Abstract
Aims: An abnormality in the glycated haemoglobin peak (Hb A1c) on Diastat (Bio-Rad) cation exchange low pressure liquid chromatography (LPLC) was found in three Punjabi patients with diabetes. The aims of this study were to identify the variant by chromatography and electrophoresis and to determine whether a DNA analysis test could be designed for confirmation that could be generally applied for the identification of any unusual abnormal haemoglobin.
Methods: The presence of an Hb variant was confirmed by cellulose acetate electrophoresis at pH 8.6. The variant was characterised further by high performance liquid chromatography (HPLC; Bio-Rad Variant) and isolelectric focusing (IEF) electrophoresis. A novel DNA analysis test based on the amplification refractory mutation system (ARMS) and the polymerase chain reaction (PCR) was developed to confirm the presence of the mutation for the uncommon variant.
Results: Comparison of the HPLC retention time and IEF band position determined the presence of the variant Hb Q-India in all three cases. Hb Q-India is caused by the mutation GAC → CAC at codon 64 of the α-1 globin gene and is clinically silent. ARMS-PCR specific primers were designed and used successfully to confirm the presence of the mutation for Hb Q-India.
Conclusions: The results show that the ARMS-PCR technique, developed previously for the diagnosis of β thalassaemia mutations, can also be adapted to provide a simple, rapid, and inexpensive approach for the identification of abnormal haemoglobins.
Keywords: Hb Q India, isolelectric focusing, high performance liquid chromatography, amplification refractory mutation system polymerase chain reaction diagnosis
Post-synthetic modification of haemoglobin occurs with the addition of glucose to the N-terminus of the β chain to form glycosylated haemoglobin (A1c). The estimation of Hb A1c is used extensively in the management of diabetes mellitus. Some systems use low pressure liquid chromatography (LPLC) for the estimation (Diastat Bio-Rad), and occasionally, previously undetected variants are discovered in patients. This report describes three patients with diabetes who were found to have the uncommon variant Hb Q-India (α64(E13)Asp → His).
“The techniques of isolelectric focusing and high performance liquid chromatography specific for haemoglobin variants are not available in many laboratories, especially in developing countries, and thus another simple approach is required”
Two cases were initially revealed during routine Hb A1c estimation and a third, during the investigation of anaemia in a patient with diabetes. To identify the variant, it was necessary to do further studies by high performance liquid chromatography (HPLC) and isoelectric focusing (IEF) electrophoresis. The Hb variant was identified initially by determining the HPLC retention time of the abnormal peak, measuring the IEF position of the variant band, and comparing the data with that of known variants. However, the techniques of IEF and HPLC specific for Hb variants are not available in many laboratories, especially in developing countries, and thus another simple approach is required. To date, mutation analysis techniques have only been developed and applied to the diagnosis of the common Hb variants such as Hb S, Hb C, and Hb E. Here, we report the use of a novel DNA test based on the amplification refractory mutation system (ARMS) for the simple, rapid, and inexpensive diagnosis of an uncommon Hb variant revealed by routine screening for diabetes, which can be applied for the routine diagnosis of all the uncommon abnormal haemoglobins.
MATERIALS AND METHODS
The estimation of glycosylated haemoglobin is carried out on large numbers of patients with diabetes in our teaching hospital in Punjab, North India. The estimation is performed on an HPLC Diastat instrument according to the manufacturer’s instructions (Bio-Rad, Hemel Hempstead, UK). A printout gives the retention time of the various fractions and also measures each fraction as a percentage of the whole of the major peaks, including glycosylated haemoglobin. Some variants such as Hb S and Hb D are also revealed as abnormal bands. Where such bands are shown, cellulose acetate electrophoresis using barbitone buffer at pH 8.6 is carried out.1
Samples with unusual findings were sent to the national haemoglobinopathy reference laboratory, institute of molecular medicine, John Radcliffe Hospital, Oxford, UK for further investigation of the abnormal Hb. Analysis there was carried out using HPLC (Bio-Rad Variant) and isoelectric focusing (IEF).2 The HPLC retention time for the abnormal peak was determined, together with the IEF position of the abnormal band in relation to the Hb A band. A comparison was then made of the retention times and IEF positions of the variants with those of known variants previously identified by DNA sequence analysis.
One of the three samples included in our study was examined further by DNA analysis for the presence of the Hb Q-India mutation using a novel diagnostic test based on the amplification refractory mutation system (ARMS).3 A mutation specific primer was designed to amplify only DNA containing the mutation for Hb Q-India in the α-1 globin gene. A mismatch was designed at the 3′ terminus of the primer to target the G → C mutation of Hb Q-India. PCR was carried out in a 25 μl reaction containing 0.75M betaine, 5% DMSO, 200μM of each dNTP, 0.9mM MgCl2, 750mM Tris/HCl at pH 9.0, 200mM (NH4)2SO4, 0.1% (wt/vol) Tween 20, 100 ng genomic DNA, 0.2 μM of each primer, 0.5 U Platinum Taq (Invitrogen, Paisley, UK). The PCR conditions were four minutes at 95°C for the initial denaturation step, then 30 cycles of denaturation for one minute at 95°C, annealing for one minute at 60°C, and extension for one minute at 72°C. After a final extension period of five minutes, the products were examined by ethidium bromide staining and agarose gel electrophoresis.4 The ARMS primer sequence (reverse and complimentary) is 5′-CACGTGCGCCACGGCGTTGGTCAGCGCGAG-3′ and the common primer sequence is 5′-CTGGTCCCCACAGACTCAGA-3′, giving a PCR product of 370 bp. The control primers chosen for this assay were from the α1 globin gene: a reverse primer with the sequence 5′-AGGCCCAAGGGGCAAGAAGCAT-3′ and the above common primer, generating a control product of 766 bp.
Routine haematological examinations were carried out by standard methods.
RESULTS
Case 1
A 55 year old man, recently diagnosed with diabetes mellitus and on diet control, presented in the diabetic clinic (Christian Medical College and Hospital, Ludhiana (CMC&H), India) for a routine evaluation. Blood was drawn for glycated haemoglobin (Hb A1c) estimation. The Hb A1c assay was carried out on a Diastat machine. The results showed an abnormal haemoglobin variant peak with a retention time of 4.42 minutes.
Case 2
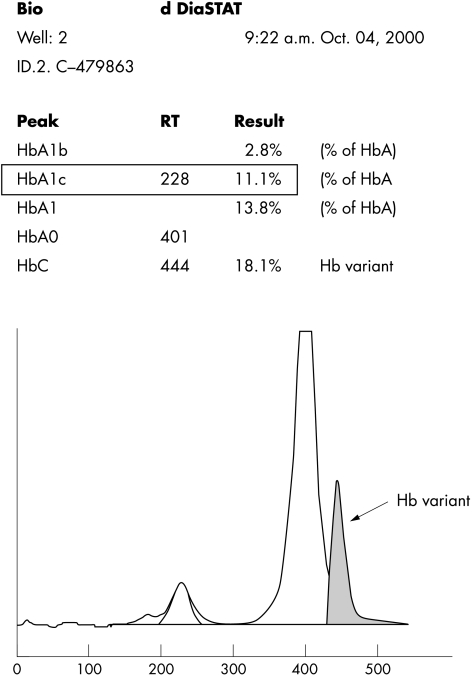
A 55 year woman, a known diabetic and hypertensive for 10 years, presented in the medical outpatients department (CMC&H) with complaints of faecal incontinence for the past eight months. A general physical examination was normal. A routine glycated haemoglobin assay was done. The assay was carried out on the Diastat machine, which showed an abnormal haemoglobin peak, with a retention time of 4.44 minutes (fig 1).
Figure 1.
Elution pattern showing the Hb Q-India variant haemoglobin with the Bio-Rad Diastat low pressure liquid chromatography system.
Case 3
A 42 year old woman, a known diabetic and hypertensive for the past five years, presented in the diabetic clinic (CMC&H) for a routine checkup. A general physical examination revealed mild pallor but no icterus or cyanosis. A routine anaemia investigation was performed, which included haemoglobin electrophoresis. Haemoglobin electrophoresis run on cellulose acetate at pH 8.6 revealed a separate variant band migrating like Hb S. The presence of the variant was confirmed on the Diastat machine, which showed an abnormal variant peak with a retention time of 4.46 minutes.
Identification of the Hb variant
Samples from the three patients were subjected to HPLC using the Bio-Rad Variant analyser. All three showed an identical variant band at a retention time of 4.76 to 4.78 minutes (fig 2). In addition, a split Hb A2 peak was revealed, indicating the presence of an α chain variant. Table 1 shows the results. IEF of the samples showed identical patterns, with the variant band focusing between Hb A and Hb S at 7.8 mm below Hb A. An abnormal Hb with a HPLC retention time of 4.76 minutes and IEF position of 7.8 mm had been previously identified as the α chain variant Hb Q-India by sequence analysis of DNA from another patient. The exact matching of both sets of data indicated that the variant was Hb Q-India.
Figure 2.
Elution pattern showing the Hb Q-India variant haemoglobin with the Bio-Rad Variant high performance liquid chromatography system.
Table 1.
Results of HPLC (Bio-Rad variant) on the 3 cases
| %Hb A2 | %Hb F | %Hb X | Retention time | |
| Case 1 | 1.3+0.5 | 0.5 | 23.6 | 4.76 |
| Case 2 | 1.2+0.5 | 1.7 | 14.2 | 4.78 |
| Case 3 | 1.0+1.2 | 1.2 | 15.4 | 4.76 |
Hb, haemoglobin; HPLC, high performance liquid chromatography.
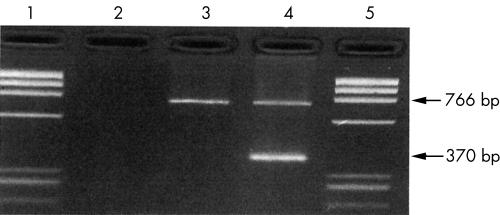
Peripheral blood DNA isolated from case 1 was subjected to molecular analysis by ARMS-PCR. An ARMS primer was developed to detect the Hb Q-India mutation using DNA known to contain the Hb Q-India mutation by DNA sequencing. The DNA from case 1 gave a positive signal with the Hb Q-India specific ARMS primer, confirming that the patient carries Hb Q-India (fig 3).
Figure 3.
Agarose gel showing the identification of the Hb Q-India mutation by amplification refractory mutation system polymerase chain reaction. Samples tested were: lane 3, DNA from a normal control; lane 4, DNA from a patient with Hb Q-India (case 1); lanes 1 and 5, φX174 DNA marker fragments; lane 2, water blank control. The amplified product of 766 bp is the control fragment and the product of 370 bp is the Hb Q-India specific product.
Table 2 provides details of the available blood sample count data. The low red blood cell indices seen in case 1 result from co-inherited α+ thalassaemia and in case 3 from iron deficiency.
Table 2.
Blood count data
| Age | Hb (g/l) | RBC (×1012/l) | MCV (Fl) | MCH (pg) | |
| Case 1 | 58 years | 131 | 5.53 | 80 | 23.7 |
| Case 2 | 55 years | Sample was haemolysed | |||
| Case 3 | 42 years | 84 | – | 83 | 26.3 |
Hb, haemoglobin; MCH, mean cell haemoglobin; MCV, mean cell volume; RBC, red blood cell count.
DISCUSSION
The first Hb Q variants to be characterised were Hb Q-Thailand (α74 Asp → His) and Hb Q-Iran (α75 Asp → His).5 However, a few cases of a different Hb Q variant have been reported from India.6 This particular Hb Q variant, Hb Q-India, is also an α chain variant: α64 Asp → His. DNA sequencing studies have shown the Hb Q India mutation to be AAG → GAG in codon 64 of the α1 gene.7 This variant haemoglobin occurs normally in the heterozygous form and is not associated with a thalassaemia phenotype. In case 1, the patient was also found to have co-inherited α+ thalassaemia by gap PCR studies (hence the increased expression of Hb Q-India seen in this case), whereas in case 3 the patient was found to have a normal α genotype and the low mean cell volume and mean cell haemoglobin values resulted from iron deficiency. Furthermore, it is probable that this variant does not cause haematological disorders because the residue involved, α64 (E 13), is on the surface of the haemoglobin tetramer and charge changes at these positions do not affect the properties of the haemoglobin molecule.
Identical findings on haemoglobin examination in the three patients with diabetes by LPLC using the Diastat machine suggested that they carried a haemoglobin variant. This was confirmed by cellulose acetate electrophoresis. However, these two results were not sufficient to confirm that each patient carried the same variant and also could not identify the variant with confidence. The association of the Hb Q-India variant with diabetes mellitus is probably fortuitous in a situation where subjects undergoing the studies are highly selected. In routine Hb Alc Diastat analysis, the presence of Hb Q-India could lead to a patient being falsely designated as having the clinically relevant variant Hb S, unless careful sickle cell screening is carried out on those found to have an abnormal band and further steps are taken to identify the variant.
A small number of Hb variants can be characterised by comparing their HPLC retention times with reference chromatograms in a library provided by the manufacturer, and also by comparing their IEF positions with those on a published chart of abnormal Hb variants.8 In general, HPLC and IEF provide reliable and reproducible data, enabling the retention time and IEF position to be used to identify variants indirectly. However, several variants are known to have identical retention times and the same applies to IEF positions. Thus, the data are limited in suggesting a candidate variant when only one technique is used. A more accurate identification is obtained when both HPLC and IEF are performed and the HPLC retention time and the IEF position match the data of a known variant, as happened in this case with Hb Q-India. However, the number of variants for which there is both a known HPLC retention time and a published IEF position is very small, and thus identification of most of the uncommon variants by this method is not possible.
“In routine glycosylated haemoglobin Diastat analysis, the presence of Hb Q-India could lead to a patient being falsely designated as having the clinically relevant variant Hb S”
The definitive method for the identification of Hb variants is characterisation by DNA sequencing of the α globin and β globin genes. This is an expensive technique and not practical for the routine identification of uncommon variants. Because most α chain and β chain variants result from a single point mutation, a simple, rapid, and inexpensive method of diagnosing point mutations is required for the definitive characterisation of the uncommon haemoglobin variants. Such a method has been developed for the routine diagnosis of β thalassaemia genes in Asian Indians by ARMS-PCR,9 and this technique has since been applied successfully in developing countries to screen for β thalassaemia mutations in other ethnic groups, using a panel of primers specific to the local spectrum of mutations.4 We have now adapted this technique to enable the quick identification of the α chain variant Hb Q-India and the successful result presented here demonstrates that in principle the technique can be used to identify any uncommon variant of the α globin or β globin genes for which the mutation is known. For the many variants that have not had their causative point mutation confirmed by DNA sequence analysis, an ARMS primer would have to be designed to detect the presumed mutation predicted by the genetic code and the amino acid change. However, all new Hb variants are now characterised by DNA sequence analysis, and the number of variants having their mutation confirmed by DNA sequencing is growing, enabling a panel of ARMS primers to be developed that would be specific to the local spectrum of Hb variants. Thus, the identification of variants by a simple mutation detection test by ARMS-PCR following preliminary characterisation by HPLC or electrophoresis is a feasible approach, and the results presented here prove that this technique can form the basis of a new routine diagnostic strategy for the uncommon abnormal haemoglobins.
Take home messages.
The amplification refractory mutation system polymerase chain reaction (ARMS-PCR) technique, which was developed for the diagnosis of β thalassaemia mutations, can be adapted for the identification of abnormal haemoglobins
The ARMS-PCR technique provides a simple, rapid, and inexpensive approach for the identification of abnormal haemoglobins, which would be suitable for use in the developing world
Abbreviations
Alc, glycated
ARMS, amplification refractory mutation system
CMC&H, Christian Medical College and Hospital
Hb, haemoglobin
HPLC, high performance liquid chromatography
IEF, isolelectric focusing
LPLC, low performance liquid chromatography
PCR, polymerase chain reaction
REFERENCES
- 1.Dacie JV, Lewis SM. Practical haematology, 9th ed. Edinburgh: Churchill-Livingston 2001:241–3.
- 2.Basset P, Beuzard Y, Garel MC, et al. Isoelectric focusing of human hemoglobin: its application to screening, to the characterization of 70 variants, and to the study of modified fractions of normal hemoglobins. Blood 1978;51:971–82. [PubMed] [Google Scholar]
- 3.Newton CR, Graham A, Heptinstall LE. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucl Acids Res 1989;17:2503–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Old J, Khan S, Verma I, et al. A multi-centre study in order to define further the molecular basis of β-thalassemia in Thailand, Pakistan, Sri Lanka, Mauritius, Syria, and India, and to the development of a simple molecular diagnostic strategy by ARMS-PCR. Hemoglobin 2001;25:397–407. [DOI] [PubMed] [Google Scholar]
- 5.Lorkin PA, Charlesworth D, Lehman H, et al. Two haemoglobins Q, alpha-74 (EF3) and alpha-75 (EF4) aspartic acid to histidine. Br J Haematol 1970;19:117–25. [DOI] [PubMed] [Google Scholar]
- 6.Sukumaran PK, Merchant SM, Desai MP, et al. Hemoglobin Q India (alpha 64(E13) aspartic acid to histidine) associated with beta-thalassemia observed in three Sindhi families. J Med Genet 1972;9:436–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molchanova TP, Pobedimskaya DD, Huisman THJ. The differences in quantities of alpha 2- and alpha1-globin gene variants in heterozygotes. Br J Haematol 1994;88:300–6. [DOI] [PubMed] [Google Scholar]
- 8.Righetti PG. Isoelectric focusing: theory, methodology and applications. Laboratory techniques in biochemistry and molecular biology, Vol. 11, Amsterdam: Elsevier Biomedical Press, 1983.
- 9.Old JM, Varawalla NY, Weatherall DJ. Rapid detection and prenatal diagnosis of β-thalassaemia: studies in Indian and Cypriot populations in the UK. Lancet 1990;ii:834–7. [DOI] [PubMed] [Google Scholar]