four of our five patients, nearly 90% of patients with CNL have normal cytogenetics, but a minority show diverse aberrations,1,2,26 probably suggesting monoclonality. Among the karyotypic abnormalities reported in CNL, 20q deletions were the most frequent.26 The finding of trisomy 9, as in our patient 4, has been described once by Di Donato et al in a patient with CNL after beginning radiotherapy and chemotherapy.27 The heterogeneity of the cytogenetic aberrations found in some of the CNL cases indicates that these aberrations probably represent secondary phenomena in the course of the disease and are not primarily involved in the pathogenesis of CNL.1
In conclusion, our HUMARA clonality studies prove the neoplastic nature of the leukaemic neutrophils in CNL, and provide evidence that CNL is a distinct myeloproliferative disorder. In female patients, HUMARA studies may help to distinguish CNL from polyclonal LR. We suggest that the monoclonality of blood neutrophils should be demonstrated for the diagnosis of “true” CNL whenever possible. The importance of skewed or monoclonal patterns of haemopoiesis in individual blood donors and in patients with clinically suspected LR should be investigated in prospective studies.
Take home messages.
Clonality studies of blood neutrophils using the human androgen receptor gene assay can help to distinguish female patients with monoclonal chronic neutrophilic leukaemia (CNL) from those who have a polyclonal, non-malignant leukaemoid reaction
This distinction is difficult using morphology alone but is important because those with CNL have a very poor prognosis
For the diagnosis of CNL, monoclonality of the neutrophils should be demonstrated whenever possible
Abbreviations
CML, chronic myelogenous leukaemia
CNL, chronic neutrophilic leukaemia
FISH, fluorescence in situ hybridisation
HPRT, hypoxanthine phosphoribosyl transferase
HUMARA, human androgen receptor gene assay
LR, leukaemoid reaction
MPS, myeloproliferative syndrome
NAP, neutrophil alkaline phosphatase
PCR, polymerase chain reaction
PHA, phytohaemagglutinin
REFERENCES
- 1.Elliott MA, Dewald GW, Tefferi A, et al. Chronic neutrophilic leukemia (CNL): a clinical, pathologic and cytogenetic study. Leukemia 2001;15:35–40. [DOI] [PubMed] [Google Scholar]
- 2.Imbert M, Bain B, Pierre R, et al. Chronic neutrophilic leukaemia. In: Jaffe ES, Harris NL, Stein H, et al, eds. World Health Organization classification of tumours. Pathology and genetics of tumours of haematopoietic and lymphoid tissues. Lyon: IARC Press, 2001:27–8.
- 3.You W, Weisbrot IM. Chronic neutrophilic leukemia. Report of two cases and review of the literature. Am J Clin Pathol 1979;72:233–42. [DOI] [PubMed] [Google Scholar]
- 4.Zittoun R, Rea D, Ngoc LH, et al. Chronic neutrophilic leukaemia. A study of four cases. Ann Hematol 1994;68:55–60. [DOI] [PubMed] [Google Scholar]
- 5.Böhm J, Schaefer HE. Chronic neutrophilic leukaemia: 14 new cases of an uncommon myeloproliferative disease. J Clin Pathol 2002;55:862–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer S, Feremans W, Cantiniaux B, et al. Successful alpha-2b-interferon therapy for chronic neutrophilic leukaemia. Am J Hematol 1993;43:307–9. [DOI] [PubMed] [Google Scholar]
- 7.Allen RC, Zoghbi HY, Moseley AB, et al. Methylation of Hpa II and Hha I sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet 1992;51:1229–39. [PMC free article] [PubMed] [Google Scholar]
- 8.Maurer J, Kinzel H, Nentwig T, et al. Molecular diagnosis of the Philadelphia chromosome in chronic myelogenous and acute lymphoblastic leukemias by PCR. Dis Markers 1990;8:211–18. [PubMed] [Google Scholar]
- 9.Gale RE, Wheadon H, Boulos P, et al. Tissue specificity of X-chromosome inactivation patterns. Blood 1994;83:2899–905. [PubMed] [Google Scholar]
- 10.Nakahara Y, Suzuki H, Ohashi H, et al. Clonality analysis of granulocytes and T lymphocytes in healthy females by the PCR-based HUMARA method. Int J Hematol 1999;69:237–43. [PubMed] [Google Scholar]
- 11.Tonon L, Bergamaschi G, Dellavecchia C, et al. Unbalanced X-chromosome inactivation in haematopoietic cells from normal women. Br J Haematol 1998;102:996–1003. [DOI] [PubMed] [Google Scholar]
- 12.Wu CD, Wickert RS, Williamson JE, et al. Using fluorescence-based human androgen receptor gene assay to analyze the clonality of microdissected dendritic cell tumors. Am J Clin Pathol 1999;111:105–10. [DOI] [PubMed] [Google Scholar]
- 13.Champion KM, Gilbert JGR, Asimakopoulos FA, et al. Clonal haematopoiesis in normal elderly women: implications for the myeloproliferative disorders and myelodysplastic syndromes. Br J Haematol 1997;97:920–6. [DOI] [PubMed] [Google Scholar]
- 14.Busque L, Mio R, Mattioli J, et al. Nonrandom X-inactivation patterns in normal females: lyonization ratios vary with age. Blood 1996;88:59–65. [PubMed] [Google Scholar]
- 15.Gale RE, Fielding AK, Harrison CN, et al. Acquired skewing of X-chromosome inactivation patterns in myeloid cells of the elderly suggests stochastic clonal loss with age. Br J Haematol 1997;98:512–19. [DOI] [PubMed] [Google Scholar]
- 16.Gale RE. Evaluation of clonality in myeloid stem-cell disorders. Semin Hematol 1999;36:361–72. [PubMed] [Google Scholar]
- 17.Lee PS, Sabbath-Solitare M, Redondo TC, et al. Molecular evidence that the stromal and epithelial cells in pleomorphic adenomas of salivary gland arise from the same origin: clonal analysis using human androgen receptor gene (HUMARA) assay. Hum Pathol 2000;31:498–503. [DOI] [PubMed] [Google Scholar]
- 18.Mitterbauer G, Winkler K, Gisslinger H, et al. Clonality analysis using X-chromosome inactivation at the human androgen receptor gene (HUMARA). Evaluation of large cohorts of patients with chronic myeloproliferative diseases, secondary neutrophilia, and reactive thrombocytosis. Am J Clin Pathol 1999;112:93–100. [DOI] [PubMed] [Google Scholar]
- 19.Delabesse E, Aral S, Kamoun P, et al. Quantitative non-radioactive clonality analysis of human leukemic cells and progenitors using the human androgen receptor (AR) gene. Leukemia 1995;9:1578–82. [PubMed] [Google Scholar]
- 20.Froberg MK, Brunning RD, Dorion P, et al. Demonstration of clonality in neutrophils using FISH in a case of chronic neutrophilic leukemia. Leukemia 1998;12:623–6. [DOI] [PubMed] [Google Scholar]
- 21.Kwong YL, Cheng G. Clonal nature of chronic neutrophilic leukemia. Blood 1993;82:1035–6. [PubMed] [Google Scholar]
- 22.Standen GR, Steers FJ, Jones L. Clonality of chronic neutrophilic leukaemia associated with myeloma: analysis using the X-linked probe M27 beta. J Clin Pathol 1993;46:297–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stevenson JP, Schwarting R, Schuster SJ. Analysis of clonality using X-linked polymorphisms in a patient with multiple myeloma and myelofibrosis. Am J Hematol 1998;59:79–82. [DOI] [PubMed] [Google Scholar]
- 24.Pascucci M, Dorion P, Makary A, et al. Chronic neutrophilic leukemia evolving from a myelodysplastic syndrome. Acta Hematol 1997;98:163–6. [DOI] [PubMed] [Google Scholar]
- 25.Takamatsu Y, Kondo S, Inoue M, et al. Chronic neutrophilic leukaemia with dysplastic features mimicking myelodysplastic syndromes. Int J Hematol 1996;63:65–9. [DOI] [PubMed] [Google Scholar]
- 26.Reilly JT. Chronic neutrophilic leukaemia: a distinct clinical entity? Br J Haematol 2002;116:10–18. [DOI] [PubMed] [Google Scholar]
- 27.Di Donato C, Croci G, Lazzari S, et al. Chronic neutrophilic leukemia: description of a new case with karyotypic abnormalities. Am J Clin Pathol 1986;85:369–71. [DOI] [PubMed] [Google Scholar]